LETTER
The prevalence of hospital- and community-acquired Clostridium difficile infection (CDI) has recently increased, particularly in the Northern Hemisphere, with spread of the hypervirulent 027/NAP/BI strain; so far, this strain is uncommon in Australia. Rapid, accurate laboratory diagnosis of CDI guides patient management and infection control; rapid identification of 027/NAP1/BI, which can cause fulminant disease, helps predict the clinical course (3, 16).
In pathogenic C. difficile, toxins A and B are encoded by tcdA and tcdB, respectively, and their expression is regulated by tcdR (positive regulator) and tcdC (negative regulator). It has been hypothesized that hypervirulence in 027/NAP1/BI is due to increased toxin A/B production as a result of a single-base deletion at nucleotide position 117 in tcdC and the production of binary toxin CDT, encoded by cdtA and cdtB (9). There is also a characteristic 18-bp deletion in tcdC (nucleotide positions 330 to 347), which alone does not affect function (9).
Molecular typing of C. difficile is generally done in reference laboratories, with resulting delays of days to weeks. The Xpert C. difficile Epi assay (Xpert; Cepheid, Sunnyvale, CA) is a real-time, multiplex PCR assay that allows presumptive identification of 027/NAP1/BI by detecting tcdB, cdt, and the single-base deletion in tcdC in stool samples or isolates (1), with reported sensitivities and specificities of 96.6% to 99.7% and 93.0% to 98.6%, respectively (1, 5, 13). Here, we describe presumptive identification of 027/NAP1/BI by Xpert, in three stool specimens, which contained toxigenic C. difficile but not 027/NAP1/BI.
In our laboratory, liquid stool specimens are tested by the C. Diff Quik Chek Complete (TechLab, Blacksburg, VA) enzyme immunoassay for glutamate dehydrogenase (GDH) and toxins A/B. GDH-positive, toxin A/B-negative and GDH-negative, toxin A/B-positive specimens are tested with Xpert to confirm the presence of toxigenic C. difficile DNA and for preliminary identification of 027/NAP1/BI. GDH- and toxin-A/B positive and Xpert-positive stool specimens are cultured on cycloserine cefoxitin fructose agar (CCFA) for 48 h at 37°C and confirmed as C. difficile using the RapID ANA II system (Remel Inc., Lenexa, KS). Moxifloxacin susceptibility testing, by disk diffusion and Etest (AB Biodisk, Solna, Sweeden), is interpreted according to CLSI breakpoints (2). Isolates are subjected to PCR ribotyping, using a modified method incorporating capillary gel electrophoresis (12).
C. difficile 027/NAP1/BI is still rare in Australia, with only two cases and a single outbreak involving five patients reported prior to 2011 (10, 11, 15). However, a more recent investigation found that 5.3% of all C. difficile isolates typed from one Area Health Service in New South Wales, between March 2009 and November 2010, were 027/NAP1/BI (6), although none had been identified elsewhere in the state. Therefore, notwithstanding the effect of low prevalence on positive predictive values (and false-positive rates) (7), we were surprised by the presumptive identification of 027/NAP1/BI by Xpert in 3 of 101 stool specimens (of which 71 were subsequently shown to contain toxigenic C. difficile) within a 16-week period. Prior to this, Xpert had not identified any presumptive 027/NAP1/BI in clinical specimens referred from within our Area Health Service or other laboratories.
Xpert testing of cultured isolates from all three specimens was positive for tcdB but not for either cdt or the tcdC deletion. PCR ribotyping identified one as similar to ribotype 078, but the others did not match any reference strains in our database. PCR ribotype 078, in common with 027/NAP1/BI, contains tcdA, tcdB, and binary toxin but, unlike the last, has a 39-bp deletion and a point mutation at position 184 in tcdC (4). Moxifloxacin resistance, which is characteristic of, but not specific for, 027/NAP1/BI, was present only in the isolate resembling ribotype 078.
Sequencing of tcdC in all three isolates showed the typical 18-bp deletion at nucleotide positions 330 to 347 (8) and an extra 21-bp deletion at positions 354 to 374 in two, including the isolate initially identified as ribotype 078. The latter did not contain the 39-bp deletion or point mutation at position 184, suggesting that it was not a typical 078 strain. Interestingly, all three isolates showed an A>T mutation at position 117 of tcdC, different from the characteristic single-base deletion in 027/NAP1/BI.
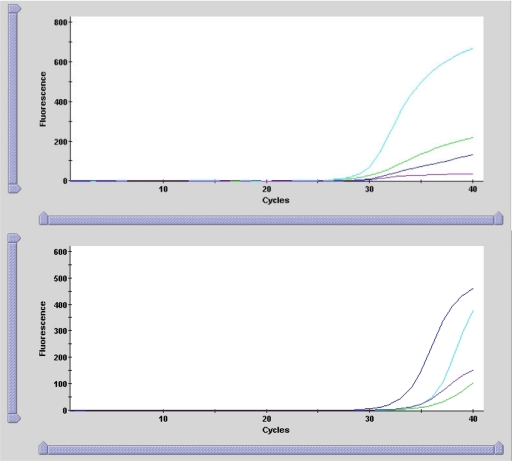
Xpert amplification curves of the three stool specimens differed from that of the positive 027/NAP1/BI control strain (a clinical isolate previously provided by another laboratory that was confirmed as 027/NAP1/BI by PCR ribotyping and tcdC sequencing) (Fig. 1) in that only the cdt curve was prominent whereas both cdt and tcdC gene deletion targets were pronounced in the control. Although the Xpert identified a single-base deletion at position 117 of tcdC, the amplification curve generated was flat compared to that of the control, possibly related to the A>T mutation at the same position. The significance of this mutation is unknown, but it may explain the false-positive Xpert result. Melt curve analysis, which is not available with Xpert at present, would have allowed further differentiation of 027/NAP1/BI and non-027/NAP1/BI strains.
Fig. 1.
Amplification curves generated by Xpert depicting fluorescence of toxin B (green), binary toxin (cdt; turquoise), single nucleotide deletion at position 117 in tcdC (purple), and positive control (blue) versus cycle number for presumptive 027/NAP1/BI. Top graph, representative pattern of three non-027/NAP1/BI stool specimens (curves for the other two isolates were similar); bottom graph, positive 027/NAP1/BI control.
Although Xpert can provide rapid laboratory diagnosis of C. difficile and presumptive 027/NAP1/BI identification in stool within 45 min (8), it is suggested that positive results be confirmed by formal typing of cultures, particularly in areas of low prevalence. However, typing of C. difficile isolates may not be available in all laboratories. Although multiple strains of C. difficile may coexist in fecal samples (14), it is unlikely that this explains our false-positive Xpert results. We prepared DNA from a sweep of each CCFA plate where C. difficile was cultured for PCR ribotyping, which should have included mixed strains if present. A North American study recently reported five stool samples in which 027/NAP1/BI was identified by Xpert but not confirmed by two or more typing methods (PCR ribotyping, pulsed-field gel electrophoresis [PFGE], and/or restriction endonuclease analysis [REA]) (13); the five isolates could not be characterized by these typing methods.
To our knowledge, this is the first report of unusual Xpert amplification curves from stool samples presumptively identified as 027/NAP1/BI but not confirmed by PCR ribotyping and tcdC gene sequencing. It is unlikely that PFGE or REA would have provided useful additional information, given the high Wallace coefficients between them and PCR ribotyping (13). Unlike the North American experience (13), we were able to characterize one of the three isolates using PCR ribotyping. We caution users of Xpert to closely examine amplification curves of presumptive 027/NAP1/BI for unusual patterns, to avoid misidentification of 027/NAP1/BI.
Nucleotide sequence accession numbers.
The three tcdC sequences were subsequently assigned GenBank accession numbers JF719678, JF719679, and JF719680.
Acknowledgments
Regarding conflicts of interest, there is nothing to declare. Also, no funding or other financial support was received in the preparation of the manuscript.
Footnotes
Published ahead of print on 17 August 2011.
REFERENCES
- 1. Babady N. E., et al. 2010. Evaluation of the Cepheid Xpert Clostridium difficile Epi assay for diagnosis of Clostridium difficile infection and typing of the NAP1 strain at a cancer hospital. J. Clin. Microbiol. 48:4519–4524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Clinical and Laboratory Standards Institute 2009. Methods for antimicrobial susceptibility testing of anaerobic bacteria. Approved standard M11-A7, 8th ed Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 3. Freeman J., et al. 2010. The changing epidemiology of Clostridium difficile infections. Clin. Microbiol. Rev. 23:529–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goorhuis A., et al. 2008. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin. Infect. Dis. 47:1162–1170 [DOI] [PubMed] [Google Scholar]
- 5. Huang H., Weintraub A., Fang H., Nord C. E. 2009. Comparison of a commercial multiplex real-time PCR to the cell cytotoxicity neutralization assay for diagnosis of Clostridium difficile infections. J. Clin. Microbiol. 47:3729–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huntington P., Darbar A., Kotsiou G. 2011. Clostridium difficile: PCR ribotype 027 in northern Sydney hospitals: a retrospective investigation. Abstr. Aust. Soc. Microbiol. Annu. Sci. Meet., Hobart, Australia http://www.asm2011.org/abstract/63.asp Accessed 8 July 2011 [Google Scholar]
- 7. Jaeschke R., Guyatt G., Sackett D. L. 1994. Users' guides to the medical literature. III. How to use an article about a diagnostic test. A. Are the results of the study valid? Evidence-Based Medicine Working Group. JAMA 271:389–391 [DOI] [PubMed] [Google Scholar]
- 8. MacCannell D. R., et al. 2006. Molecular analysis of Clostridium difficile PCR ribotype 027 isolates from Eastern and Western Canada. J. Clin. Microbiol. 44:2147–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. O'Connor J. R., Johnson S., Gerding D. N. 2009. Clostridium difficile infection caused by the epidemic BI/NAP1/027 strain. Gastroenterology 136:1913–1924 [DOI] [PubMed] [Google Scholar]
- 10. Richards M., et al. 2011. Severe infection with Clostridium difficile PCR ribotype 027 acquired in Melbourne, Australia. Med. J. Aust. 194:369–371 [DOI] [PubMed] [Google Scholar]
- 11. Riley T. V., Thean S., Hool S., Golledge C. L. 2009. First Australian isolation of epidemic Clostridium difficile PCR ribotype 027. Med. J. Aust. 190:706–708 [DOI] [PubMed] [Google Scholar]
- 12. Stubbs S. L., Brazier J., O'Neill G., Duerden B. I. 1999. PCR targeted to the 16S-23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J. Clin. Microbiol. 37:461–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tenover F. C., et al. 2011. Comparison of strain typing results for Clostridium difficile isolates from North America. J. Clin. Microbiol. 49:1831–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van den Berg R. J., et al. 2005. Coexistence of multiple PCR-ribotype strains of Clostridium difficile in faecal samples limits epidemiological studies. J. Med. Microbiol. 54:173–179 [DOI] [PubMed] [Google Scholar]
- 15. Wallace L. 2010. Hospital circular 15/2010: identification of a hypervirulent strain of Clostridium difficile. Department of Health, Melbourne, Victoria, Australia: http://www.health.vic.gov.au/hospitalcirculars/circ10/circ1510.htm Accessed 8 July 2011 [Google Scholar]
- 16. Warny M., et al. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079–1084 [DOI] [PubMed] [Google Scholar]