Abstract
Type-specific detection of human papillomavirus (HPV) is indicated for better risk stratification and clinical management of women testing positive for HPV and for epidemiologic surveillance. MassARRAY spectrometry (MassARRAY; Sequenom) is a novel method for type-specific detection of 15 high-risk oncogenic HPV types: HPV-16, -18, -31, -33, -35, -39, -45, -51, -52, -56, -58, -59, -66, -68, and -73. PreTect HPV-Proofer (Proofer; Norchip) is a type-specific assay that detects E6/E7 mRNA from five high-risk oncogenic HPV types: HPV-16, -18, -31, -33, and -45. The performance of these tests for type-specific identification of HPV was assessed with cervical specimens from 192 cases of cervical cancer in comparison with consensus MY09/MY11 PCR followed by nucleotide sequencing (consensus PCR). The overall HPV detection rates were 94.8% (95% confidence interval [CI], 91.7, 97.9), 83.3% (95% CI, 78.1, 88.5), and 86.5% (95% CI, 81.7, 91.3) for MassARRAY, Proofer, and consensus PCR, respectively. All tests were negative in six (3.1%) of the 192 cases. Considering only the specimens that contained at least one of the five types targeted by Proofer, the detection rates were 96.6%, 91.4%, and 86.9% for MassARRAY, Proofer, and consensus PCR, respectively. MassARRAY detected multiple infections in 14.1%, Proofer detected multiple infections in 3.6%, and consensus PCR failed to detect any multiple infections. The agreement was highest at 86.0% (kappa = 0.76) between MassARRAY and Proofer and lowest at 81.8% (kappa = 0.69) between Proofer and consensus PCR. In conclusion, MassARRAY is a highly sensitive and accurate method for type-specific detection of oncogenic HPV in cervical cancer, with Proofer showing impressive performance.
INTRODUCTION
Based on cervical cancer case-control studies, 15 human papillomavirus (HPV) types, HPV-16, -18, -31, -33, -35, -39, -45, -51, -52, -56, -58, -59, -68, -73, and -82, are classified as high-risk oncogenic types, and an additional three types, HPV-26, -53, and -66, are classified as probable oncogenic types associated with cervical cancer (6, 18). A recent report from the International Agency for Cancer Research lists 12 of these types, HPV-16, -18, -31, -33, -35, -39, -45, -51, -52, -56, -58, and -59, as having sufficient evidence for causing cervical cancer (3). Regardless, just eight of these types, HPV-16, -18, -31, -33, -35, -45, -52, and -58, account for 95% of HPV-positive cervical cancers worldwide with HPV-16 and -18 accounting for about 70% of cervical cancers (2, 18). In recent years, evidence has mounted for the type-specific identification of HPV-16 and -18 to aid risk stratification and better clinical management of women testing positive for HPV, as these two types are associated with persistent infection and lesion progression, thus conferring a significantly higher risk for cervical cancer than other oncogenic types even in the absence of cytological abnormalities (8, 14). Further, type-specific detection of HPV could increase the overall specificity in routine screening and posttreatment follow-up (13). However, the currently available HPV tests generally do not provide information on individual genotypes (9, 11, 12).
MassARRAY spectrometry (MassARRAY, Sequenom Inc.) is a novel method for type specific detection of 15 high-risk oncogenic types, HPV-16, -18, -31, -33, -35, -39, -45, -51, -52, -56, -58, -59, -66, -68, and -73. The test utilizes a three-step process composed of competitive PCR, primer extension, and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) separation of products on a matrix-loaded silicon chip array. MassARRAY is a fully automated high-throughput method which can process up to 2,000 samples in about 11 h. In addition, this test incorporates an internal oligomer standard for quality assurance which also allows for viral load quantitation (24). The latter option has not been previously available in commercial kits and may have useful clinical application. While these features are appealing, the efficacy and clinical utilization of MassARRAY have not been fully assessed.
PreTect HPV-Proofer (Proofer; Norchip) is a unique type-specific test in that it is the only test of its kind which detects E6/E7 mRNA from the five most common oncogenic types, HPV-16, -18, -31, -33, and -45, individually. Several studies have shown this test to be significantly more specific than other HPV tests for detecting high-grade cervical intraepithelial neoplasia or worse (CIN 2+) (15, 16, 22, 25). This assay is also highly specific for identifying the five types targeted by the test (16, 22). The performance of this test for the detection of HPV in cervical cancer was recently determined, in comparison with the consensus MY09/MY11 PCR followed by nucleotide sequencing of PCR products (consensus PCR), in a study we carried out to ascertain the prevalent genotypes in cervical cancer in the Indian subcontinent (1).
The aim of the present study was to compare the performance of MassARRAY with that of Proofer and consensus PCR for type-specific identification of HPV in cervical cancer. This study utilized cervical specimens that had been characterized by the Proofer and the consensus PCR methods in our previous study (1).
MATERIALS AND METHODS
Study population and sample collection.
This study utilized archived cervical specimens obtained from a total of 193 histologically proven cervical cancer cases. These were enrolled from four tertiary care cancer centers in India for a study which ascertained the prevalent genotypes in cervical cancer in the Indian subcontinent by using consensus PCR and the Proofer assay (1). The histology slides were reviewed to confirm the diagnosis. Each patient signed a written informed consent. The study was approved by the institutional ethics review boards of all participating centers.
Cervical scraping was obtained with a Cervex brush (Rovers Medical Devices, Netherlands) in a PreservCyt vial (ThinPrep, Hologic) containing a methanol-based fixative solution. All samples from the study sites were couriered in ice packs to Reliance Life Sciences Pvt. Ltd., Mumbai, India, where the initial tests were carried out. Three aliquots were made from each sample and stored at −80°C until testing. The consensus PCR was performed within a month of obtaining the samples by using one of the aliquots. The Proofer assay was carried out after all samples were accumulated and within 12 months of the collection of the first sample by using the second aliquot. MassARRAY was performed nearly 2 years later using the third aliquot at the School of Life Sciences, Jawaharlal Nehru University, New Delhi, India. MassARRAY testing was carried out under code and blinded to the results of both the consensus PCR and Proofer assays. The test procedures of the three methods are described below.
MassARRAY.
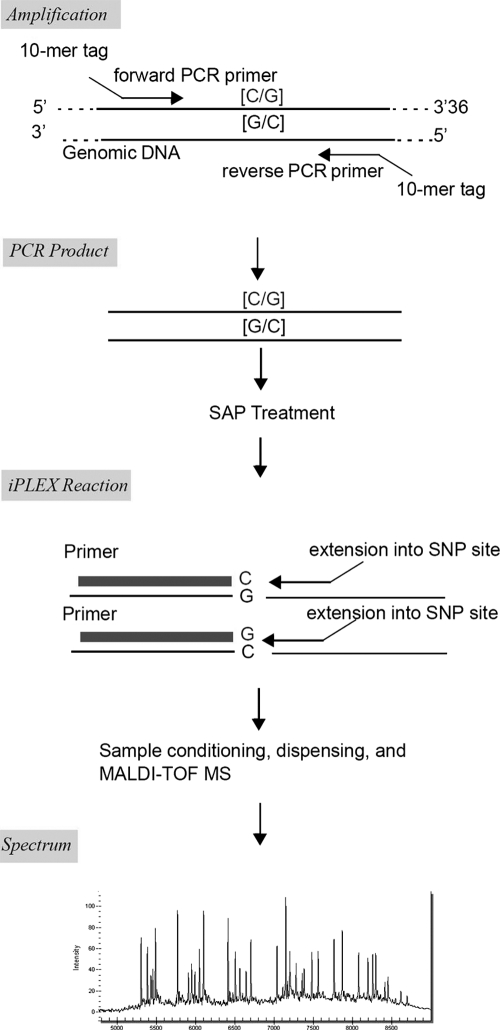
For MassARRAY (Sequenom Inc., San Diego, CA), PCR and mass spectrometer primer pairs were used for 15 high-risk HPV genotypes (HPV-16, -18, -31, -33, -35, -39, -45, -51, -52, -56, -58, -59, -66, -68, and -73). Following DNA extraction and PCR amplification, iPLEX single-base extension was performed per the manufacturer's instructions. The iPLEX assay is based on multiplex PCR followed by a single-base primer extension reaction. After the PCR, remaining nucleotides were deactivated by shrimp alkaline phosphatase (SAP) treatment, the single-base primer extension step was performed, and the primer extension products were analyzed using MALDI-TOF MS. Individual competitors for 15 HPV types were designed and mixed with extension primers of each HPV type targeted. As each HPV type had its own respective competitor into one multiplexed reaction, analysis confirmed that there were only the peaks of competitors for each type that was not present in the sample and thus excluded any possibility of contamination across the group of types targeted in the multiplexed reactions performed. The scheme of a single iPLEX assay is shown in Fig. 1 and information about the regions and the sequences selected in the design is shown in Table 1.
Fig. 1.
Scheme of a single iPLEX assay. SNP, single-nucleotide polymorphism.
Table 1.
Sequences and molecular weights of unextended (UEP) and extended primers to detect 15 high-risk HPV types in a single multiplex reaction
| HPV type | Forward primera/competitora,c | Reverse primera | Extension primera | No. of basesb; or Dad |
|---|---|---|---|---|
| 16 | ACGTTGGATGAACAGTTACTGCGACGTGAG/AGCATATGGATTCCCATCTCTATATACTATCCATAAATCCCGAAAAGCAAAGTCATATACCTCACGTCGCAGTAACTGTT | ACGTTGGATGAGCATATGGATTCCCATCTC | TTGCTTTTCGGGATTTATG | 73; 5,831 |
| 18 | ACGTTGGATGATAGCTGGGCACTATAGAGG/TGTGTTTCTCTGCGTCGTTGGAGTCGTTCCTGTCGTGCTCCGTTGCAGCACGAATGGCACTGGCCTCTATAGTGCCCAGCTAT | ACGTTGGATGTGTGTTTCTCTGCGTCGTTG | GCCATTCGTGCTGCAAC | 83; 5,146 |
| 31 | ACGTTGGATGTGGTGAACCGAAAACGGTTG/CAGGATTTTTGAACATGGCGTCTGTAGGTTTCCACAAAATACTATGTGCTTTATATACCAACCGTTTTCGGTTCACCA | ACGTTGGATGCAGGATTTTTGAACATGGCG | ATGGCGTCTGTAGGTTT | 78; 5,247 |
| 33 | ACGTTGGATGCGATTTCATAATATTTCGGG/TCACAGTGCAGTTTCTCTACGTCGGGACCTCCAACACGCCGCACAGCGCCCTCCCCAACGACCCGAAATATTATGAAATCG | ACGTTGGATGCACAGTGCAGTTTCTCTACG | ACACGCCGCACAGCGCCCT | 81; 5,704 |
| 35 | ACGTTGGATGCGCTGTGTCCAGTTGAAAAG/TTCCAACAGGACATACACCGACCTGTCCACCGTCCACGGATGTTATGGAATCGTTTTTTTTCTTCTAAATGTCTTTGCTTTTCAACTGGACACAGCG | ACGTTGGATGTTCCAACAGGACATACACCG | ACCTGTCCACCGTCCAC | 97; 5,051 |
| 39 | ACGTTGGATGGCCATACAAATTGCCAGACC/TCGTCTGCAATAGACACAGGCTATTGTAATGTCCTGCAAGGTGGTGTCCAGGGTTGTGCACAGGTCTGGCAATTTGTATGGC | ACGTTGGATGTCGTCTGCAATAGACACAGG | TTGCCAGACCTGTGCACAAC | 82; 6,062 |
| 45 | ACGTTGGATGAAGTGCATTACAGGATGGCG/CTGTGCACAAATCTGGTAGCTTGTAGGGTCGTTCCTTTGGATCGTCAAAGCGCGCCATCCTGTAATGCACTT | ACGTTGGATGCTGTGCACAAATCTGGTAGC | AAATCTGGTAGCTTGTAGGGTCGTT | 72; 7,743 |
| 51 | ACGTTGGATGGGTTATGACCGAAAACGGTG/GTCTTTCCCTCTTGTCTTCGAACATGGTGTTCTTCTATACTTTTAGCACTGCACTTTTATATGCACCGTTTTCGGTCATAACC | ACGTTGGATGGTCTTTCCCTCTTGTCTTCG | CGGTGCATATAAAAGTGCAGTG | 83; 6,824 |
| 52 | ACGTTGGATGATTGTGTGAGGTGCTGGAAG/ACCTCTCTTCGTTGTAGCTCTTTTTTGCACTGCACACACTGCAGCCTTATTTCATCCACCGATTCTTCCAGCACCTCACACAAT | ACGTTGGATGACCTCTCTTCGTTGTAGCTC | GTGCTGGAAGAATCGGTG | 84; 5,620 |
| 56 | ACGTTGGATGGTTAACTCCGGAGGAAAAGC/CAAACATGACCCGGTCCAACCATGTGCTATTAGATGAAATGGTCTTTTTCTGTCACAATGCAATTGCTTTTCCTCCGGAGTTAAC | ACGTTGGATGCAAACATGACCCGGTCCAAC | CAACCATGTGCTATTAGATGAAAT | 85; 7,360 |
| 58 | ACGTTGGATGTGATTTGTGTCAGGCGTTGG/TACCTCAGATCGCTGCAAAGTCTTTTTGCATTCAACGCATTTCAATTCGATTTCATGCACACATGTCTCCAACGCCTGACACAAATCA | ACGTTGGATGTACCTCAGATCGCTGCAAAG | GTGTCAGGCGTTGGAGACAT | 88; 6,213 |
| 59 | ACGTTGGATGCTGCCTGATTTGAGCACAAC/GCAGTTCCCCTTTGCAAAACACACAATTGATGGGAATATCATGCAGAGGAATATTCAATGTTGTGCTCAAATCAGGCAG | ACGTTGGATGGCAGTTCCCCTTTGCAAAAC | TTGCAAAACACACAATTGATG | 79; 6,422 |
| 66 | ACGTTGGATGCGGAGGAAAAACAATTGCAC/CCGGTCCATGCATATGCTATATAATGAAATCGTCTTTTATATTCACAGTGCAATTGTTTTTCCTCCG | ACGTTGGATGCCGGTCCATGCATATGCTAT | AGGAAAAACAATTGCACTGTGAA | 87; 7,113.7 |
| 68 | ACGTTGGATGTGCAGAAGGCAACTACAACG/ACCCCGTCCCTATATACTACATTTAAGTCAGCAAAGGCAAATTCATATACCTCTGTCCGTTGTAGTTGCCTTCTGCA | ACGTTGGATGACCCCGTCCCTATATACTAC | CCCTATATACTACATTTAAGTCA | 77; 6,942 |
| 73 | ACGTTGGATGTGTACAGCGTCCGGTCCAC/TGTACAGCGTCCGGTCCACTGTTCTCCTATTTGATGAAACCGTTTTTTTTCATCTACATGCTTTTGCTTTTCCAGTGGA | ACGTTGGATGTCCACTGGAAAAGCAAAAGC | GTCCGGTCCACTGTTCT | 99; 5,128.3 |
All primers are listed 5′ - 3′. The sequence of the competitor primer for each HPV type is shown in a separate row immediately below the corresponding set of forward, reverse, and extension primers.
Length of HPV type-specific PCR products amplified with forward and reverse primers.
Bold and underlined C, G, or A indicates the base of the competitor sequence that is the complement to the wild-type HPV type-specific sequence.
Mass in Da of extension primers.
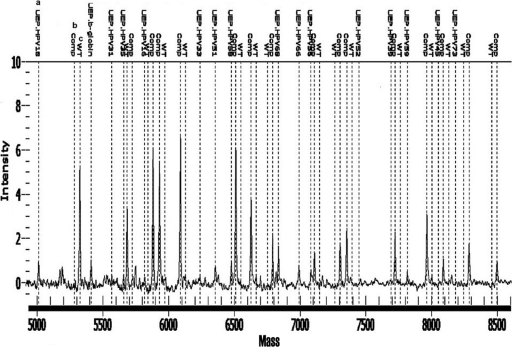
The HPV genotyping reagents obtained from the manufacturer were prestandardized to determine HPV DNA levels by real-competitive PCR (rc-PCR) technique as previously described (20). Briefly, rc-PCR involved incorporation of an oligomer within the PCRs at a final concentration of 250 attomolar, correlating to 500 copies per final reaction. This oligonucleotide was identical in sequence to the sample-amplified 53- to 97-bp product, except for the substitution of one centrally located C or G nucleotide with its complementary base. The competitor provides semiquantitation of the target HPV DNA load into 3 categories: HPV DNA greater than 1,250 aM or 2,500 copies per final reaction (target DNA product observed and no competitor product observed), HPV DNA between 50 aM and 1,250 aM or 100 to 2,500 copies (both target DNA and competitor products observed with varied frequencies relative to input copy number), and HPV DNA less than 50 aM or 100 copies (no target and only competitor product observed). Inclusion of the competitor also provides an internal quality control to ensure reaction efficiency in the absence of target HPV DNA as well as suppressing low-level environmental contamination, if present. The assay design was achieved using Sequenom assay design software as described previously (19). The primer extension products were analyzed using MALDI-TOF MS and the genotypes differentiated on the basis of the mass of each allele. Figure 2 shows an example of the assay positive for HPV type 18. A 384-well format was utilized with four 96-well plates to test in quadruplicate up to 93 patient samples per plate. Each plate consisted of three controls, comprising a positive HeLa cell line control, a negative control, and a blank. All reactions were performed in the same plate from start to finish.
Fig. 2.
Spectra of Single 15 Plex reaction confirming presence of HPV 18 type. The illustration shows wild-type (WT) calls for HPV 18 and β-globin standard type and no WT calls for other HPV types, which have only competitor peaks. Footnotes: a, UEP, unextended primer peak for individual HPV type. Low intensity confirms extension specific for type present, and high intensity confirms that no HPV type-specific material is present in the reaction. b, Comp., competitor oligomer peak. High intensity confirms that no specific HPV type is present. c, WT call confirms the presence of specific HPV type in the sample.
Proofer assay.
Proofer is a real-time multiplex nucleic acid sequence-based amplification assay (NASBA) for isothermal detection, amplification, and detection of E6/E7 mRNA from oncogenic HPV types 16, 18, 31, 33, and 45, by using molecular beacon probes. Five milliliters of cervical specimen in PreservCyt was processed for the extraction of HPV RNA by using RNeasy columns (Qiagen) following the manufacturer's instructions, and type-specific E6/E7 mRNA was detected as previously described (16). FLx800I fluorescence reader (Bio-Tek) was used for real-time detection of the accumulated mRNA product. PreTect analysis software (Norchip) was used to analyze the fluorescence profiles. To verify the integrity of RNA in the sample, the test includes a primer set and probe directed against the human U1 small nuclear ribonucleoprotein (snRNP)-specific mRNA. Standardized artificial oligonucleotides corresponding to the respective viral sequences were included as positive controls for each HPV type. Water was used as the negative control.
Consensus PCR.
DNA amplification of the HPV L1 region was performed with the consensus MY09 and MY11 PCR primers. The process involved 30 cycles of denaturation, amplification, and extension to yield a final product of 451 bp that was visualized by agarose gel electrophoresis. Molecular weight markers were used to size the specific fragment. Positive and negative controls were included, and the specimen adequacy was assessed by verifying the presence of human genomic DNA by PCR amplification of the human β-globin gene. All HPV DNA-negative samples were rechecked for conclusive results. For HPV DNA genotyping, the PCR products were subjected to nucleotide sequencing. The PCR products were purified using the Millipore Montage SEQ96 sequencing reaction cleanup kit. Nucleotide sequencing was performed by Sanger's dideoxy nucleotide sequencing method with the help of a BigDye Terminator sequencing kit (Applied Biosystems) using 5 pmol of MY09/MY11 as the sequencing primers. The sequence obtained was analyzed in silico on an ABI 3100 genetic analyzer (Applied Biosystems). For the determination of HPV type, the results were compared with documented virus sequences available in the GenBank database by using the National Center for Biotechnology Information (NCBI) BLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
Statistical analysis.
The descriptive statistics for the continuous variables were given as means with standard deviations while those for categorical data were given as frequency distributions. Fisher's exact test was used to evaluate the significance of difference between proportions. Significance was defined at P < 0.05 and P was two-tailed. The concordance between the test methods for the detection of HPV types was calculated by means of kappa statistics.
RESULTS
The mean age of the 193 patients enrolled in the study was 51.5 (±11.0) years. Information on the FIGO (Federation of International Gynecology and Obstetrics) clinical stage of cervical cancer was available for 186 patients. The stage distribution was as follows: stage I, 20 patients (10.8%); stage II, 55 patients (29.6%); stage III, 108 patients (58.0%); and stage IV, 3 patients (1.6%). Of the 193 patients, 186 (96.4%) had invasive squamous cell carcinoma on histology. Adenocarcinoma was detected in 5 (2.6%) and undifferentiated carcinoma was diagnosed in the remaining 2 (1.0%) patients.
HPV typing by MassARRAY was invalid in one sample in quadruplicate runs. The mass peak was seen only for the unextended PCR primer, thus confirming that the PCR had failed during the amplification step. This failure was attributed to the starting DNA material being of inferior quality, due to which a proper extension of the desired regions post-PCR was not achieved. This sample was excluded from further analysis, with the remaining 192 samples representing unique patients serving as the study samples.
HPV was detected by one or the other method in 186 cases (96.9%), and 6 cases (3.1%) tested negative by all three methods. MassARRAY showed the highest detection rate at 94.8% (182/192; 95% confidence interval [CI], 91.7, 97.9), followed by consensus PCR at 86.5% (166/192; 95% CI, 81.7, 91.3) and Proofer at 83.3% (160/192; 95% CI, 78.1, 88.5). The detection rate of MassARRAY was significantly different from that of either consensus PCR (P = 0.005) or Proofer (P = 0.0004). Since the Proofer assay targets only five types, i.e., HPV-16, -18, -31, -33, and -45, the detection rates were separately determined based on samples containing at least one of these types. This yielded 175 specimens which tested positive for any one of the five types by any of the methods. Based on this, the detection rate of MassARRAY to identify any of the five types was 96.6% (169/175) compared with Proofer at 91.4% (160/175) and consensus PCR at 86.9% (152/175). There were 32 (16.7%) samples containing multiple genotypes as determined by any of the methods. MassARRAY detected 27 of these (84.4%) with Proofer detecting 7 (21.9%); consensus PCR failed to detect any multiple infections (Table 2). Throughout the MassARRAY testing, there were no calls either in the negative or blank controls which contained all reagents but the DNA template, with consistent calls on the positive control, and this ensured the validity of the results.
Table 2.
Comparison of multiple HPV genotypes obtained with MassARRAY, PreTect HPV-Proofer, and consensus PCR
| Specimen serial no. | HPV genotype(s) or other result detected by: |
||
|---|---|---|---|
| MassARRAY | PreTect HPV-Proofer | Consensus PCR | |
| 1 | 16, 58 | 33 | Negative |
| 2 | 16, 39 | 16 | 16 |
| 3 | 16, 18 | 18 | 18 |
| 4 | 16, 18 | 18 | 18 |
| 5 | 16, 56 | 16 | 16 |
| 6 | 16, 18 | 18 | 16 |
| 7 | 16, 18 | 16 | 16 |
| 8 | 16 | 16, 31 | 16 |
| 9 | 16, 35 | Negative | 16 |
| 10 | 16, 18 | 18 | 18 |
| 11 | 16, 31 | 16 | Negative |
| 12 | 16, 18 | 18 | 18 |
| 13 | 16, 33 | 16 | 16 |
| 14 | 16, 18 | 18 | 18 |
| 15 | 16, 52 | Negative | 52 |
| 16 | 16, 35 | Negative | 16 |
| 17 | 16, 18 | 16 | 16 |
| 18 | 16, 18 | 16 | 16 |
| 19 | 16, 73 | Negative | 73 |
| 20 | 16 | 16, 31 | 16 |
| 21 | 16, 33 | 16 | 16 |
| 22 | 16 | 16, 18 | 16 |
| 23 | 16, 18 | 18 | 18 |
| 24 | 16, 68 | 16 | 16 |
| 25 | 16, 52 | Negative | 52 |
| 26 | 16, 18 | 16, 18, 45 | 18 |
| 27 | 16, 18 | 18 | 18 |
| 28 | 16, 18 | 18 | 18 |
| 29 | 16 | 16, 18 | 16 |
| 30 | 16, 18 | 16 | Positive; sequence uninformative |
| 31 | 16 | 16, 18 | 16 |
| 32 | 18, 35 | 16, 18 | 18 |
To calculate the agreement in genotyping results, the test results were considered to be concordant if at least one HPV type was common among the types identified by the tests in the same sample. Based on this, overall concordant results were obtained for 149 (77.6%) of the 192 samples. The observed agreement between MassARRAY and Proofer was 86.0% (165/192) (kappa = 0.76; 95% CI, 0.67, 0.84), and that of MassARRAY and consensus PCR was 84.4% (162/192) (kappa = 0.73; 95% CI, 0.64, 0.82). The lowest agreement was observed between Proofer and consensus PCR (157/192; 81.8%) (kappa = 0.69; 95% CI, 0.60, 0.78). The three techniques compared in this study were fully concordant in 65.1% (125/192), in that all genotypes present in test samples were identified by all three methods or in instances of HPV-negative results, all methods were negative for HPV. A total of 43 samples were considered discordant, as the results were not the same, with at least one HPV type being common across the three methods (Table 3). Of these, the HPV type detected by MassARRAY was also detected by one of the other two methods in 26 samples. MassARRAY also detected oncogenic HPV types in 9 samples that were HPV negative on both Proofer and PCR tests. Consensus PCR detected nononcogenic HPV types 53 and 69 in two samples that were negative on MassARRAY.
Table 3.
Comparison of discordant HPV genotype results obtained with MassARRAY, PreTect HPV-Proofer, and consensus PCR
| Specimen serial no. | Discordant HPV genotype(s) or other result obtained with: |
||
|---|---|---|---|
| MassARRAY | PreTect HPV-Proofer | Consensus PCR | |
| 1 | 16 | 16 | 18 |
| 2 | 18 | 18 | Negative |
| 3 | 16, 58a | 33 | Negative |
| 4 | 16 | 16 | Negative |
| 5 | 58a | Negative | Negative |
| 6 | Negative | 16 | 16 |
| 7 | 58a | 33 | 58a |
| 8 | 59a | Negative | Negative |
| 9 | 16 | 16 | Negative |
| 10 | 39a | Negative | Negative |
| 11 | 16 | 16 | Negative |
| 12 | 16 | Negative | Negative |
| 13 | 56a | Negative | 56a |
| 14 | 16 | 16 | 18 |
| 15 | 59a | Negative | Negative |
| 16 | Negative | Negative | 16 |
| 17 | 59a | Negative | 16 |
| 18 | 16 | 16 | Negative |
| 19 | 16 | Negative | Negative |
| 20 | 16, 35a | Negative | 16 |
| 21 | 16 | Negative | 16 |
| 22 | 16 | Negative | Negative |
| 23 | 16, 31 | 16 | Negative |
| 24 | 16, 52a | Negative | 52a |
| 25 | 16, 35a | Negative | 16 |
| 26 | 33 | 33 | 62a |
| 27 | 58a | 33 | 58a |
| 28 | Negative | Negative | 69a |
| 29 | 56a | Negative | 56a |
| 30 | 16 | Negative | 16 |
| 31 | 59a | Negative | 16 |
| 32 | 16, 73a | Negative | 73a |
| 33 | 16 | Negative | Negative |
| 34 | 56a | Negative | 56a |
| 35 | 52a | Negative | 52a |
| 36 | 16 | 16 | Negative |
| 37 | 18 | 18 | Negative |
| 38 | 16, 52a | Negative | 52a |
| 39 | 16 | 16 | 52a |
| 40 | 52a | Negative | 52a |
| 41 | Negative | Negative | 53a |
| 42 | 16 | 16 | Negative |
| 43 | 45 | Negative | Negative |
HPV genotype not targeted by PreTect HPV-Proofer.
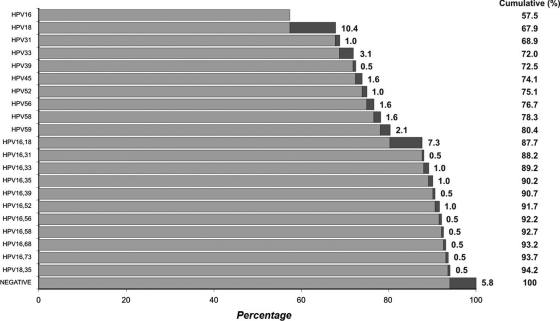
The high-risk oncogenic types detected in the 186 HPV-positive cases by at least one of the methods were limited to types 16, 18, 31, 33, 39, 45, 52, 56, 58, 59, and 68. Based on MassARRAY, type 16 alone was detected in 57.5% of cervical cancers, and in association with other oncogenic types in an additional 13.3%. Type 18 alone was detected in 10.4%, and in association with type 16 in 7.3% and with type 35 in 0.5%. The prevalence of HPV genotypes as determined by the MassARRAY method is shown in Fig. 3.
Fig. 3.
Cumulative percentages of 192 invasive cervical cancer cases attributed to the HPV types as detected by MassARRAY. The top bar indicates the attributable percentage of type 16. Subsequent to that, black bars indicate the attributable percentages of the HPV type(s) shown on the y axis.
DISCUSSION
The currently available HPV tests, including the widely used Hybrid Capture 2 assay (HC2; Qiagen), typically detect 13 or 14 high-risk oncogenic types collectively and do not provide information of individual HPV genotypes (9, 11, 12). While the sensitivity and negative predictive value of HPV tests for the detection of CIN 2+ are higher than those of cervical cytology, the specificity is lower due to the high prevalence of transient infection with a variety of HPV types (8, 9). The lower specificity could also be attributable to cross-reactivity of an HPV test with nononcogenic types (4, 22). Type-specific detection of oncogenic HPV will improve the specificity and positive predictive value of HPV tests. High-throughput HPV genotyping will also be useful for monitoring the effectiveness of HPV vaccination programs by documenting decreasing prevalence of the vaccine-targeted HPV types or by identifying possible type replacement or cross-protection after vaccination (10).
A number of different methods have been developed for HPV genotyping with some inherent differences in their performance (13). Target PCR amplification of a conserved region of the HPV L1 gene with the consensus MY09/MY11 and GP5+/GP6+ primers, or their equivalent, and subsequent genotyping with direct DNA sequencing or type-specific PCR are generally accepted scientific tools in research. However, they are too labor-intensive for routine high-throughput applications. Further, L1 gene-specific consensus PCR may fail to detect HPV in high-grade cervical lesions and cancers due to the loss of the L1 gene during the process of integration. In our study, consensus PCR failed to detect any HPV type in 26 cervical cancer samples. The presence of oncogenic HPV was confirmed by MassARRAY or Proofer in 19 of these samples, indicating an overall false-negative rate of 9.9% (19/192) for consensus PCR. Several researchers have reported the inefficiency of consensus PCR followed by sequencing to identify multiple infections in samples where viral sequences overlap and the difficulty of distinguishing the various types (5, 23, 26). There were 32 samples with multiple infections detected by either MassARRAY or Proofer assays, and consensus PCR failed to detect them in any of these samples. These data highlight the limitations of consensus PCR for HPV genotyping.
HPV E6/E7 oncoproteins are overexpressed in infected cervical epithelium during oncogenic process and cervical malignancy, and hence the detection of E6/E7 mRNA transcripts of high-risk HPV types may serve as a better risk identifier for disease progression than mere detection of DNA. We observed that the sensitivity of Proofer to detect HPV in invasive cervical cancer was better than that of consensus PCR in spite of the fact that the assay targets only 5 of the 13 or so of high-risk oncogenic HPV types. This is attributable to the preponderance of the five genotypes, especially types 16 and 18, in the cervical cancers we studied.
Mass spectrometry has been previously indicated to have high sensitivity and accuracy for detecting oncogenic HPV types. In an initial validation study, this method was found to have a better clinical sensitivity to detect CIN 2+ compared to reverse dot blot hybridization, with an overall concordance of 0.945 for detecting HPV genotypes (24). In another study, a prototype PCR- and mass spectrometry-based method to detect and quantitate 13 high-risk HPV types showed a sensitivity of 98.7% in cervical cancer tissue samples (20). A recent study which evaluated the performance of MALDI-TOF MS and surface plasmon resonance (SPR) methods reported HPV genotyping accuracy of 100% for MALDI-TOF MS and 94.8% for SPR and concluded that MALDI-TOF MS is a sensitive, specific, and feasible method for HPV detection (21). There are also indications that MassARRAY spectrometry can be successfully used to screen for oncogenic HPV types in specimens containing very low quantities of the virus, such as plasma or urine (27). Besides being a fully automated high-throughput method, this test incorporates an internal oligomer standard which allows for test validation and quality assurance. Further, MassARRAY can be used for viral load quantitation, but in the present study we did not assess this feature.
In our study, MassARRAY showed the highest HPV detection rate at 94.8%. Meticulous efforts were made to avoid contamination; all samples were tested in quadruplicate to rule out any preanalytical contamination or experimental artifacts, and negative controls were satisfactory in all runs, thus ruling out false-positive reactions. PCR failures due to allelic dropouts were excluded, as the DNA isolations were quantified and found to have a sufficient concentration of template DNA per the manufacturer's instructions. Competitors were designed and mixed with extension primers of each HPV type, and this excluded any possibility of contamination across the group of the 15 types in the multiplexed reactions performed. However, some positive results in MassARRAY, especially for type 16, were not confirmed by either of the other tests, and we did not have recourse to any other method to verify these results independently. While we attribute this to the higher sensitivity of MassARRAY, it is possible that these represent false-positive results. Although adequate safeguards and controls were used to minimize the chance of contamination as a source of false positives, we acknowledge that this risk can not be completely eliminated.
The MassARRAY assay had a high concordance with Proofer in the type-specific identification of HPV. The slightly reduced sensitivity of Proofer could be explained by low transcriptional activity, the occurrence of mutations in the regions covered by the Proofer primers or probes, or the difference in targets between the two assays. It is also likely to depend on the Proofer test's cutoff for detection as previously reported (22). MassARRAY also identified the highest number of multiple infections that may have practical implications. While multiple oncogenic HPV infection does not necessarily constitute a higher risk for the development of cervical neoplasias compared with single oncogenic HPV infection (7), a recent study has indicated that treatment failure in cervical cancer was nearly 5-fold higher in multiple HPV infections (17). Our study data based on MassARRAY indicated that types 16 and 18 alone or in association with other oncogenic types accounted for 81.7% of cervical cancers assessed in the Indian subcontinent (Fig. 3), and this has implications for HPV vaccination programs using vaccines that target HPV types 16 and 18.
In conclusion, the MassARRAY assay is a highly sensitive and accurate method for the type-specific detection of 15 high-risk oncogenic HPV in cervical cancer. Its potential capacity to detect HPV even in small quantities and to handle large number of test samples needs to be further explored in the context of cervical cancer screening and posttreatment assessment.
ACKNOWLEDGMENTS
The study was supported by a research grant through the Public Health Laboratory, St. John's, Newfoundland and Labrador, Canada, and Cancer Foundation of India, India.
We thank the following for their contributions to the study: Sutapa Biswas, Cancer Foundation of India, Kolkata, India; Uttam Das Bafna, Kidwai Memorial Institute of Cancer, Bangalore, India; Sarita Kothari, RST Regional Cancer Center, Nagpur, India; Rupinder Sekhon, Rajiv Gandhi Institute of Cancer, New Delhi, India; and Darryl Irwin, Sequenom Inc., Herston, Australia. We also thank Laura Gilbert, Public Health Laboratory, St. John's, Canada, for assistance with the preparation of the manuscript.
Footnotes
Published ahead of print on 3 August 2011.
REFERENCES
- 1. Basu P., et al. 2009. Human papillomavirus genotype distribution in cervical cancer in India: results from a multi-center study. Asian Pac. J. Cancer Prev. 10:27–34 [PubMed] [Google Scholar]
- 2. Bosch F. X., et al. 2008. Epidemiology and natural history of human papillomavirus infections and type-specific implications in cervical neoplasia. Vaccine 26S:K1–K16 [DOI] [PubMed] [Google Scholar]
- 3. Bouvard V., et al. 2009. Special report: policy. A review of human carcinogens – Part B: biological agents. Lancet 10:321–322 [DOI] [PubMed] [Google Scholar]
- 4. Castle P. E., et al. 2008. Human papillomavirus genotype specificity of Hybrid Capture 2. J. Clin. Microbiol. 46:2595–2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Choi Y. D., Jung W. W., Nam J. H., Choi H. S., Park C. S. 2005. Detection of HPV genotypes in cervical lesions by the HPV DNA chip and sequencing. Gynecol. Oncol. 98:369–375 [DOI] [PubMed] [Google Scholar]
- 6. Cogliano V., et al. 2005. Carcinogenicity of human papillomaviruses. Lancet Oncol. 6:204. [DOI] [PubMed] [Google Scholar]
- 7. Cuschieri K. S., et al. 2005. Persistent high risk HPV infection associated with development of cervical neoplasia in a prospective population study. J. Clin. Pathol. 58:946–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cuzick J., et al. 2008. Overview of human papillomavirus-based and other novel options for cervical cancer screening in developed and developing countries. Vaccine 26S:K29–K41 [DOI] [PubMed] [Google Scholar]
- 9. Cuzick J., et al. 2006. Overview of the European and North American studies on HPV testing in primary cervical cancer screening. Int. J. Cancer 119:1095–1101 [DOI] [PubMed] [Google Scholar]
- 10. Dillner J., Arbyn M., Dillner L. 2007. Translational mini review series on vaccines: monitoring of human papillomavirus vaccination. Clin. Exp. Immunol. 148:199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dockter J., et al. 2009. Clinical performance of the APTIMA® HPV Assay for the detection of high-risk HPV and high-grade cervical lesions. J. Clin. Virol. 45:S55–S61 [DOI] [PubMed] [Google Scholar]
- 12. Ginocchio C. C., Barth D., Zhang F. 2008. Comparison of the third wave invader human papillomavirus (HPV) assay and the Digene HPV Hybrid Capture 2 assay for detection of high-risk HPV DNA. J. Clin. Microbiol. 46:1641–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gravitt P. E., et al. 2008. New technologies in cervical cancer screening. Vaccine 26:K42–K52 [DOI] [PubMed] [Google Scholar]
- 14. Khan M. J., et al. 2005. The elevated 10-year risk of cervical precancer and cancer in women with human papillomavirus (HPV) type 16 or 18 and the possible utility of type-specific HPV testing in clinical practice. J. Natl. Cancer Inst. 97:1072–1079 [DOI] [PubMed] [Google Scholar]
- 15. Lie A. K., et al. 2005. DNA-versus RNA-based methods for human papillomavirus detection in cervical neoplasia. Gynecol. Oncol. 97:908–915 [DOI] [PubMed] [Google Scholar]
- 16. Molden T., Kraus I., Skomedal H., Nordstrøm T., Karlsen F. 2007. PreTect™ HPV-Proofer: real-time detection and typing of E6/E7 mRNA from carcinogenic human papillomaviruses. J. Virol. Methods 142:204–212 [DOI] [PubMed] [Google Scholar]
- 17. Munagala R., et al. 2009. Significance of multiple HPV infection in cervical cancer patients and its impact on treatment response. Int. J. Oncol. 34:263–271 [PubMed] [Google Scholar]
- 18. Muñoz N., et al. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348:518–527 [DOI] [PubMed] [Google Scholar]
- 19. Nygren A. O., et al. 2010. Quantification of fetal DNA by use of methylation-based DNA discrimination. Clin. Chem. 56:1627–1635 [DOI] [PubMed] [Google Scholar]
- 20. Patel D. A., et al. 2009. Development and evaluation of a PCR and mass spectroscopy-based (PCR-MS) method for quantitative, type-specific detection of human papillomavirus. J. Virol. Methods 160:78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qu S., et al. 15 January 2011, posting date A comparison of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and surface plasmon resonance for genotyping of high-risk human papillomaviruses. Intervirology. doi:10.1159/000322722 [DOI] [PubMed] [Google Scholar]
- 22. Ratnam S., et al. 2010. Clinical performance of the PreTect HPV-Proofer E6/E7 mRNA assay in comparison with that of Hybrid Capture 2 test for identification of women at risk of cervical cancer. J. Clin. Microbiol. 48:2779–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Serrano M. L., Correa M., Medina O., Melgarejo D., Bravo M. M. 2003. Tipificación de vírus del papiloma humano mediante secuencia directa em mujeres com citologia normal. Rev. Colomb. Cancer 7:18–24 [Google Scholar]
- 24. Soderlund-Strand A., Dillner J., Carlson J. 2008. High-throughput genotyping of oncogenic human papillomaviruses with MALDI-TOF mass spectrometry. Clin. Chem. 54:86–92 [DOI] [PubMed] [Google Scholar]
- 25. Szarewski A., et al. 2008. Comparison of predictors for high-grade cervical intraepithelial neoplasia in women with abnormal smears. Cancer Epidemiol. Biomarkers Prev. 17:3033–3042 [DOI] [PubMed] [Google Scholar]
- 26. Vernon S. D., Unger E. R., Williams D. 2000. Comparison of human papillomavirus detection and typing by cycle sequencing, line blotting, and hybrid capture. J. Clin. Microbiol. 38:651–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang H., et al. 2005. Sensitive detection of human papillomavirus in cervical, head/neck, and schistosomiasis-associated bladder malignancies. Proc. Natl. Acad. Sci. U. S. A. 102:7683–7688 [DOI] [PMC free article] [PubMed] [Google Scholar]