Abstract
Ninety-five colonizing isolates and 74 invasive isolates of Streptococcus agalactiae from Kenyan adults were characterized by using capsular serotyping and multilocus sequence typing. Twenty-two sequence types clustering into five clonal complexes were found. Data support the view that S. agalactiae isolates belonging to a limited number of clonal complexes are invasive in adults worldwide.
TEXT
Streptococcus agalactiae, or group B streptococcus (GBS), regarded mainly as a pathogen of neonates and pregnant women (15), is increasingly affecting nonpregnant adults (19). In adults, S. agalactiae causes skin and soft tissue infections, bacteremia, urinary tract infections, pneumonia, osteomyelitis, meningitis, endocarditis, and streptococcal toxic shock syndrome (2, 6, 18, 20). Risk factors associated with invasive GBS in adults are old age, diabetes mellitus, neurologic diseases, cirrhosis or other liver diseases, stroke, breast cancer, and renal failure (7, 22, 23). S. agalactiae colonizes the lower gastrointestinal and genitourinary tracts of 30 to 50% of healthy adults (26), and an estimated 20 to 30% of all pregnant women are carriers (25). S. agalactiae can be isolated from vaginal or rectal swabs, and prenatal screening for colonization of pregnant women is recommended (27). The objective of this study was to analyze the epidemiology of GBS in East African adults by using multilocus sequence typing (MLST) (13) and capsular serotyping (10), with a focus on investigating possible correlations between clonal complexes of GBS in invasive or colonizing isolates.
The Aga Khan University Hospital in Nairobi (AKUH,N) is a tertiary care, referral university hospital in Nairobi, Kenya. GBS were cultivated at the Division of Microbiology of AKUH,N from specimens derived from both inpatients and outpatients between January 2007 and June 2010. Swabs from outreach collection points were transported in Stuart's transport medium to the hospital. Blood cultures were routinely performed using Plus-aerobic/F, Plus-anaerobic/F, and Peds Plus Bactec 9120 media (Becton Dickinson) in combination with commercially available biphasic culture medium bottles (Roche Diagnostics, France). Single GBS colonies were picked from 5% sheep blood agar plates (Oxoid, United Kingdom), and identification of GBS was performed using colony morphology, Gram staining, CAMP test, the API Streptococcus identification kit (Bio-Merieux, France), and the latex agglutination test using a bacterial meningitis kit with specific group B latex (Wellcogen kit from Remel Europe Ltd.). Antimicrobial susceptibility testing was performed, and results were interpreted according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (5). Streptococcus pneumoniae ATCC 49619 was used for quality control. All GBS isolates were found to be susceptible to penicillin and cephalosporin. GBS isolates analyzed here were selected according to availability of stocked isolates and accessibility of patient files. One isolate per patient was included in this study. Clinical information was obtained by review of patient files. Ethical clearance for this study was obtained from the Ethikkommission beider Basel (EKBB) and the AKUH scientific and ethical committee.
For MLST, GBS isolates were grown overnight at 37°C on plates with Columbia agar enriched with 5% sheep blood (Oxoid). Genomic DNA extraction, MLST PCR amplification, and sequencing of seven housekeeping genes were performed as described previously (13). Novel alleles of the housekeeping genes were amplified and sequenced twice and submitted to Nicola Jones, Oxford, United Kingdom, for allele assignment. The sequence types (STs) were determined by the S. agalactiae MLST website (http://pubmlst.org/sagalactiae/) (12). The neighbor-joining tree was generated from the allelic profiles obtained using Start2 (11). The multiplex PCR assay for identification of capsular serotypes was performed as described previously (10). To assess if any clonal complex is associated with colonizing or invasive isolates, the distribution of each clonal complex was examined against random distribution by using the Pearson chi-square test. Association between clonal complex and capsular serotype distributions was assessed by using Fisher's exact test. Statistical analyses were performed using STATA version 10 (Stata Corporation, College Station, TX).
Ninety-five colonizing GBS isolates were derived from vaginal swabs collected during routine prenatal screening of pregnant women. Out of the 74 invasive isolates, 40 (54%) were derived from urine, 10 (13.5%) from blood, 8 (10.8%) from pus, and 16 (21.6%) from other body locations. Thirty-seven (50%) of the GBS from urine samples originated from women. Twenty-one of these nonpregnant female patients had repeated attacks of symptomatic urinary tract infection with significant pyuria and heavy vaginal colonization. Nine patients were antenatal symptomatic women with significant pyuria and bacteriuria. Two male patients with chronic GBS urinary tract infection suffered from prostatic hypertrophy. One elderly male patient suffered from catheter-associated GBS urinary tract infection due to neurogenic bladder. This patient developed Pseudomonas aeruginosa bacteriuria after GBS subsided under antibiotic treatment (8, 21). Five of the patients with positive blood cultures were elderly adults with comorbidities, including diabetes, renal failure, cardiac asthma, and malignancies with primary GBS bacteremia. One adult male developed GBS bacteremia secondary to peritonitis and later progressed to renal failure subsequent to toxic shock. Out of the 8 pus isolates, 4 came from cases of osteomyelitis and one each came from a diabetic foot, liver abscess, pelvic abscess aspirate, and joint fluid. A fat tissue isolate was cultured from postmortem samples from an elderly woman. Five isolates from urethral swabs came from patients with nongonococcal urethritis (16).
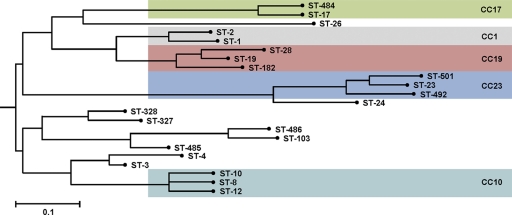
In total, 22 STs could be distinguished in this collection, with the colonizing and invasive GBS isolates grouped into 16 STs and 19 STs, respectively (Table 1). Isolates were grouped into clonal complexes (CCs) by eBURST, relaxing the group definition to six out of seven shared alleles. Thirteen STs were clustered in five CCs, namely, CC23, CC19, CC17, CC10, and CC1 (Fig. 1). The most prevalent CC was CC23 (27.2%), followed by CC17 (21.3%), CC10 (16.6%), CC19 (13.6%), and CC1 (12.4%). Eighty-eight percent of the isolates could be grouped into one of these five CCs. These five CCs are also highly prevalent in other GBS collections representing either worldwide collections (24) or investigations performed in Sweden (17), Italy (9), England (14), the United States (3), the Central African Republic, and Senegal (4) as well as Israel (1). ST-26, which represented 15% of all identified GBS isolates in the Central African Republic and Senegal (4), was found three times in our collection only. Singletons identified were ST-3, ST-4, ST-24, ST-26 ST-103, ST-327, ST-328, ST-485, and ST-486 (Table 1). Five novel STs named ST-484, ST-485, ST-486, ST-492, and ST-501 were detected. ST-485 is a double-locus variant and ST-486 is a single-locus variant of ST-103. ST-484 is a single-locus variant of ST-17, while ST-492 and ST-501 are single-locus variants of ST-23. All novel STs were found at least once in clinical isolates, and ST-484 and ST-486 were additionally found in colonizing isolates. The most prevalent novel ST was ST-484, found in 6 case isolates and 2 colonizing isolates. ST-4, ST-103, ST-327, ST-485, ST-492, and ST-501 were found exclusively in invasive GBS isolates. ST-3, ST-24, and ST-8 could be detected only among the colonizing GBS bacteria (Table 1). Interestingly, out of the 10 isolates obtained from blood, the novel ST-484, ST-485, ST-486, and ST-492 represented 30% (Table 2), supporting their high invasive potential. The possible relationship between invasive or colonizing GBS and the five CCs was analyzed (Table 2). No statistically significant correlation between any of the five CCs and invasive or colonizing GBS isolates could be described by using Pearson's chi-square test (P = 0.926). However, here we compare colonizing isolates from pregnant woman with invasive isolates from nonpregnant adults.
Table 1.
Overview of the GBS sequence types and capsular serotypes obtained in the current study
| CC and/or ST | No. of isolates |
Capsular serotype(s) (no. of isolates) |
||||
|---|---|---|---|---|---|---|
| Total | Colonizing | Invasive | Total | Colonizing | Invasive | |
| CC1a | ||||||
| ST-1 | 16 | 9 | 7 | V (15), Ia (1) | V (9) | V (6), Ia (1) |
| ST-2 | 5 | 4 | 1 | II (4), V (1) | II (4) | V (1) |
| CC10a | ||||||
| ST-8 | 5 | 5 | 0 | Ib (5) | Ib (5) | |
| ST-10 | 20 | 10 | 10 | Ib (18), II (2) | Ib (9), II (1) | Ib (9), II (1) |
| ST-12 | 3 | 1 | 2 | V (2), Ib (1) | V (1) | V (1), Ib (1) |
| CC17a | ||||||
| ST-17 | 28 | 20 | 8 | III (28) | III (20) | III (8) |
| ST-484 | 8 | 2 | 6 | III (8) | III (2) | III (6) |
| CC19a | ||||||
| ST-19 | 8 | 6 | 2 | III (5), V (3) | III (5), V (1) | V (2) |
| ST-28 | 6 | 3 | 3 | II (6) | II (3) | II (3) |
| ST-182 | 9 | 4 | 5 | III (9) | III (4) | III (5) |
| CC23a | ||||||
| ST-23 | 44 | 24 | 20 | Ia (42), III (2) | Ia (24) | Ia (18), III (2) |
| ST-492 | 1 | 0 | 1 | Ia (1) | Ia (1) | |
| ST-501 | 1 | 0 | 1 | Ia (1) | Ia (1) | |
| Singletons | ||||||
| ST-3 | 1 | 1 | 0 | IV (1) | IV (1) | |
| ST-4 | 1 | 0 | 1 | Ia (1) | Ia (1) | |
| ST-24 | 1 | 1 | 0 | Ia (1) | Ia (1) | |
| ST-26 | 3 | 2 | 1 | V (3) | V (2) | V (1) |
| ST-103 | 1 | 0 | 1 | Ia (1) | Ia (1) | |
| ST-327 | 1 | 0 | 1 | V (1) | V (1) | |
| ST-328 | 4 | 2 | 2 | V (4) | V (2) | V (2) |
| ST-485 | 1 | 0 | 1 | Ia (1) | Ia (1) | |
| ST-486 | 2 | 1 | 1 | Ia (2) | Ia (1) | Ia (1) |
Fisher's exact test P value, 0.000 (comparison among CCs).
Fig. 1.
Neighbor-joining tree demonstrating the relationship between the 22 STs of the GBS identified in the current study. The phylogenetic tree is based on the alleles of the seven housekeeping alleles sequenced and was constructed using the Start2 program (http://pubmlst.org/software/analysis/start2/). The scale bar indicates distance as calculated by the number of mismatched loci.
Table 2.
Sequence types and capsular serotypes of the GBS clinical isolates according to location of origin
| CC and/or ST | No. of isolates |
Capsular serotype(s) (no. of isolates) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | Urine | Blood | Pus | Other | Total | Urine | Blood | Pus | Other | |
| CC1a | ||||||||||
| ST-1 | 7 | 4 | 1 | 2 | V (6), Ia (1) | V (4) | Ia (1) | V (2) | ||
| ST-2 | 1 | 1 | V (1) | V (1) | ||||||
| CC10a | ||||||||||
| ST-10 | 10 | 5 | 1 | 4 | Ib (9), II (1) | Ib (5) | II (1) | Ib (4) | ||
| ST-12 | 2 | 1 | 1 | V (1), Ib (1) | V (1) | Ib (1) | ||||
| CC17a | ||||||||||
| ST-17 | 8 | 7 | 1 | III (8) | III (7) | III (1) | ||||
| ST-484 | 6 | 3 | 2 | 1 | III (6) | III (3) | III (2) | III (1) | ||
| CC19a | ||||||||||
| ST-19 | 2 | 2 | V (2) | V (2) | ||||||
| ST-28 | 3 | 3 | II (3) | II (3) | ||||||
| ST-182 | 5 | 2 | 2 | 1 | III (5) | III (2) | III (2) | III (1) | ||
| CC23a | ||||||||||
| ST-23 | 20 | 10 | 2 | 3 | 5 | Ia (18), III (2) | Ia (9), III (1) | Ia (2) | Ia (2), III (1) | Ia (5) |
| ST-492 | 1 | 1 | Ia (1) | Ia (1) | ||||||
| ST-501 | 1 | 1 | Ia (1) | Ia (1) | ||||||
| Singletons | ||||||||||
| ST-4 | 1 | 1 | Ia (1) | Ia (1) | ||||||
| ST-26 | 1 | 1 | V (1) | V (1) | ||||||
| ST-103 | 1 | 1 | Ia (1) | Ia (1) | ||||||
| ST-327 | 1 | 1 | V (1) | V (1) | ||||||
| ST-328 | 2 | 2 | V (2) | V (2) | ||||||
| ST-485 | 1 | 1 | Ia (1) | Ia (1) | ||||||
| ST-486 | 1 | 1 | Ia (1) | Ia (1) | ||||||
Pearson's chi-square test P value, 0.926 (comparison among CCs).
The capsular serotypes detected here included Ia, Ib, II, III, IV, and V. The most common serotype was serotype III (30.8%), followed by serotype Ia (30.2%), serotype V (17.2%), serotype Ib (14.2%), and serotype II (7.1%); one isolate was of serotype IV (0.6%). Out of the 44 isolates identified as ST-23, 42 were serotype Ia. All ST-17 isolates carried serotype III, 18 out of 20 ST-10 isolates had serotype Ib, 15 out of 16 ST-1 isolates expressed V, and all ST-484 and ST-182 isolates had serotype III. Hence, a tight relationship between distinct capsular serotypes and the CC could be observed (Fisher's exact test P = 0.000) (Table 1). Of note, invasive GBS typed as ST-23, ST-12, and ST-1 showed a higher variability in their capsular serotypes than colonizing isolates (Table 1). In summary, our data show that the population structure of GBS isolates in East African adults is similar to the GBS population structure reported in industrialized countries. A number of novel STs that share alleles with housekeeping genes of widespread known STs were detected.
Acknowledgments
We are thankful to the team in the Division of Microbiology, Department of Pathology, at the AKUH,N. The excellent assistance rendered by Patricia Shivachi with data management is gratefully acknowledged. The contributions made by Patricia Muthaura, Timona Obura, Alfred Murage, Maria Carvaloh, K. K. Bal, Elizabeth Njoroge, Jyothi Saha, and P. S. Patel are gratefully acknowledged. We are thankful to Penelope Vounatsou for statistical support.
Footnotes
Published ahead of print on 24 August 2011.
REFERENCES
- 1. Bisharat N., et al. 2005. Population structure of group B streptococcus from a low-incidence region for invasive neonatal disease. Microbiology (Reading, Engl.). 151:1875–1881 [DOI] [PubMed] [Google Scholar]
- 2. Blancas D., et al. 2004. Group B streptococcal disease in nonpregnant adults: incidence, clinical characteristics, and outcome. Eur. J. Clin. Microbiol. Infect. Dis. 23:168–173 [DOI] [PubMed] [Google Scholar]
- 3. Bohnsack J. F., et al. 2008. Population structure of invasive and colonizing strains of Streptococcus agalactiae from neonates of six U.S. academic centers from 1995 to 1999. J. Clin. Microbiol. 46:1285–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brochet M., Couvé E., Bercion R., Sire J.-M., Glaser P. 2009. Population structure of human isolates of Streptococcus agalactiae from Dakar and Bangui. J. Clin. Microbiol. 47:800–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clinical and Laboratory Standards Institute 2007. Performance standards for antimicrobial susceptibility testing; 16th informational supplement. M100–S17. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 6. Corvec S., et al. 2011. Clinical features of group B Streptococcus prosthetic joint infections and molecular characterization of isolates. J. Clin. Microbiol. 49:380–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Edwards M. S., Baker C. J. 2005. Group B streptococcal infections in elderly adults. Clin. Infect. Dis. 41:839–847 [DOI] [PubMed] [Google Scholar]
- 8. Farley M. M., et al. 1993. A population-based assessment of invasive disease due to group B Streptococcus in nonpregnant adults. N. Engl. J. Med. 328:1807–1811 [DOI] [PubMed] [Google Scholar]
- 9. Gherardi G., et al. 2007. Molecular epidemiology and distribution of serotypes, surface proteins, and antibiotic resistance among group B streptococci in Italy. J. Clin. Microbiol. 45:2909–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Imperi M., et al. 2010. A multiplex PCR assay for the direct identification of the capsular type (Ia to IX) of Streptococcus agalactiae. J. Microbiol. Methods 80:212–214 [DOI] [PubMed] [Google Scholar]
- 11. Jolley K. A., Feil E. J., Chan M. S., Maiden M. C. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230–1231 [DOI] [PubMed] [Google Scholar]
- 12. Jolley K., Chan M.-S., Maiden M. 2004. mlstdbNet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 5:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jones N., et al. 2003. Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 41:2530–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jones N., et al. 2006. Enhanced invasiveness of bovine-derived neonatal sequence type 17 group B streptococcus is independent of capsular serotype. Clin. Infect. Dis. 42:915–924 [DOI] [PubMed] [Google Scholar]
- 15. Koenig J. M., Keenan W. J. 2009. Group B streptococcus and early-onset sepsis in the era of maternal prophylaxis. Pediatr. Clin. North Am. 56:689–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lefevre J. C., Lepargneur J. P., Bauriaud R., Bertrand M. A., Blanc C. 1991. Clinical and microbiologic features of urethritis in men in Toulouse, France. Sex. Transm. Dis. 18:76–79 [DOI] [PubMed] [Google Scholar]
- 17. Luan S.-L., et al. 2005. Multilocus sequence typing of Swedish invasive group B streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. J. Clin. Microbiol. 43:3727–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Phares C. R., et al. 2008. Epidemiology of invasive group B streptococcal disease in the United States, 1999-2005. JAMA 299:2056–2065 [DOI] [PubMed] [Google Scholar]
- 19. Schrag S. J., et al. 2000. Group B streptococcal disease in the era of intrapartum antibiotic prophylaxis. N. Engl. J. Med. 342:15–20 [DOI] [PubMed] [Google Scholar]
- 20. Schuchat A. 1999. Group B streptococcus. Lancet 353:51–56 [DOI] [PubMed] [Google Scholar]
- 21. Schwartz B., et al. 1991. Invasive group B streptococcal disease in adults. A population-based study in metropolitan Atlanta. JAMA 266:1112–1114 [PubMed] [Google Scholar]
- 22. Sendi P., Johansson L., Norrby-Teglund A. 2008. Invasive group B streptococcal disease in non-pregnant adults. Infection 36:100–111 [DOI] [PubMed] [Google Scholar]
- 23. Skoff T. H., et al. 2009. Increasing burden of invasive group B streptococcal disease in nonpregnant adults, 1990-2007. Clin. Infect. Dis. 49:85–92 [DOI] [PubMed] [Google Scholar]
- 24. Sørensen U. B. S., Poulsen K., Ghezzo C., Margarit I., Kilian M. 2010. Emergence and global dissemination of host-specific Streptococcus agalactiae clones. mBio 1:e00178–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Valkenburg-van den Berg A. W., et al. 2010. Timing of group B streptococcus screening in pregnancy: a systematic review. Gynecol. Obstet. Invest. 69:174–183 [DOI] [PubMed] [Google Scholar]
- 26. van der Mee-Marquet N., et al. 2008. Molecular characterization of human-colonizing Streptococcus agalactiae strains isolated from throat, skin, anal margin, and genital body sites. J. Clin. Microbiol. 46:2906–2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Verani J. R., McGee L., Schrag S. J. 2010. Prevention of perinatal group B streptococcal disease—revised guidelines from CDC, 2010. MMWR Recomm. Rep. 59:1–36 [PubMed] [Google Scholar]