Abstract
Long-distance population dispersal leaves its characteristic signature in genomes, namely, reduced diversity and increased linkage between genetic markers. This signature enables historical patterns of range expansion to be traced. Herein, we use microsatellite loci from the human pathogen Coccidioides immitis to show that genetic diversity in this fungus is geographically partitioned throughout North America. In contrast, analyses of South American C. immitis show that this population is genetically depauperate and was founded from a single North American population centered in Texas. Variances of allele distributions show that South American C. immitis have undergone rapid population growth, consistent with an epidemic increase in postcolonization population size. Herein, we estimate the introduction into South America to have occurred within the last 9,000–140,000 years. This range increase parallels that of Homo sapiens. Because of known associations between Amerindians and this fungus, we suggest that the colonization of South America by C. immitis represents a relatively recent and rapid codispersal of a host and its pathogen.
Disease epidemics resulting from the introduction of fungal pathogens into previously unoccupied, unexposed environments are an unfortunate feature of globalization. Such introductions have been well documented, for instance, Phytophthora infestans into Irish potato agriculture (1), Cryphonectria parasitica in the American Chestnut tree (2), and Magnaporthe grisea in non-Asian rice (3). Introductions are associated with founder effects that dramatically alter a pathogen's genomic structure by reducing variability and increasing linkage (4, 5). These genetic signatures persist over historical time as has been ably demonstrated by studies of species in postglacially colonized regions (5). Detecting such signatures provides a key for describing historical changes in a pathogen's distribution and demography.
Coccidioides immitis, the etiologic agent of coccidioidomycosis, is endemic to arid soils of the New World, principally the lower sonoran life zone (6). Infection is caused by the inhalation of asexual propagules (arthroconidia) produced by the saprobic form of C. immitis. Within the host, a dimorphic shift causes the formation of spherules and endospores, a process that can result in a systemic mycosis in humans and other vertebrates (7). Death and decay of the host results in the fungus reverting to its environmental, infectious, morphology (8). The high virulence of this pathogen has led to fears of its biohazard potential, culminating in it being the only eukaryote regulated under the United States Anti-Terrorism and Effective Death Penalty Act (9). Herein, we describe the worldwide population genetic structure of C. immitis by using microsatellite loci. These data were used to infer historical changes in the distribution of this fungus and the potential role that early human migration may have played in spreading this pathogen.
Methods and Analyses
We obtained 161 clinical and two environmental isolates from clinicians, covering the known geographical range of C. immitis. These isolates represent eight geographical populations (Fig. 1): central California; southern California; Arizona; Texas; northern, central, and southern Mexico; and South America. Liquid cultures of each isolate were grown in a BL3 containment facility, and total genomic DNA was extracted from lyophilized mycelia according to the protocol described by Burt et al. (10). PCR amplification of nine microsatellite-containing loci (GAC, 621, GA37, GA1, ACJ, KO3, KO7, KO1, and KO9) was performed for each isolate with the fluorescently labeled primers and conditions described previously (11). The alleles present at each locus were determined by electrophoresis with an automated sequencer (Applied Biosystems) and reference against a fluorescently labeled internal size standard.
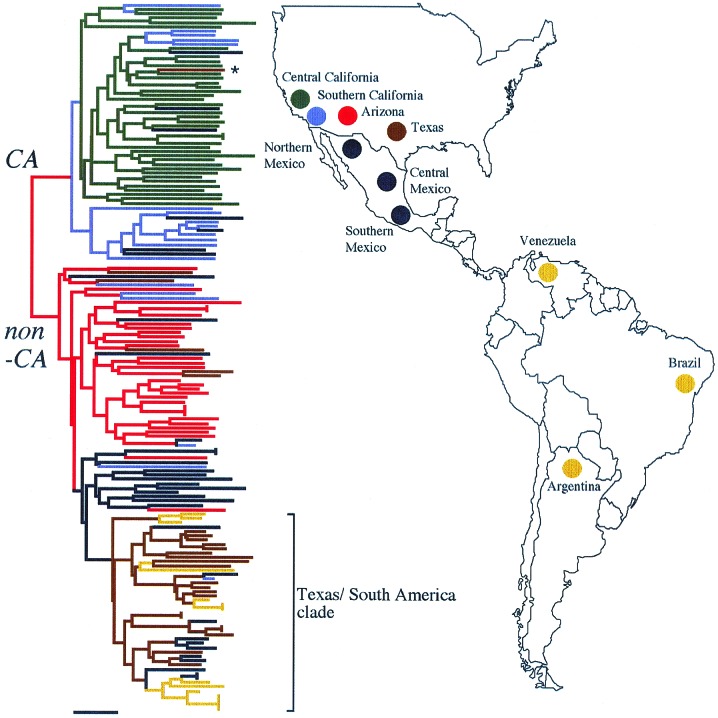
Figure 1.
Neighbor-joining tree of pairwise allele-sharing genetic distances calculated with the program MICROSAT. Tree construction was performed with programs in the PHYLIP package (36). The isolate marked with an asterisk signifies a patient who was diagnosed in Texas but was subsequently found to have acquired the infection in California (42). The tree is mid-point rooted, and the scale bar signifies 0.1 changes. CA, Californian; non-CA, non-Californian.
Analyses of Microsatellite Diversity.
We used two genetic distances. The first, DAS = 1 − (the total number of shared alleles at all loci/n), where n is the number of loci compared (12, 13). Genetic distances between populations were assessed by using the microsatellite distance (δμ) (2, 14). This distance was specifically developed for microsatellites and is linear with respect to time as long as size constraints do not operate on alleles (15). Although the assumption of linearity does not hold over long periods (16, 17), we have shown that (δμ)2 works well in dating recent divergences in C. immitis by using the loci described in this study (18). Confidence intervals for (δμ)2 were calculated by bootstrapping over loci with the program microsat (19). Demographic bottlenecks are associated with reduced genetic variation and increased interlocus associations, measured for this study by comparing the numbers of alleles between populations and the coefficients of disequilibria (D) between loci within populations (20). Rapid population growth alters allele distributions by effectively decreasing the variance of allele distributions relative to expectations from constant-sized populations (21). In this study, two sample F tests with unequal sample size were used to compare locus variances between populations, and the interlocus g test was used to test for signs of population expansion within populations (21, 22).
Analyses of Gene Genealogies.
Three genes were amplified by PCR and sequenced as described (23), with the exception that BIGDYE terminators (Applied Biosystems) were used with the following primer combinations: deoxygenase, DO7 GAGAAGATCCTCGGATTCCA, DO10 GCCCTGAAGTTGCCCGC; serine proteinase, SP3 CCAGGCACCGACAAGCAGTA, SP6 TAGCGTGTCCACCTTCATCG; and chitinase, CT31 CTCCAAACTCTTGTCCAGGC, CT4 TCAGCGAATTTCTTCCTGCC. The sequences were aligned with the clustal v sequence alignment algorithm (24). Distance analyses were performed by neighbor-joining in PAUP* 4.0b2a (25). Because of the closely related nature of these sequences, correcting for multiple hits was not necessary and an uncorrected p distance measure used. Stability of the individual branches was assessed by 1,000 bootstrap replicates of the data.
Results
North American Microsatellite Diversity.
Allele distributions at the nine microsatellite loci were sampled from eight geographical populations. From this data set of 1,424 alleles, DAS was used to group isolates phylogenetically (Fig. 1). The resulting tree shows that isolates occur within one of two major clades. Previous studies of multilocus gene genealogies have resulted in these clades being recognized as the CA and non-CA phylogenetic species (23, 26). We have previously estimated the time of genetic isolation between these two groups as 12.8 million years (SEM = 8.0 million years; refs. 18 and 23). Fig. 1 shows that CA and non-CA are largely allopatric, except in southern California and Mexico where regions of sympatry occur. Within CA and non-CA, there is a strong tendency for isolates to cluster according to where they were isolated, showing that geographically distinct populations occur. The deepest divergence in the CA clade corresponds to a geographical division between the Central Valley and the rest of southern California, delineated by the Tehachapi mountain range. Here, (δμ)2 is significantly greater than zero, demonstrating that genetic drift has occurred between these populations. A similar pattern of differentiation is seen for the non-CA species. Arizona isolates cluster independently from Mexico, and South American isolates group with those from Texas in a subclade, as had been previously recognized (42). Pairwise values for (δμ)2 are significant between all seven non-CA populations.
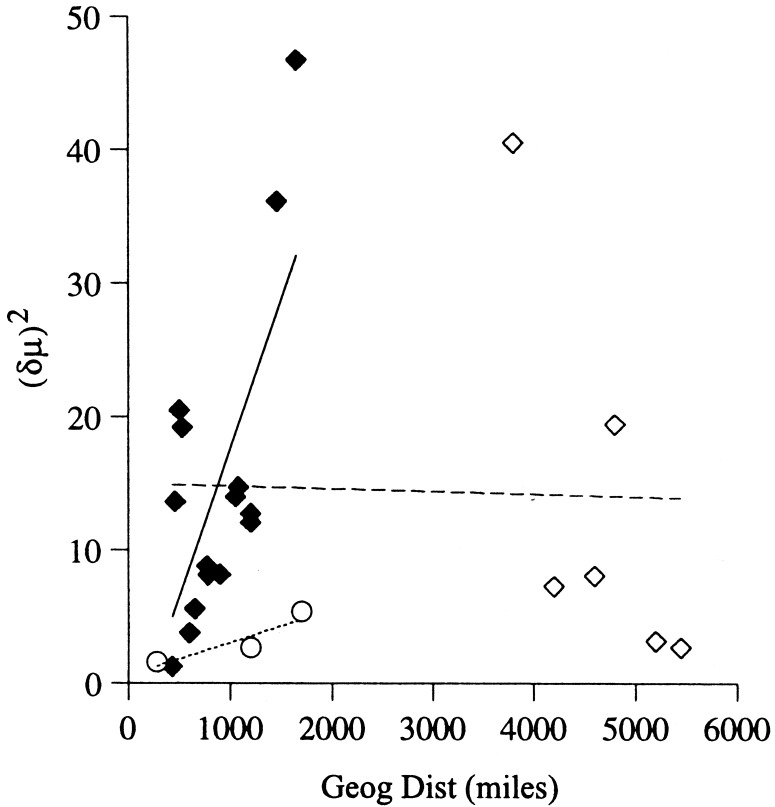
One mode of dispersal in this fungus is by wind-blown arthroconidia, a fact that has been demonstrated by observing an association between dust storms and epidemics of coccidioidomycosis (27). That both species of C. immitis are genetically structured in North America suggests that the distances over which wind-dispersal operates is not great. This hypothesis can be tested by observing correlations between geographical and genetic distances (28). For North American C. immitis, mantel tests (29) showed positive correlations between geographical distance and (δμ)2 within CA and non-CA (CA, r = 0.916, P = 0.16; non-CA, r = 0.694, P = 0.017; Fig. 2). However, the pattern of isolation by distance seen in non-CA C. immitis is lost when South American isolates are considered (r = −0.029 P = 0.362). Despite the large physical distance separating the South American population from the others, little genetic distance has accrued.
Figure 2.
Unrooted phylogenetic tree of the 21 isolates sequenced at the following three loci: deoxygenase, serine proteinase, and chitinase. Distance trees were calculated with PAUP* 4.0 (35). The scale bar signifies 0.05% nucleotide divergence.
Low Genetic Differentiation of South American C. immitis.
Several explanations exist as to why there is low divergence between these South and North American populations. Firstly, South American migrant agricultural workers may have acquired coccidioidomycosis in North America and been diagnosed subsequently in their home countries. However, that two of the Brazilian isolates were acquired from armadillo burrows and that none of the South American patients had a history of travel argue against this case. Secondly, genetic constraints may have caused genetic distances to reach an asymptotic value at these microsatellites (16). To test this scenario, we partially sequenced 1,566 bases from three genes for 11 of the 14 South American isolates and compared them to sequences deposited in GenBank. Although these loci had been shown to be polymorphic between and within non-CA and CA (23), in this study, no genetic variation was found among the South American isolates. Further, these isolates showed identical genotypes to several C. immitis from Texas and Mexico (Fig. 3). It seems that, over the time scale represented herein, genetic differentiation has not had a chance to accrue at these three genes. This comparison proves that the microsatellites are not behaving anomalously. Finally, we examined the hypothesis that South American C. immitis represents an introduction from North America.
Figure 3.
Scatterplots of genetic and geographical distances separating each pairwise combination of populations. CA populations are signified by circles; North American non-CA populations by filled diamonds; and South American non-CA populations by open diamonds. The regression line is dotted for CA (P = 0.16, not significant), solid for non-CA excluding the South American isolates (P = 0.017, significant), and dashed for non-CA including the South American isolates (P = 0.362, not significant).
Origin and Demography of the South American Population.
The relative amount and structure of genetic variation within a population contains information about its recent evolutionary history. Although the South American population contained alleles that were representative of all North American populations, allele frequencies most closely matched those of Texas and all South American isolates grouped within this clade (Fig. 1). Of the eight loci that were polymorphic in non-CA, seven were allelically more diverse in the north, compared with the South American isolates (Table 1). This result is corroborated by the low levels of polymorphism observed in the three sequenced loci from South American isolates. Within South America, levels of linkage disequilibria were high with over 28% pairwise comparisons of loci being significant, compared with 7.14% for the highest levels seen in a North American population (Texas; Table 1). These data are all consistent with South American isolates being descended from a bottlenecked population that originated in Texas—recently enough that mutation has not restored genetic variation to the levels seen in the parent population. Populations that have undergone rapid expansions have reduced locus variances when compared against constant-sized populations. Pairwise comparisons of the variances in allele size at each locus showed that all had reduced variances in the south compared with the North American isolates, and of these 32 comparisons, 26 (81%) were significant (P < 0.05; Table 1). Reich and Goldstein's (21) interlocus g ratio tests for population expansion by examining the variance of the locus variances against the locus variance expected from a constant-sized population, low values of g being associated with population growth. Our data show that g for South America is two orders of magnitude less than for any other population and that this difference is highly significant (Table 1). These data are consistent with the patterns of genetic diversity expected from a species that has undergone a range expansion into a previously unoccupied territory (30).
Table 1.
Variances of allele-length distributions and numbers of alleles indicate a bottleneck and subsequent population growth in South American non-CA C. immitis
| Population | n | Locus†
|
Linkage disequilibria, %‡ | g ratio | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 621 | GA37 | GA1 | ACJ | KO3 | KO7 | KO1 | KO9 | ||||
| South America§ | 14 | 0.18 (2) | 3.80 (3) | 0.40 (3) | 0.00 (1) | 0.29 (2) | 1.79 (2) | 3.19 (3) | 2.18 (3) | 28.5 | 0.006* |
| Southern California | 6 | 0.67* (3) | 5.10 (4) | 1.10 (4) | 1.20* (2) | 29.8* (4) | 11.8* (3) | 19.9* (4) | 15.1* (5) | 0.00 | 0.309 |
| Texas | 24 | 0.20 (2) | 96.0* (7) | 0.70 (4) | 0.30* (2) | 37.0* (5) | 47.0* (4) | 6.90* (4) | 7.10* (5) | 7.14 | 3.036 |
| Arizona | 31 | 0.20 (3) | 35.9* (8) | 1.10* (5) | 18.0* (3) | 52.0* (7) | 8.00* (6) | 12.5* (8) | 6.60* (7) | 0.00 | 0.903 |
| Mexico¶ | 38 | 0.70* (2) | 178* (11) | 1.20* (5) | 0.40* (3) | 98.0* (11) | 3.70* (5) | 12.2* (8) | 12.9* (11) | 3.57 | 10.92 |
, P < 0.05 for locus variances that are significantly greater in comparisons against South American in F tests.
The locus GAC2 is omitted, because it is monomorphic in non-CA C. immitis.
Proportion of loci in significant linkage disequilibria after sequential Bonferroni corrections for multiple pairwise comparisons.
Variances and numbers of alleles (in parentheses) within South American are shown in bold. For the interlocus g test for population expansion, significance is assessed by using the table of 0.05 significance level cutoffs published previously (22) and is shown with an asterisk.
Northern, central, and southern Mexico populations are considered together in order to increase sample size.
Date of Colonization.
Assuming no current migration, we estimated the coalescence time between these populations from (δμ)2 (15). Owing to facts (i) that the generation time in C. immitis is unknown and (ii) that mutation rates vary within and between microsatellites according to their size (31), we could calculate only an approximate value. Assuming a generation time of 1 year and a mutation rate between 4.5 × 10−4 and 3.0 × 10−5, from the values observed in humans and yeast, respectively (32, 33), we estimate the arrival of non-CA C. immitis in South America to be in the range 8,940–134,000 years ago (standard errors based on 1,000 bootstraps over the nine loci; 2,700–228,000 years).
Discussion
We infer from our data and analysis that C. immitis arrived in South America at some point during the late Pleistocene era. Population genetic theory shows that low levels of gene flow are enough to homogenize population structure (34). The strong population structure that exists in North America therefore argues strongly against long-range wind dispersal of arthroconidia. Rather, it seems more likely that C. immitis was carried southwards from North America as encysted spherules within mammalian hosts.
Potential Carriers of C. immitis.
Observations (i) that human patients may carry latent C. immitis infections for up to 12 years (35) and (ii) that spherules have been found in the ≈9,000-year-old skeletal remains of bison from Nebraska (Bison antiquus; W. Morrow, unpublished data) illustrate the capacity of C. immitis to comigrate in association with a host. The southward dispersal of C. immitis seems to have occurred long after the initial Great American Biotic Interchange (GABI) across the Panamanian land bridge 2.5 million to 3.5 million years ago (36). Our data indict a host species that underwent a recent range expansion as the agent of dispersal of C. immitis. Moreover, this host species must have migrated rapidly enough that sequence divergence did not accumulate between the widely spatially separated sites seen in our South American samples of C. immitis. The observation that C. immitis is found highly concentrated in many ancient Amerindian middens shows that the disease was prevalent among these people (37). Archaeological remains from Monte Verde in southern Chile show that the arrival of Amerindians in South America occurred at least 12,500 years ago (38, 39). This date falls within our time frame for the southward movement of C. immitis. The human migration throughout South America was also fast, probably occurring within 800 years, satisfying our criteria for a rapidly dispersing vector of C. immitis. Herein, we postulate that the current population genetic structure of C. immitis reflects the invasion of South America by Homo sapiens. Successful dispersal into South America by C. immitis would have occurred if a strong cycle of infection was maintained between migrating tribes and the environment around their settlements. Support for our hypothesis will come from the detailed analyses of chronological series of human and animal remains, dated precisely by the use of radioisotopes. Studies of the GABI have shown that, other than humans, several small mammal taxa capable of carrying C. immitis arrived from North America within the last 100,000 years: kangaroo rats, squirrels, shrews, and rabbits (36). In particular, kangaroo rats (Heteromyidae) are known reservoirs of C. immitis in North America (40). Analyzing chronologically organized small mammal bone series for the characteristic sequelae resulting from coccidioidal osteomyelitis will reveal the signature of infection and show whether the arrival of South American C. immitis preceded the arrival of the Amerindian settlers, thus disproving our hypothesis. Further, that fungal DNA has been found in the bones of the giant ground sloth Mylodon darwinii (41) suggests that PCR of ancient DNA may be useful to date the arrival of C. immitis in South America. Use of these techniques will help determine whether the arrival of C. immitis in South America represents the earliest known example of a disease spread through the action of human migration.
Acknowledgments
This work was supported by National Institute of Allergy and Infectious Diseases Grant AI37232 and National Heart, Lung, and Blood Institute Grant HL55953 from the National Institutes of Health and the Miller Institute for Basic Research in Science, University of California, Berkeley.
Abbreviations
- CA
Californian
- non-CA
non-Californian
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Goodwin S B, Cohen B A, Fry W E. Proc Natl Acad Sci USA. 1994;91:11591–11595. doi: 10.1073/pnas.91.24.11591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Milgroom M G, Lipari S E. Phytopathology. 1995;85:155–160. [Google Scholar]
- 3.Zeigler R S. Annu Rev Phytopathol. 1998;36:249–275. doi: 10.1146/annurev.phyto.36.1.249. [DOI] [PubMed] [Google Scholar]
- 4.Taylor J W, Jacobson D J, Fisher M C. Annu Rev Phytopathol. 1999;37:197–246. doi: 10.1146/annurev.phyto.37.1.197. [DOI] [PubMed] [Google Scholar]
- 5.Hewitt G M. Nature (London) 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- 6.Rippon J W. Medical Mycology. Philadelphia: Saunders; 1988. [Google Scholar]
- 7.Saubolle M. In: Coccidioidomycosis. Einstein H E, Catenzaro A, editors. Washington, DC: Natl. Found. Infect. Dis.; 1996. pp. 1–8. [Google Scholar]
- 8.Maddy K T, Crecelius T. In: Coccidioidomycosis. Ajello L, editor. Tucson, AZ: Univ. of Arizona Press; 1967. pp. 309–312. [Google Scholar]
- 9.61 Federal Register 207 (1996), p. 55190.
- 10.Burt A, Carter D A, Koenig G L, White T J, Taylor J W. Fungal Genet Newsletter. 1995;42:23. [Google Scholar]
- 11.Fisher M C, Koenig G L, White T J, Taylor J W. Mol Ecol. 1999;8:1082–1084. doi: 10.1046/j.1365-294x.1999.00655_5.x. [DOI] [PubMed] [Google Scholar]
- 12.Stephens J C, Gilbert D A, Yuhki N, O'Brien S J. Mol Biol Evol. 1992;9:729–743. doi: 10.1093/oxfordjournals.molbev.a040755. [DOI] [PubMed] [Google Scholar]
- 13.Bowcock A M, Ruiz-Linares A, Tomfohrde J, Minch E K, Kidd J R, Cavalli-Sforza L L. Nature (London) 1994;368:455–457. doi: 10.1038/368455a0. [DOI] [PubMed] [Google Scholar]
- 14.Goldstein D B, Ruiz Linares A, Cavalli-Sforza L L, Feldman M W. Proc Natl Acad Sci USA. 1995;92:6723–6727. doi: 10.1073/pnas.92.15.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldstein D B, Ruiz Linares A, Cavalli-Sforza L L, Feldman M W. Genetics. 1995;139:463–471. doi: 10.1093/genetics/139.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garza J C, Slatkin M, Freimer N B. Mol Biol Evol. 1995;12:594–603. doi: 10.1093/oxfordjournals.molbev.a040239. [DOI] [PubMed] [Google Scholar]
- 17.Lehmann T, Hawley W A, Collins F H. Genetics. 1996;144:1155–1163. doi: 10.1093/genetics/144.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher M C, Koenig G L, White T J, Taylor J W. Mol Biol Evol. 2000;17:1164–1174. doi: 10.1093/oxfordjournals.molbev.a026399. [DOI] [PubMed] [Google Scholar]
- 19.Minch E, Ruiz-Linares A, Goldstein D, Feldman M, Cavalli-Sforza L L. microsat, The Microsatellite Distance Program. Stanford, CA: Stanford Univ. Press; 1995. [Google Scholar]
- 20.Hartl D L, Clark A G. Principles of Population Genetics. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 21.Reich D E, Goldstein D B. Proc Natl Acad Sci USA. 1998;95:8119–8123. doi: 10.1073/pnas.95.14.8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reich D E, Feldman M W, Goldstein D B. Mol Biol Evol. 1999;16:453–466. [Google Scholar]
- 23.Koufopanou V, Burt A, Taylor J W. Proc Natl Acad Sci USA. 1997;94:5478–5482. doi: 10.1073/pnas.94.10.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgins D G, Bleasby A J, Fuchs R. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 25.Swofford D L. paup*, Phylogenetic Analysis Using Parsimony (* and Other Methods) Sunderland, MA: Sinauer; 1998. , Version 4. [Google Scholar]
- 26.Koufopanou V, Burt A, Taylor J W. Proc Natl Acad Sci USA. 1998;95:8414–8414. [Google Scholar]
- 27.Pappagianis D, Einstein H. West J Med. 1978;129:527–530. [PMC free article] [PubMed] [Google Scholar]
- 28.Nei M. Am Nat. 1972;106:283–292. [Google Scholar]
- 29.Mantel N. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- 30.Ibrahim K M, Nichols R A, Hewitt G M. Heredity. 1996;77:282–291. [Google Scholar]
- 31.Rubinsztein D C, Amos W, Leggo J, Goodburn S, Jain S, Li S H, Margolis R L, Ross C A, Ferguson-Smith M A. Nat Genet. 1995;10:337–343. doi: 10.1038/ng0795-337. [DOI] [PubMed] [Google Scholar]
- 32.Kwiatkowski D J, Henske E P, Weimer K, Ozelius L, Gusella J F, Haines J. Genomics. 1992;12:229–240. doi: 10.1016/0888-7543(92)90370-8. [DOI] [PubMed] [Google Scholar]
- 33.Henderson S T, Petes T D. Mol Cell Biol. 1992;12:2749–2757. doi: 10.1128/mcb.12.6.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slatkin M. Science. 1987;236:787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]
- 35.Hernández J L, Echevarría S, García-Valtuille A, Mazorra F, Salesa R. Eur J Clin Microbiol Infect Dis. 1997;16:592–594. doi: 10.1007/BF02447922. [DOI] [PubMed] [Google Scholar]
- 36.Marshall L G, Webb S D, Sepkoski J J, Raup D M. Science. 1982;215:1351–1357. doi: 10.1126/science.215.4538.1351. [DOI] [PubMed] [Google Scholar]
- 37.Lacey G H, Swatek F. Appl Microbiol. 1974;27:379–388. doi: 10.1128/am.27.2.379-388.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meltzer D J. Science. 1997;276:754–755. [Google Scholar]
- 39.Dillehay T. Monte Verde: A Late Pleistocene Settlement in Chile: The Archaeological Context and Interpretation. Vol. 2. Washington, DC: Smithisonian; 1997. [Google Scholar]
- 40.Pappagianis D. Current Topics in Medical Mycology. Berlin: Springer; 1988. pp. 199–238. [DOI] [PubMed] [Google Scholar]
- 41.Hoss M, Dilling A, Currant A, Päabo S. Proc Natl Acad Sci USA. 1996;93:181–185. doi: 10.1073/pnas.93.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burt A, Dechairo B M, Koenig G L, Carter D A, White T J, Taylor J W. Mol Ecol. 1997;6:781–786. doi: 10.1046/j.1365-294x.1997.00245.x. [DOI] [PubMed] [Google Scholar]