Abstract
Diagnosis of opportunistic infections in HIV-infected individuals remains a major public health challenge, particularly in resource-limited settings. Here, we describe a rapid diagnostic system that delivers a panel of serologic immunoassay results using a single drop of blood, serum, or plasma. The system consists of disposable cartridges and a simple reader instrument, based on an innovative implementation of planar waveguide imaging technology. The cartridge incorporates a microarray of recombinant antigens and antibody controls in a fluidic channel, providing multiple parallel fluorescence immunoassay results for a single sample. This study demonstrates system performance by delivering antibody (Ab) reactivity results simultaneously for multiple antigens of HIV-1, Treponema pallidum (syphilis), and hepatitis C virus (HCV) in a collection of clinical serum, plasma, and whole-blood samples. By plotting antibody reactivity (fluorescence intensity) for known positive and negative samples, empirical reactivity cutoff values were defined. The HIV-1 assay shows 100% agreement with known seroreactivity for a collection of 82 HIV Ab-positive and 142 HIV Ab-negative samples, including multiple samples with HCV and syphilis coinfection. The treponema-specific syphilis assay correctly identifies 67 of 68 T. pallidum Ab-positive and 100 of 102 T. pallidum Ab-negative samples, and the HCV assay correctly identifies 59 of 60 HCV Ab-positive and 120 of 121 HCV Ab-negative samples. Multiplexed assay performance for whole-blood samples is also demonstrated. The ability to diagnose HIV and opportunistic infections simultaneously at the point of care should lead to more effective therapy decisions and improved linkage to care.
INTRODUCTION
Decades of effort have gone into developing a host of HIV screening and diagnostic techniques, ranging from simple single-analyte blood tests to more complex multianalyte clinical laboratory analyzers. A recurring challenge is to diagnose the diverse coinfections that account for a significant fraction of HIV-associated morbidity and mortality (14). Multianalyte testing for AIDS and its opportunistic infections is essential for the development of individualized management of HIV infections and its common copathogens. At the time of HIV diagnosis, the standard of care includes testing for related infections, such as those caused by hepatitis C virus (HCV), hepatitis B virus (HBV), Treponema pallidum (syphilis), and human herpesvirus 8 (HHV-8) (11, 14). These multiple diagnoses typically require extensive use of serological diagnostic tools, often in diverse test formats. Unfortunately, coinfection testing using current technology is costly and functionally too complex for most point-of-care (POC) environments, particularly in resource-limited settings where the disease burden is high. The ability to rapidly and inexpensively discriminate HIV monoinfection from more complex coinfections using a single, multianalyte platform would be a significant advance in the field.
Rapid diagnostic tests (RDTs) have had an enormous impact on infectious disease screening programs worldwide over the last decade and are the backbone of HIV screening efforts. While RDTs provide the advantages of low per-test cost, simple operation, and no required instrumentation, there are also significant limitations. Most RDTs are configured for only a single pathogen, so multiple RDTs are needed to support coinfection testing, which can be prohibitive from test cost, personnel training, and results management perspectives. RDTs are generally based on immunochromatographic or lateral flow technology, and many RDTs give good performance at a low per-test cost (3, 7, 20, 22, 26). But issues with lateral flow rapid tests include the subjective nature of result interpretation by visual inspection and a narrow read time window, both of which require rigorous staff training and quality assurance. RDTs not requiring instrumentation present cost and simplicity advantages but also present disadvantages, including no link to electronic medical records and no automated quality control features, such as untrained user lockout and expired lot rejection.
Here, we describe a simple diagnostic system that solves many of the problems outlined above. System utility is demonstrated for a multiplexed HIV-1/syphilis/HCV assay using a combination of clinical sample collections.
MATERIALS AND METHODS
Biological reagents.
Assays demonstrated here were all based on commercially available recombinant proteins. The HIV-1 assay demonstration utilizes envelope glycoprotein 41 (gp41) and capsid antigen p24. The syphilis treponemal assay (19) was based on treponemal proteins Tp47 and Tp17. Recombinant proteins were sourced through Meridian Life Sciences, Inc. (Memphis, TN), Fitzgerald Industries International (Acton, MA), and CTK Biotech, Inc. (San Diego, CA).
Hepatitis C virus serodiagnosis is challenging due to the high level of genomic and antigenic variability associated with the virus (2, 8), and anti-HCV antibody (Ab) screening depends on multiple antigenic targets (1, 5). FDA-approved enzyme immunoassays, for example, rely on combinations of recombinant proteins and peptides (e.g., see the package inserts for Abbott HCV enzyme immunoassay [EIA] 2.0 and Ortho HCV version 3.0 enzyme-linked immunosorbent assay [ELISA]). Consistent with the need for HCV antigen multiplexing, we have used four commercially available HCV recombinant proteins in this demonstration, including recombinant core protein (nucleocapsid, p22 fusion protein), full-length NS3 (c33c), a mosaic recombinant comprising the NS4 immunodominant regions, and a recombinant that contained HCV nucleocapsid, NS3, NS4, and NS5 immunodominant regions. The last molecule is referred to here as the multiple-epitope antigen. HCV antigens were sourced through Meridian Life Sciences and US Biological.
Assay reagents.
Other biological reagents include purified human IgG (Sigma, St. Louis, MO), goat anti-human IgG (Thermo Scientific, Rockford, IL), and goat anti-human IgG conjugated with fluorescent dye (DyLight649; KPL, Inc.). Assay reagents include bovine serum albumin (BSA; Sigma Life Science, St. Louis, MO), phosphate-buffered saline (PBS; Fisher Scientific, Rockford, IL), blocker casein in PBS (Thermo Scientific, Rockford, IL), and Tween 20 (Thermo Scientific, Rockford, IL).
Clinical samples.
Five sets of clinical samples were used to characterize the MBio system. A total of 251 different clinical samples were processed in this study.
Commercial controls.
Well-characterized human plasma samples with known antibody reactivity for each of the three pathogens were purchased from SeraCare Life Sciences (Milford, MA). These samples included four with known HIV-1 antibody reactivity, four with known T. pallidum antibody reactivity, and four with known HCV antibody reactivity.
HIV-1 antibody-reactive samples.
A total of 25 human serum samples with known HIV-1 Western blot reactivity were provided under an Institutional Review Board (IRB)-approved protocol by Susan Little of the University of California, San Diego (UCSD) Medical Center. Coinfection status is not known for the majority of these samples. Due to limited available sample volumes, coinfection reference testing was not performed for this collection at MBio Diagnostics.
Syphilis samples.
A collection of 30 deidentified sera known to be positive for syphilis infection were sourced from the Colorado Department of Public Health and Environment (CDPHE; Denver, CO). Syphilis reactivity was determined at the CDPHE laboratory using rapid plasma reagin (RPR) and T. pallidum particle agglutination (TPPA). The HIV serostatus of these samples was not known upon receipt from the CDPHE. All 30 were therefore also characterized at MBio Diagnostics with an FDA-approved HIV-1/-2 RDT (Trinity Uni-Gold Recombigen HIV).
Coinfection samples.
A collection of deidentified clinical samples from existing sample archives were coordinated with IRB approval by Sharon Reed (UCSD). Samples were selected for likely HIV, HCV, HBV, and/or syphilis infection. UCSD clinical samples were all characterized for HIV and HCV infection by using a Siemens Centaur clinical analyzer. Syphilis samples were tested by RPR with confirmation of RPR-positive samples by TPPA (Fujirebio Inc., Tokyo, Japan). The UCSD coinfection collection includes a large number of highly complex pathogen antibody and antigen reactivities, as commonly encountered in HIV- and HCV-infected individuals. In addition to their being positive for HIV, HCV, HBV, and/or syphilis infection, many of these samples have positive reactivities for Toxoplasma gondii, cytomegalovirus (CMV), Epstein-Barr virus, and various human herpesviruses. Detailed stratification of the collection based on coinfection status is beyond the scope of this paper.
Negative control sera.
Human serum controls were sourced through Valley Biomedical (Winchester, VA). These were vendor-certified as HIV, HCV, and RPR negative. T. pallidum antibody reactivities were not provided by the vendor for this collection. In the event of a positive T. pallidum antibody result with the MBio assay, reference testing was performed for those specific samples. T. pallidum tests run at MBio Diagnostics included a TPPA (Fujirebio, Malvern, PA), a syphilis RDT (SD Bioline, South Korea), and a treponemal ELISA (Trep-Sure, Phoenix Biotech, Ontario, CA). T. pallidum reference testing was not performed with samples that were negative by RPR and negative by the MBio assay.
Whole-blood samples.
Because the ultimate use of this technology will be in point-of-care settings, it is important to demonstrate performance with whole-blood samples. Whole blood was sourced under an IRB-approved protocol from HIV-positive donors at the UCSD Antiviral Research Center under the direction of Constance Benson. Venipuncture samples were collected in EDTA blood collection tubes (Lavender Cap BD Vacutainer) and shipped overnight to the MBio Diagnostics laboratory, where they were run within 2 h of receipt (i.e., within 30 h of draw). Negative whole-blood samples (i.e., HIV-1 and HIV-2 antibody negative, HIV-1 nucleic acid negative) were sourced through Valley Biomedical.
Assay cartridge and instrument.
The system described here combines single-use disposable assay cartridges with a simple reader instrument, as illustrated in Fig. 1. The instrument operates as a universal serial bus (USB) peripheral device, drawing power and communication via a laptop computer. Assay results are compiled as electronic digital files and can be presented to the user via computer monitor or printer or can be exported to some other data management system.
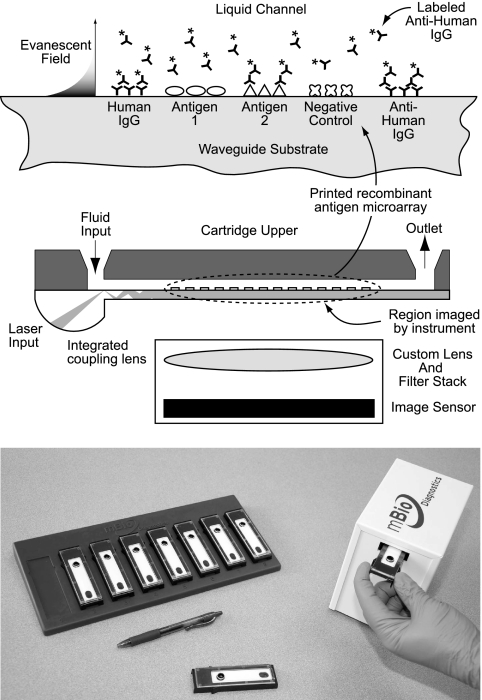
Fig. 1.
(Top) Schematic representation of the multiplexed fluorescence immunoassay and disposable assay cartridge. A protein microarray is printed to a plastic planar waveguide which is bonded to a plastic upper component to define a flow channel. Sample, wash, and detect reagents are introduced via a fluid inlet. Illumination of the assay surface is via the evanescent field generated down the length of the multimode waveguide. The array is imaged in a single field of view through the plane of the waveguide. (Bottom) MBio instrument and rack of assay cartridges.
Fluorescence immunoassays are illuminated and imaged using a novel multimode planar waveguide technology. Planar waveguides have been used in biosensor and immunoassay applications for decades and are the subject of several technical reviews (15, 16, 18). Briefly, laser light is directed into a waveguide substrate, where it propagates by total internal reflection (TIR) at the interface between the high index of refraction waveguide and the surrounding medium (air or aqueous solution) (Fig. 1). At the TIR interface, an evanescent field that decays exponentially into the aqueous medium is generated. The decay length of the evanescent field is on the order of 100 nm for visible light. For fluorescence immunoassay applications, the advantage is localization of the illumination source precisely at the solid-liquid assay interface, limiting effects of the bulk solution, line of sight, light scattering, etc.
The cartridge is based on a thick (∼1-mm), multimode planar waveguide fabricated by injection molding of a low-autofluorescence plastic. The major innovation is the incorporation of a coupling lens into the molded waveguide (Fig. 1). Building on a patented design introduced by Herron and colleagues (6, 12, 13, 17), the MBio lens design overcomes the fundamental challenge of reproducible light coupling to the waveguide. The lens design creates a diverging beam; modes mix down the length of the waveguide, eventually creating a spatially uniform illumination field along the axial length of the waveguide.
The plastic waveguides were activated with a surface chemistry treatment to render them amine-reactive. Details of the surface activation protocol are beyond the scope of this paper. Methods for surface activation are described in the literature (10).
A protein microarray was printed to the planar waveguide prior to assembly into the disposable cartridge. Details of the array features and layout are provided in Results. Arrays were printed with a Bio-Dot AD3200 robotic arrayer equipped with a Bio-Jet print head dispensing 28-nanoliter droplets. Spot diameters were approximately 0.5 mm, and the arrays were printed on a grid with 1.25-mm centers. The length of the 30-feature (2-row by 15-column) array was approximately 15 mm. After printing, the waveguide arrays were rinsed with a protein-based blocking agent, spin-dried, and then coated with a sugar-based stabilizer for storage.
Printed waveguides were assembled into an injection-molded cartridge to form a 5-mm-wide fluidic channel with a volume of approximately 30 μl. The cartridge inlet port provides a reservoir for introduction of assay fluids. The exit port provides a fluidic contact to an absorbent pad that serves as a waste reservoir. The cartridge has been engineered to provide reproducible passive fluid flow, driven by a combination of capillary action and hydrostatic pressure. All fluids stay on board the cartridge, minimizing biohazard.
Assay procedure.
Samples were processed in cartridges on the benchtop at ambient temperature (20 to 25°C). Because the assay is independent of the reader instrument, sample cartridges can be batch processed, with up to 30 run in parallel. Sample volumes for the disposable cartridge were 6 μl of serum or plasma or 12 μl of whole blood, making the cartridge compatible with finger-stick capillary samples. The larger volume for the whole-blood samples is to compensate for the cell volume. Results presented here were based on the following procedure. A 6-μl aliquot of serum or plasma was diluted in 194 μl of diluent (PBS, 0.5% casein, 0.05% Tween 20). For whole blood, 12 μl was added to 188 μl of diluent. One hundred seventy-five microliters of this diluted sample mixture was then loaded into the cartridge input port by transfer pipet. Passive flow through the cartridge during a 15-min incubation occurs independently of any user interaction. One hundred seventy-five microliters of wash buffer (PBS, 0.1% Tween 20) was then added to the input port and allowed to flow through the cartridge for 3 min, followed by the addition of 175 μl of dye-conjugated anti-human IgG in diluent (PBS, 1 mg/ml BSA, and 0.05% Tween 20) and incubation for 10 min. The total per-cartridge assay time was approximately 28 min. The cartridge can be read on the instrument any time within an hour of processing without affecting results. Read time and data processing in the instrument were approximately 30 s per cartridge.
Custom image processing software has been developed by MBio Diagnostics for spot finding, intraspot fluorescence signal intensity measurement, and normalization. After the reporting of results, the cartridge is removed and disposed of as biohazardous waste, and the next processed cartridge is inserted. The combination of parallel cartridge processing, a large read window, and rapid analysis allows more than 100 samples per work shift to be processed.
RESULTS
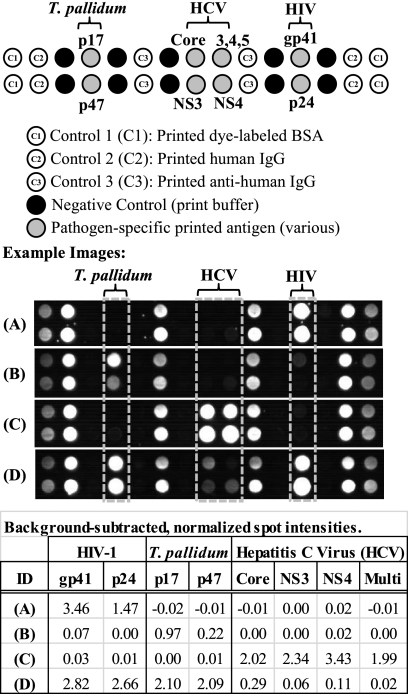
Figure 2 provides the layout of the HIV-1/syphilis (T. pallidum)/HCV array map along with images from a representative set of clinical samples. In-array control features include printed human IgG, anti-human IgG, and print buffer controls. The printed human IgG serves as a procedural control for the fluorescently labeled “detect” antibody. Printed anti-human IgG serves as a sample control. These features must yield fluorescence signal in an acceptable range for a test to be considered valid, ensuring that sample and fluorescently labeled detect antibody were added to the cartridge and that the cartridge was properly illuminated. The print buffer spots serve as negative controls and monitor any unusual nonspecific binding. Excessive fluorescence signal on the negative-control spots yields an invalid test result.
Fig. 2.
Representative images and array layout for the multiplex HIV/syphilis/HCV assay. The 30-feature array (2 rows by 15 columns) includes pathogen-specific printed antigens as well as multiple in-assay controls. Dye-labeled BSA spots serve as corner markers (C1) for imaging. These spots are not used in the analysis. Printed human IgG (C2) serves as a control for the dye-labeled detect antibody. Printed anti-human IgG (C3) serves as a sample control. Print buffer spots serve as a nonspecific binding negative control. Images from four clinical plasma samples are shown, along with background-subtracted spot intensities for the pathogen-specific antigens. Based on reference test methods, samples A to C are each monoinfected, with reactivity to HIV Ab (A), T. pallidum Ab (B), and HCV Ab (C). Sample D has Ab reactivity to all three pathogens by both the reference methods and the MBio assay. See the text for additional details.
We note that the human IgG and anti-human IgG control spots have been designed to give fluorescence signal comparable to that of a typical seropositive sample. Obviously, both total human IgG and the goat anti-human IgG dye conjugate are present in great excess relative to the amounts of specific antibodies reacting with each of the antigen spots. We adjust the IgG print concentration to bring positive-control signals into an appropriate range.
The clinical sample images in Fig. 2 are a representative sampling of images generated in the 28-min cartridge assay. Positive and negative controls are present in all assays, and the printed antigen spots show different intensities for different individual samples. Based on reference test methods, samples A through C are each reactive for one pathogen and negative for the other two. Sample D is antibody reactive for all pathogens, as indicated by all array antigen spots yielding a positive signal.
The fluorescence arrays are also quantitative. The table in Fig. 2 provides quantitative antibody-antigen signal intensities associated with each spot. Results for each spot in the array are reported as background-subtracted and normalized fluorescence signal intensity. Background signal is an average of the signal on negative reference spots adjacent to the antigen spot of interest. The intensity of the printed human IgG spot signal is utilized for normalization, and the reported intensity is thus a dimensionless number. Slightly negative numbers result when the signal on the antigen spot is lower in intensity than that of the adjacent negative control. All negative numbers encountered are invariably close to zero. The dynamic range of the current instrument is approximately 3.5 logs. A detailed analysis of dynamic range, assay kinetics, and limits of detection is beyond the scope of this paper and will be presented in a separate manuscript.
With the combination of commercial positive controls, the UCSD HIV-1 samples, the CDPHE syphilis samples, the UCSD coinfection samples, and the negative controls, results of a total of 251 different samples (serum and plasma) are presented here.
HIV-1 antibody assay results.
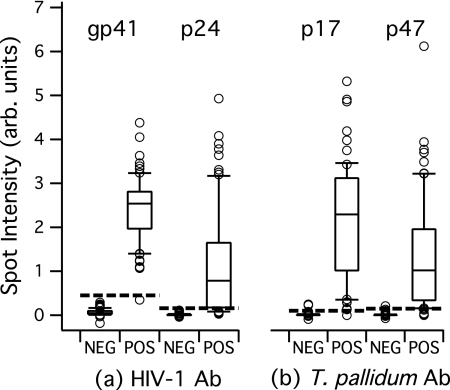
A total of 224 samples in the collection had known HIV-1 seroreactivity status, with 82 being HIV-1 Ab positive and 142 being HIV-1 Ab negative. Antibody reactivity (signal as described above) against the two HIV antigens in the array (gp41 and p24) are linked to the known HIV Ab reactivity status and plotted in Fig. 3a. Ideally, samples that are known to be HIV Ab negative should show little or no intensity on the HIV antigen spots. As expected, gp41 and p24 signal results are clustered near zero for these negative samples. We note that the gp41 spots do show some cross-reactivity, with normalized signals of between 0 and 0.4 on this scale. For the HIV Ab-positive samples, we see a distribution of intensities. A strong gp41 antibody response is expected for the seroconverted individuals represented in this collection, and the Fig. 3 gp41 results are consistent with this expectation. One known HIV Ab-positive sample does show low signal on the gp41 spot. This particular sample yielded robust signal on the p24 spot. For the collection, reactivity against the p24 antigen is varied, as seen in Fig. 3a.
Fig. 3.
Antibody reactivity in the MBio system for clinical samples with known HIV-1 and T. pallidum serostatus. Spot intensity is the background-subtracted, normalized intensity described in the text. Antibody reactivity against all antigens in Fig. 3 and 4 is measured simultaneously for each sample. The horizontal solid bar in each box represents the median; upper and lower boundaries of the boxes are the 75th and 25th percentiles; and the upper and lower whisker bars are the 90th and 10th percentiles, respectively. The open circles represent samples with values above and below the 90th and 10th percentiles. The dashed lines are empirically derived cutoffs. (a) Antibody reactivity results for a total of 224 samples with known HIV-1 Ab reactivity status (82 positive and 142 negative). (b) Antibody reactivity results for a total of 170 samples with known treponemal Ab reactivity status (68 positive and 102 negative). arb. units, arbitrary units; NEG, negative; POS, positive.
The data plots of Fig. 3a are used to establish empirical cutoff values that will be used for subsequent signal/cutoff (S/CO) calculations. Cutoffs are set near the highest-signal negative sample in the collection. Increasing the cutoff value will yield a more specific assay (fewer false positives). Decreasing the cutoff will yield a more sensitive assay (fewer false negatives). Given that the antibody reactivity status of the samples in this collection was known, individual antigen cutoffs were adjusted empirically to optimize “effective” sensitivity and specificity. We note that this is a self-referential data set, i.e., we are applying the cutoff to the same data as those used to generate the cutoff. We therefore do not report results in terms of sensitivity and specificity. On the scale shown in Fig. 3a, the gp41 and p24 cutoffs are set at a value above the highest signal-negative sample in the collection (cutoffs are 0.45 and 0.16, respectively). If it is assumed that a S/CO of >1 for either antigen constitutes overall HIV-1 Ab reactivity for that sample, then the cutoffs, as defined, yield 100% agreement between the MBio multiplex assay and the reference result.
Syphilis assay results.
A total of 170 samples in the collection had known T. pallidum antibody reactivity status, including 68 treponema-positive and 102 treponema-negative samples. Results for the treponemal antigens p17 and p47 are provided in Fig. 3b. As described above, cutoffs were empirically determined and are indicated on the plots. Applying these cutoffs to the 170 samples, we report treponemal Ab reactivity on 67 of 68 known treponemal Ab-reactive samples (one false negative). The assay correctly identifies 100 of 102 treponemal negatives (two false positives). Next steps will be to improve specific activity and minimize nonspecific binding to the treponemal antigen spots. Approaches for assay improvement include screening alternative recombinant antigens and alternative blocking strategies.
Hepatitis C assay results.
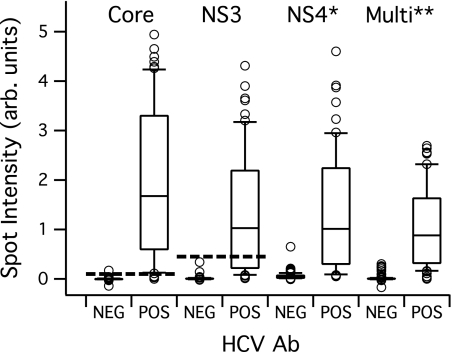
A total of 181 samples had known HCV antibody reactivity, including 60 HCV Ab positive and 121 HCV Ab negative. Results for the four recombinant antigens are provided in Fig. 4. A large spread of antibody reactivity signals is observed, as expected (2). The core and NS3 antigens provide the best overall performance in terms of distinguishing positive from negative. The two multiple-epitope antigens (NS4* and Multi**) did not offer a benefit over the core and NS3 antigens in this assay. Using these results, cutoffs were set for the core and NS3 antigen as described above. For the purpose of this study, antibody reactivity above the cutoff on one or more of the HCV antigens renders the sample HCV Ab positive. With this definition, the assay correctly identifies 59 of 60 HCV-positive and 120 of 121 HCV-negative samples. Figure 4 shows a large number of samples (positive and negative) near the cutoff values. Approaches for assay improvement include screening additional recombinant antigens and print condition optimization, as well as optimization of alternative protein and peptide blocking strategies. Antigen selection and activity improvements are required to improve robustness of the HCV assay.
Fig. 4.
Antibody reactivity in the MBio assay for 181 clinical samples with known HCV antibody serostatus (60 positive and 121 negative). Spot intensity is the background-subtracted, normalized intensity described in the text. Antibody reactivity against all antigens is measured simultaneously for each sample. Box plot details are as described in the legend to Fig. 3. The dashed lines are empirically derived cutoffs for the core and NS3 recombinant antigens. The synthetic NS4* and Multi** multiple-epitope antigens showed relatively high signals on the HCV Ab negative samples, so cutoffs for these two antigens were not used in subsequent analyses.
Whole-blood assay results.
Because point of care is the target application of the MBio system, whole-blood performance is an important demonstration. Lateral flow-based RDTs typically incorporate a sample pad material designed to capture red blood cells (RBCs) without promoting hemolysis, as the RBCs and hemoglobin can interfere with the colorimetric read of the device. The MBio system has no such requirement; 6% whole blood in buffer is added directly to the device with no separation of cellular components prior to running the standard assay protocol described above.
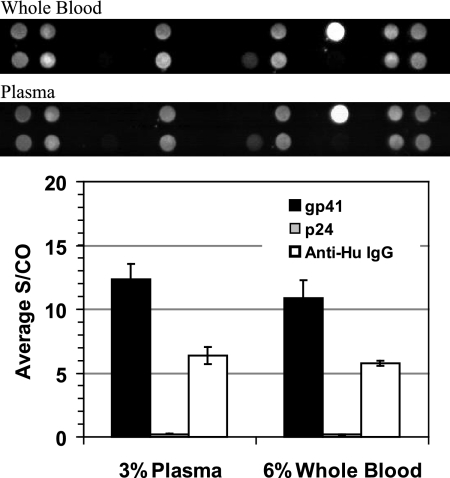
Seven whole-blood samples from HIV-positive donors and 13 whole-blood samples from HIV-negative donors were processed as part of this study. For a subset of these samples, a performance comparison between whole blood and plasma from the same EDTA-Vacutainer blood tube was performed. Results from a sample representative of all 7 HIV-positive samples are provided in Fig. 5. The image comparison shows that the whole-blood and plasma sample array profiles are effectively the same. Importantly, no residual interference from the cellular blood components is observed. The bar graph in Fig. 5 shows the same quantitative signal profile for the whole-blood and plasma samples. This particular sample had strong reactivity to the gp41 antigen but very little reactivity to p24.
Fig. 5.
Comparison of whole blood and plasma results performed with the MBio system. Cartridges were processed with 6% whole blood or 3% plasma from the same venipuncture sample approximately 24 h after draw. The larger volume of whole blood was to approximately compensate for cell volume. The array layout was as in Fig. 2. Images for the whole blood and plasma from this HIV-positive sample were essentially identical, demonstrating that there is no cellular or hemoglobin-induced interference in the system. The bar graph provides quantitative output for the gp41, p24, and anti-human IgG spots on the arrays. Note that the anti-human IgG is printed at a very low concentration in the array, with activity tuned to give signal on the same scale as that of positive antigen spots. Total IgG is in large excess in the sample. Data presented are mean values for three replicate cartridges for each sample type. Error bars represent one standard deviation. See the text for a detailed discussion.
Assay reproducibility was determined for the blood sample presented in Fig. 5. Three aliquots of whole blood from one blood tube were processed using three cartridges. The tube contents were then spun down, and three aliquots of plasma were processed using three more cartridges. Figure 5 included results from the two sets of triplicate measurements. Defining the percent coefficient of variation (%CV) as the standard deviation of the three measurements divided by the mean, the following results were observed. For the plasma samples, the gp41 S/CO %CV was 10%. For the whole-blood samples, it was 13%. For the anti-human IgG spots, the plasma samples gave an 11% CV and the whole blood gave a 4% CV.
None of the negative whole-blood samples showed signal above the cutoff for the HIV antigen. All seven HIV-positive whole-blood samples showed strong antibody reactivity, comparable to those in Fig. 4. This initial set of whole-blood samples therefore showed 100% correlation with expected results.
DISCUSSION
Multianalyte testing for AIDS and its coinfections is essential for the development of individualized management of HIV-1 infection and its common copathogens. At the time of HIV diagnosis, the standard of care includes determinations of related infections, such as those with hepatitis C virus (HCV), hepatitis B virus (HBV), Toxoplasma gondii, Treponema pallidum, and cytomegalovirus (CMV) (11, 14). Coinfection information is used for treatment (as in the case of T. pallidum), vaccinations (as in the case of HBV), and prophylaxis (in the case of T. gondii). The multiplexed system described here has the potential to offer a combination of critical tests which detect multiple pathogens in a single assay, meeting the recognized desire for combination test platforms (23).
Increased access to antiretroviral therapy in resource-limited settings, and in particular sub-Saharan Africa, has had a major impact on morbidity and mortality from AIDS. By the end of 2009, over 5 million people living in low- and middle-income countries were receiving antiretroviral therapy (25). By most estimates, even before treatment recommendations were revised to encourage the initiation of antiretroviral therapy at higher CD4 cell counts, contemporary antiretroviral therapy was reaching only 30 to 40% of those needing therapy (24). In all likelihood, there will continue to be a substantial gap between the number of people needing antiretroviral therapy and the resources available to treat them. In order to maximize the benefits from the resources available, it is essential that antiretroviral therapy be delivered as efficiently as possible to those most likely to benefit. A multiplex platform that provides rapid and accurate information about critical coinfections could help to better prioritize those who should be treated and could provide guidance about antiretroviral drug selection.
In addition to antiviral treatment decisions based on improved coinfection information, the ability to simultaneously detect markers for multiple pathogens in the same sample offers diagnostic advantages. It is well known that HIV infection complicates HCV serodiagnosis (4, 9, 21). An HIV/HCV coinfection test can help identify difficult to characterize infections at the time of initial screening.
The system presented here offers several potential technical advantages over existing technology. Most significant is the ability to perform quantitative multiplexed immunoassays on whole-blood samples at the point of care in a cost-effective manner. While some RDTs do offer a degree of multiplexing (see, for example, the 5-band dual-path platform HIV-1/-2 test in development by ChemBio, Inc.), the MBio system can readily be configured to simultaneously measure up to 30 markers.
Some in the global health field argue against any type of instrumented test in a point-of-care setting. Arguments against instrumentation hinge primarily on instrument procurement costs and servicing requirements that are not a factor in visually read tests. There is increasing awareness, however, of systemic costs associated with visually read tests. In particular, maintaining staff trained in the nuances of different RDT protocols is a challenge, and the simultaneous management of multiple RDTs (for HIV, syphilis, malaria, etc.) can become logistically intractable. The system described here provides the advantages of multiple RDTs in a single-protocol, disposable cartridge with automatic quality control features. Further, the system is capable of quantitative output more analogous to that of laboratory analyzers or EIAs.
From a cost perspective, the MBio instrument is engineered to have a cost structure that will effectively price it as a supply rather than as equipment. Component costs and assembly labor place the current prototype instrument cost at a few hundred US dollars. With no moving parts, the MBio system falls in the “minimally instrumented” category recently described in the literature as being robust, simple to operate, self-powered, and affordable both to acquire and to maintain (23).
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant AI068543 from the National Institute of Allergy and Infectious Diseases (NIAID). Initial system development was also performed under a subaward to the Rocky Mountain Regional Center of Excellence (RMRCE) for Biodefense and Emerging Infectious Diseases Research, Public Health Service grant AI065357 from NIAID. Waveguide component research was supported by the Department of Commerce, National Institute of Standards and Technology (NIST) Advanced Technology Program award 70NANB7H7053.
We are grateful to Susan Little, UCSD Division of Infectious Diseases, for supplying the characterized HIV-1 samples. We are grateful to Laura Gillim-Ross and Hugh Maguire, Colorado Department of Public Health and Environment, for access to the syphilis samples.
We are grateful to Susan Radka and Idit Pe'er of Precision Photonics Corporation for initial work on the multiplexed array concept.
Footnotes
Published ahead of print on 24 August 2011.
REFERENCES
- 1. Aach R. D., et al. 1991. Hepatitis C virus infection in post-transfusion hepatitis: an analysis with first- and second-generation assays. N. Engl. J. Med. 325:1325–1329 [DOI] [PubMed] [Google Scholar]
- 2. Allain J.-P. 1998. The status of hepatitis C virus screening. Transfus. Med. Rev. 12:46–55 [DOI] [PubMed] [Google Scholar]
- 3. Anderson D. A., Crowe S. M., Garcia M. 2011. Point-of-care testing. Curr. HIV/AIDS Rep. 8:31–37 [DOI] [PubMed] [Google Scholar]
- 4. Chamot E., et al. 1990. Loss of antibodies against hepatitis C virus in HIV-seropositive intravenous drug users. AIDS 4:1275–1277 [DOI] [PubMed] [Google Scholar]
- 5. Chien D. Y., et al. 1992. Diagnosis of hepatitis C virus (HCV) infection using an immunodominant chimeric polyprotein to capture circulating antibodies: reevaluation of the role of HCV in liver disease. Proc. Natl. Acad. Sci. U. S. A. 89:10011–10015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Christensen D. A., Herron J. N. 1996. Optical immunoassay systems based upon evanescent wave interactions. Proc. SPIE 2680:58–67 [Google Scholar]
- 7. Clerc O., Greub G. 2010. Routine use of point-of-care tests: usefulness and application in clinical microbiology. Clin. Microbiol. Infect. 16:1054–1061 [DOI] [PubMed] [Google Scholar]
- 8. Colin C., et al. 2001. Sensitivity and specificity of third-generation hepatitis C virus antibody detection assays: an analysis of the literature. J. Viral Hepat. 8:87–95 [DOI] [PubMed] [Google Scholar]
- 9. Ghany M. G., Strader D. B., Thomas D. L., Seeff L. B. 2009. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology 49:1335–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grainger D. W., Greef C. H., Gong P., Lochhead M. J. 2007. Current microarray surface chemistries. Methods Mol. Biol. 381:37–57 [DOI] [PubMed] [Google Scholar]
- 11. Hammer S. M., et al. 2008. Antiretroviral treatment of adult HIV infection: 2008 recommendations of the International AIDS Society—USA panel. JAMA 300:555–570 [DOI] [PubMed] [Google Scholar]
- 12. Herron J. N., et al. 1993. Fluorescent immunosensors using planar waveguides. Proc. SPIE 1885:28–39 [Google Scholar]
- 13. Herron J. N., et al. 2006. Planar waveguide biosensors for point-of-care clinical and molecular diagnostics. In Thompson R. B. (ed.), Fluorescence sensors and biosensors. CRC Press Taylor & Francis Group, Boca Raton, FL [Google Scholar]
- 14. Kaplan J. E., et al. 2009. Guidelines for prevention and treatment of opportunistic infections in HIV-infected adults and adolescents: recommendations from CDC, the National Institutes of Health, and the HIV Medicine Association of the Infectious Diseases Society of America. MMWR Recomm. Rep. 58(RR-4):1–207 [PubMed] [Google Scholar]
- 15. Ligler F. S. 2009. Perspective on optical biosensors and integrated sensor systems. Anal. Chem. 81:519–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mukundan J., et al. 2009. Waveguide-based biosensors for pathogen detection. Sensors 9:5783–5809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Plowman T. E., et al. 1999. Multiple-analyte fluoroimmunoassay using an integrated optical waveguide sensor. Anal. Chem. 71:4344–4352 [DOI] [PubMed] [Google Scholar]
- 18. Taitt C. R., Anderson G. P., Ligler F. S. 2005. Evanescent wave fluorescence biosensors. Biosens. Bioelectron. 20:2470–2487 [DOI] [PubMed] [Google Scholar]
- 19. Sena A. C., White B. L., Sparling P. F. 2010. Novel Treponema pallidum serologic tests: a paradigm shift in syphilis screening for the 21st century. Clin. Infect. Dis. 51:700–708 [DOI] [PubMed] [Google Scholar]
- 20. Sturenburg E., Junker R. 2009. Point-of-care testing in microbiology: the advantages and disadvantages of immunochromatographic test strips. Dtsch. Arztebl. Int. 106:48–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thio C. L., et al. 2000. Screening for hepatitis C virus in human immunodeficiency virus-infected individuals. J. Clin. Microbiol. 38:575–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tucker J. D., et al. 2010. Accelerating worldwide syphilis screening through rapid testing: a systematic review. Lancet Infect. Dis. 10:381–386 [DOI] [PubMed] [Google Scholar]
- 23. Urdea M., et al. 2006. Requirements for high impact diagnostics in the developing world. Nature 444(Suppl. 1):73–79 [DOI] [PubMed] [Google Scholar]
- 24. WHO 2010, posting date Antiretroviral therapy for HIV infection in adults and adolescents: recommendations for a public health approach. 2010 revision. http://www.who.int/hiv/pub/arv/adult2010/en/index.html [PubMed]
- 25. WHO 2010, posting date Towards universal access: scaling up priority HIV/AIDS interventions in the health sector. Progress report 2010. http://www.who.int/hiv/pub/2010progressreport/report/en/index.html
- 26. Wong R., Tse H. (ed.). 2009. Lateral flow immunoassay, vol. XII Humana Press, New York, NY [Google Scholar]