Abstract
Human adenovirus C (HAdV-C) species are a common cause of respiratory infections and can occasionally produce severe clinical manifestations. A deeper understanding of the variation and evolution in species HAdV-C is especially important since these viruses, including HAdV-C6, are used as gene delivery vectors for human gene therapy and in other biotechnological applications. Here, the full-genome analysis of the prototype HAdV-C6 and a recently identified virus provisionally termed HAdV-C57 are reported. Although the genomes of all species HAdV-C members are very similar to each other, the E3 region, hexon and fiber (ten proteins total) present a wide range of identity values at the amino acid level. Studies of these viruses in comparison to the other three HAdV-C prototypes (1, 2, and 5) comprise a comprehensive analysis of the diversity and conservation within HAdV-C species. HAdV-C6 contains a recombination event within the constant region of the hexon gene. HAdV-C57 is a recombinant virus with a fiber gene nearly identical to HAdV-C6 and a unique hexon distinguished by its loop 2 motif.
INTRODUCTION
Human adenovirus species C type 6 (HAdV-C6) was one of the first viruses identified and assigned a name among a group of respiratory viral pathogens isolated and recognized as HAdVs in the 1950s (33, 34). Within a short period, all of the original species C members (HAdV-C1, -C2, -C5, and -C6) were characterized and grouped together, based on their biological similarities and serotyping characteristics. In the intervening years, despite the isolation and identification of many types of HAdV and numerous circulating species C field strains using serum neutralization (SN) methods, no novel species C type has been observed until recently (23). The present study provides the genomic and computational analyses of the HAdV-C6 prototype, as well as the emergent HAdV-C57.
Although less commonly reported in the literature than other HAdVs, species C members are in fact the most prevalent HAdV species in some samplings (15–17, 23). They are important human pathogens, particularly in the immunocompromised (20, 21), and are reported primarily as respiratory agents; HAdV-C infections may be mild or asymptomatic (7) or cause serious acute respiratory disease (5, 7, 15–17, 32). HAdV-C species members may also establish as latent infections and are capable of long-term persistence, presumably evading the major histocompatibility complex class I immune response (10). As an example, species C members have constituted up to 28.5% of the typed isolates in a epidemiologic survey of HAdV acute respiratory infections of children in South America in a 4-year period (17), indicating a significant presence in that population. Another report observed that 23% of the pathogens causing acute respiratory disease in 100 Mexican children were HAdVs, with all of the isolates identified as species C exclusively (32), including HAdV-C6. This was also observed in a recent survey of Malaysian HAdV respiratory tract infections, which noted that species C were the most commonly isolated HAdV among pediatric patients in a 7-year period (1).
HAdV-C strains are used as gene delivery vectors for human gene therapy protocols (18, 36). The less prevalent HAdV-C6 is an alternative that may circumvent preexisting anti-HAdV-C5 immunity (4). As an indication of their importance as pathogens and as biotechnology tools, HAdV-C2 and -C5 were among the first five HAdV genomes sequenced (6, 30), and a resource and reference strain of HAdV-C5 has been established (37). We describe here a genomic and bioinformatic analysis of the remaining member of this group, HAdV-C6, along with the analysis of emergent HAdV-C57, an isolate formerly known as “strain 16700,” which has been noted as representing “a novel serotype of AdC” based on its earlier characterization by SN analysis and limited phylogenetic analysis (23). This is now supported by additional data and analysis using a genomics-based method that is applied as a new algorithm for understanding, defining and naming novel HAdVs (31, 40, 41).
MATERIALS AND METHODS
Virus stocks and growth.
HAdV-C6 was obtained from the American Type Culture Collection (Manassas, VA) as stock number VR-1083. It was originally isolated in the early 1950s from “spontaneously degenerating tissue culture of tonsil tissue cultures” and designated Tonsil 99 (34). This prototype was characterized and grouped into species HAdV-C using a variety of characteristics, including SN (13, 33). Growth of HADV-C6 in A549 cells and DNA production were outsourced to Virapur, LLC (San Diego, CA), using methods described earlier (29).
Isolation, growth, and preparation of HAdV-C57 (formerly designated strain 16700) were performed as described previously (23). HAdV-C57 was isolated from the feces of a healthy child as part of an acute flaccid paralysis surveillance program (12 December 2001; Baku, Azerbaijan). It was serotyped by SN and provided an ambiguous typing result.
Genome sequencing.
Commonwealth Biotechnologies, Inc. (Richmond, VA), provided the DNA sequencing of HAdV-C6, using protocols and strategies for a series of HAdV genomes reported earlier (29). In brief, Sanger-based DNA sequencing reactions following PCR amplification of the genome using DYEnamic ET terminator cycle sequencing kits (Amersham Biosciences, Piscataway, NJ) generated sequence ladders. These were resolved on an ABI Prism 377 sequencer (Applied Biosystems, Foster City, CA). This provided an average coverage of 5-fold with a minimum of 3-fold redundancy and with both strands sequenced for higher quality data. Regions yielding unreliable data, such as base-call discrepancies, were resequenced for better resolution. Additional sequencing quality control was provided by genome annotation, resulting in additional PCR-based, primer-driven resequencing to clarify any ambiguities. HAdV-C57 was sequenced using a similar strategy, with the sequencing ladders resolved on an ABI 3130x. Eightfold redundancy with both strands sequenced ensured high-quality sequence data. The GenBank accession numbers for the sequences are FJ349096 (HAdV-C6) and HQ003817 (HAdV-C57).
Genome sequence analysis.
Computational analyses were performed using publicly accessible software tools as described in earlier publications (40). All of the archived HAdVs and some simian adenoviruses were used in the analyses, with accession numbers available from an AdenovirusWiki site (http://www.binf.gmu.edu/wiki/index.php/databases). For detailed comparisons, the following species C genomes were used: HAdV-C1 (FJ349096), HAdV-C2 (FJ349096), and HAdV-C5 (AC_000008).
Whole-genome alignments were performed using Multiple Alignment, which utilizes Fast Fourier Transforms (MAFFT) (19). This program is available online (http://www.ebi.ac.uk/Tools/mafft/) and was applied using the default gap parameters in all alignments. Alignments, comparisons and visualization of genomes were performed using the zPicture software (http://zpicture.dcode.org).
Sequence percent identity analysis.
Protein and noncoding annotations were completed as described previously (29, 40). Global alignments of the sequences from HAdV-C6 and -C57 were performed using the Needle program of EMBOSS (28). As noted by Madisch et al. (24), a BLOSUM62 matrix was used for the amino acid sequence analysis, and a DNA full matrix was used for nucleotide sequence analysis. For the hexon loop analysis, the primer sequences and protocols were from Madisch et al. (24). The coding sequences of HAdV-C6 and -C57 were compared to homologs found in all other HAdV-C genomes, with the percent identities for the proteins calculated as part of the EMBOSS analysis.
Genome and gene recombination analysis.
Hexon and fiber genes from the HAdV-C genomes were by first aligned with MAFFT. SimPlot (22) was used to complete a Bootscan (22) analysis of the aligned hexon and fiber genes. Default settings were used for the window size (200 nucleotides [nt]), step size (20 nt), replicates used (n = 100), gap stripping (on), distance model (Kimura), and tree model (neighbor joining).
Whole genomes were analyzed similarly, starting with an alignment using MAFFT and following with recombination analysis using SimPlot. Only the window size and step size were altered (1,000 and 200, respectively), with the other default parameters left unchanged.
Phylogenetic analysis.
Sequence alignments for phylogenetic trees were constructed using MAFFT. Selected portions of the alignments of hexon and fiber were extracted according to genome regions used by Madisch et al. (24). Bootstrapped, neighbor-joining trees with 1,000 replicates were constructed using MEGA4 software via the maximum-composite-likelihood method (38). All of the other parameters used were set by default.
RESULTS
Genome analysis.
The genome lengths of HAdV-C6 and -C57 are 35,758 and 35,818 bp, respectively, with GC contents of 55.35 and 55.25%, respectively. The ca. 50 putative coding regions that were identified are organized in a similar manner as the genomes of other mastadenoviruses (data not shown).
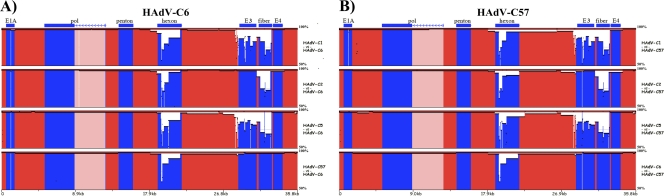
Pairwise whole-genome alignment visualizations of HAdV-C6 and -C57 were compared to the other members of species HAdV-C using the zPicture software (Fig. 1). HAdV-C6 had the greatest similarity to HAdV-C57 across the entire genome, with >95% similarity in most genome regions. The major difference between the two genomes occurred in the hexon gene, specifically within the SN epitope region, and probably accounted for its novel SN properties (23). This may be a response to selection pressures, for example, “immune escape.” Both HAdV-C6 and HAdV-C57 both displayed elevated similarity to HAdV-C2. The major differences between the three genomes were found at the hexon and fiber genes (Fig. 1).
Fig. 1.
Pairwise whole-genome comparative analysis. zPicture software was used to compare the genome sequences of HAdV-C6 (A) and HAdV-C57 (B) to other members of species C, with a default search window of 100. The y axis (scale, 50 to 100%) provides the percent identities between regions of HAdV genomes, with the x axis representing the genome. The colors are arbitrary and are used to provide contrast: blue regions highlight select genes or genome regions, also noted above the alignments, with the pink regions denoting the intron present in the DNA polymerase gene. The red regions include both coding and noncoding sequences.
Proteome analysis.
Analysis of the in silico proteomes of the HAdV-C species is informative in comparing the amino acid differences between the members. Coding regions for HAdV-C6 and HAdV-C57 genomes were compiled and presented in Table 1, along with re-annotation of the other species HAdV-C members (HAdV-C1, -C2, and -C5) (data not shown). All contained homologous open reading frames (ORFs) at similar genome locations (data not shown), reflecting the high degree of similarity among these viruses. A graphical presentation of identities between HAdV-C6 and other HAdV-C proteins is provided in Fig. 2. The proteome of HAdV-C6 was most similar to that of HAdV-C2 and only slightly less similar to HAdV-C57. This pattern was unexpected since the nucleotide sequence of HAdV-C6 was more similar to that of HAdV-C57. Several of the proteins encoded by the 5′ portion of the genome of HAdV-C6 were identical to their homologs in HAdV-C2 (Fig. 2, top portion of y axis). The lowest identity value between these two viruses was observed in the fiber protein.
Table 1.
Coding annotations of HAdV-C6 and -C57a
| Gene | Product | Coding annotation |
|
|---|---|---|---|
| HAdV-C6 | HAdV-C57 | ||
| E1A | E1A 6-kDa protein | (560 … 637, 1229 … 1318) | (560 … 637, 1227 … 1316) |
| E1A 32-kDa protein | (560 … 1112, 1229 … 1545) | (560 … 974, 1227 … 1543) | |
| E1A 26-kDa protein | (560 … 974, 1229 … 1545) | (560 … 1112, 1227 … 1543) | |
| E1B | E1B small T-antigen | (1714 … 2241) | (1714 … 2256) |
| Hypothetical protein | (2005 … 2293)c | (2006 … 2308)c | |
| Hypothetical protein | (2019 … 2252, 3215 … 3259) | (2019 … 2267, 3230 … 3274) | |
| E1B | E1B large T-antigen | (2019 … 3506) | (2019 … 3521) |
| Hypothetical protein | (2019 … 2252, 3273 … 3506) | (2019 … 2267, 3288 … 3521) | |
| pIX | IX protein | (3603 … 4025) | (3618 … 4040) |
| pIVa2 | IVa2 protein | (4084 … 5420, 5699 … 5711)c | (4099 … 5435, 5714 … 5726)c |
| E2B | DNA polymerase | (5190 … 8777, 14100 … 14108)c | (5205 … 8792, 14119 … 14127)c |
| Hypothetical protein | (5330 … 5677)c | (5345 … 5692)c | |
| Hypothetical protein | (6283 … 6603) | (6298 … 6618) | |
| Hypothetical protein | (6447 … 6783)c | (6460 … 6798)c | |
| Hypothetical protein | (7971 … 8420) | (7986 … 8435) | |
| Hypothetical protein | (8386 … 9033)c | (8401 … 9048)c | |
| Terminal protein | (8576 … 10582, 14100 … 14108)c | (8591 … 10597, 14119 … 14127)c | |
| Hypothetical protein | (9297 … 9803) | (9312 … 9818) | |
| Hypothetical protein | (10583 … 11113)c | (10762 … 11127)c | |
| L1 | 52-kDa protein | (11044 … 12291) | (11058 … 12305) |
| IIIa protein | (12312 … 14069) | (12326 … 14083) | |
| L2 | Penton base | (14145 … 15869) | (14164 … 15888) |
| VII protein | (15876 … 16472) | (15895 … 16491) | |
| V protein | (16542 … 17651) | (16561 … 17667) | |
| X protein | (17679 … 17921) | (17695 … 17937) | |
| L3 | IV | (18003 … 18755) | (18019 … 18771) |
| Hexon | (18841 … 21777) | (18857 … 21736) | |
| Protease | (21764 … 22378) | (21769 … 22383) | |
| E2A | DNA binding protein | (22475 … 24064)c | (22481 … 24070)c |
| L4 | 100-kDa protein | (24093 … 26513) | (24099 … 26522) |
| 22-kDa protein | (26224 … 26808) | (26233 … 26817) | |
| 33-kDa protein | (26224 … 26538, 26741 … 27109) | (26233 … 26548, 26751 … 27118) | |
| VIII protein | (27197 … 27880) | (27206 … 27889) | |
| E3 | 12.5-kDa protein | (27881 … 28117) | (27890 … 28213) |
| CR1-α | (28608 … 28793) | (28625 … 28810) | |
| Glycoprotein | (28790 … 29269) | (28807 … 29286) | |
| CR1-beta | (29446 … 29751) | (29508 … 29813) | |
| RID-alpha | (29759 … 30034) | (29821 … 30096) | |
| RID-beta | (30037 … 30429) | (30099 … 30491) | |
| 14.7-kDa protein | (30422 … 30808) | (30484 … 30870) | |
| U exon | U protein | (30834 … 30998)c | (30893 … 31060)c |
| L5 | Fiber | (31009 … 32595) | (31071 … 32657) |
| E4 | ORF6/7 protein | (32731 … 33009, 33721 … 33894)c | (32793 … 33071, 33783 … 33956)c |
| 34-kDa protein | (33010 … 33894)c | (33072 … 33956)c | |
| ORF4 protein | (33815 … 34159)c | (33877 … 34221)c | |
| ORF3 protein | (34171 … 34521)c | (34232 … 34582)c | |
| Hypothetical protein | (34285 … 34356, 34614 … 34946) | (34346 … 34417, 34675 … 35007) | |
| ORF2 protein | (34518 … 34910)c | (34579 … 34971)c | |
| Hypothetical protein | (34518 … 34922, 34991 … 35350)c | (34579 … 34983, 35051 … 35410)c | |
| 24-kDa protein | (34518 … 34808, 34988 … 35350)c | (34579 … 34869, 35048 … 35410)c | |
| 17-kDa protein | (34518 … 34619, 34988 … 35350)c | (34579 … 34680, 35048 … 35410)c | |
| ORF1 protein | (34964 … 35350)c | (35024 … 35410)c | |
The genomes of HAdV-C6 and -C57 are annotated with respect to the protein coding regions and listed along with their genome locations. Both genomes present homologs at similar locations and reflect the coding patterns of all species C members (data not shown).
Fig. 2.
Dot plot of the percent identity values (x axis) of the predicted proteins of HAdV-C6 versus their homologs in the other members of HAdV-C species. Amino acid comparisons were performed, with gaps considered to be mismatches. Alignments were based on a Needleman-Wunsch algorithm with the percent identification equal to the number of matches between the sequences divided by the length of the alignment. The plot was created using the R statistical computing environment (http://www.R-project.org/). The proteins appear in the order of their location within the genome (y axis), with the top of the axis representing the 5′ end. Data for HAdV-C1 (diamond), HAdV-C2 (cross), HAdV-C5 (square), and HAdV-C57 (dot) are indicated.
Hexon loop 2 motif analysis.
Quantitative relationships between hexon loops 1 and 2 regions for the viruses in species HAdV-C were generated based on sequence alignments of divergent sequences that are bracketed by conserved regions (primers) as defined by Madisch et al. (24). These authors calculated the values for pairs of closely related hexon loop 2 motif sequences in order to define the percent amino acids difference defining a new prototype as ≥1.2%. This value was calculated for HAdV-D39 and -D43. In the present study, the hexon loop 2 motif amino acid percent identity differences for HAdV-C57 and the two closest types, HAdV-C2 and -C6, are 10.9 and 13.3%, respectively. These values clearly establish a new type. The corresponding minimal nucleotide identity difference is 2.5% for HAdV-D39 and -D43 (reported by Madisch et al. [24]), and the same analysis yields 16.7 and 19.4%, respectively, for HAdV-C2 and -C6 (against HAV-C57). These metrics were calculated by using the EMBOSS Needle software with a BLOSUM62 matrix for the amino acid percent identity analysis and DNA full matrix for the nucleotide percent identity analysis. The data reported originally for HAdV-D39 and -D43 hexon loop 2 motif (24) were reconfirmed, as a control.
Recombination analysis.
Recombination analysis was performed using SimPlot, which includes the Bootscan software for two different types of analysis. SimPlots are based on any given alignment method was used to align the input sequences; in the present study we used MAFFT. MAFFT is more discriminatory than Bootscan analysis. Bootscan analyses are based on a phylogeny algorithm (unweighted pair-group method with arithmetic averages). The presence of recombination in both analyses suggests an event has occurred. Although many suggestive recombination events were detectable in the genome analysis, the high similarity of HAdV-C members in most genome regions resulted in low phylogenetic signal and made it difficult to state conclusively the origin of these regions. Since it is difficult to interpret the recombination data with absolute certainty, one conservative criterion for reliably defining recombination is the presence of a plateau rather than a series of peaks. Under this more stringent criterion, one possible recombination event was contained within each of the genomes of HAdV-C6 and -C57 in the hexon gene (Fig. 3A and B). The whole-genome analysis of HAdV-C6 (Fig. 3A) identified a recombination event between HAdV-C6 and -C2. This is supported in a higher-resolution analysis (Fig. 3C), as involving the third conserved region (C3) of the hexon gene and by the Simplot analysis (data not shown). The putative recombination in the conserved region of the HAdV-C57 hexon gene is less convincing (Fig. 3D). The higher-resolution analysis suggests that HAdV-C57 contains a portion of the conserved hexon region similar to HAdV-C1. However, the presence of multiple peaks rather than a single plateau renders this inconclusive and reflects the highly similar sequences among species HAdV-C members or multiple recombination events with an unresolvable pattern. This may be interpreted also as an “ancient” recombination event that has undergone subsequent genetic drift.
Fig. 3.
Genome recombination analysis of HAdV-C6 and -C57. Whole-genome (A) and hexon (C) Bootscan analyses of HAdV-C6 and whole-genome (B) and hexon (D) Bootscan analyses of HAdV-C57 are presented. For the hexon Simplot/Bootscan graphs, the default settings were used: window size, 200; a step size 20; 100 replicates used; gap stripping, on; Kimura distance model; and neighbor-joining tree model. The window size and step size were to increased 1,000 and 200, respectively, for the whole-genome scans. Genome nucleotide positions are noted along the x axis, and the percentages of permutated trees that supported grouping are marked along the y axis. For reference, gene-specific and selected genome landmarks are noted above each graph. The hexon genes of HAdV-C6 and C-57 are highly similar. To avoid signal competition and resultant artifacts, in the analysis, HAdV-C57 was masked in the final HAdV-C6 hexon Bootscan plot. Colors: red, HAdV-C1; green, HAdV-C2; black, HAdV-C5; blue, HAdV-C6; orange, HAdV-C57.
Another recombination event was found in the fiber gene between HAdV-C6 and -C57, detectable in a whole-genome analysis (Fig. 3A and B). A closer inspection of the fiber gene shows a plateau spanning the entire gene (see Fig. S1 in the supplemental material). If HAdV-C57 is a recently emerging type, then the parent of this sequence is a HAdV-C6-like virus.
Phylogenomic analysis.
The phylogenomic examination of HAdV-C57 is shown in Fig. 4, with partial views of the complete phylogenetic trees presented for brevity. A full analysis of all sequenced HAdVs is available at AdenovirusWiki (http://www.binf.gmu.edu/wiki/index.php/databases). The whole-genome phylogenetic analysis clearly demonstrates that the members of species HAdV-C are closely related to each other, forming a clade. Within this, the grouping of HAdV-C2, -C6 and -C57 together as a subclade, and with high confidence values, underscores a potential lineage, as well as reflects the recombination events that transferred genome fragments between ancestors of these viruses.
Fig. 4.
Phylogenomic analysis of HAdV-C57. Phylogenetic trees of the whole-genome, penton base, and fiber knob regions are presented in the top row with the hexon and its parts, “loop 1” and “conserved,” presented in the bottom row. All were generated after sequence alignments. Sequences for fiber knob and the hexon loop 1 and conserved region (C3) were the same those as reported by Madisch et al. (24). All alignments were completed using MAFFT software (http://www.ebi.ac.uk/tools/mafft) and default parameters. Bootstrapped, neighbor-joining trees with 1,000 replicates were constructed using MEGA4 software with a maximum-composite-likelihood model. Bootstrap numbers shown at the branching points indicate the percentages of 1,000 replications producing the clade.
Examination of select individual genes across the genome provides additional support for the uniqueness of HAdV-C57. The penton base gene phylogenetic tree analysis depicts a subclade that contains HAdV-C57 and -C1 and is supported by a robust bootstrap value of 96. The fiber knob region (hemagglutinin [HA] epitope) analysis shows that HAdV-C57 forms a group with HAdV-C6 with a reliable score of 100 and confirms the recombination analysis that shows both sharing a highly similar fiber gene (see above). Of particular interest are the hexon loop 1 and 2 regions, which contain the SN epitope. These regions are responsible for the serotype differentiation by serological tests, which were the gold standards in HAdV type identification. Phylogenomic analysis of the species hexon loop 2 (see Fig. S2 in the supplemental material) provides a graphical view of the amino acid and nucleotide percent identities supporting HAdV-C57 as a new type, as defined by metrics published by Madisch et al. (24) and as noted above.
DISCUSSION
General species-scale trends may be noted from the analysis of the genomes and the in vitro proteomes (Fig. 2). First, the majority of the proteins within HAdV-C species were highly similar between viruses of different types. Second, the hexon, E3 region and fiber proteins (total of 10 proteins) showed a wide range of identity values, indicating a higher degree of variability at the amino acid level. These proteins are involved in interaction with cellular receptors and host immune system (3). High variability among the major immunogenic proteins, hexons, and fibers can be explained by immune pressure. A comparable degree of divergence among E3 proteins suggests a similar degree of evolutionary pressure on these viruses and implies that selection and conservation in these proteins is markedly different from other adenovirus nonstructural proteins. It is conceivable that these proteins are involved in host cell adaptation and correspond in function and evolution to virus “security proteins,” which are proposed to form “a distinct class” and are “dedicated specifically to counteracting host defenses” (2).
The in silico analysis presented in Fig. 2 suggests that other genome regions, in addition to the ones corresponding to the major coat proteins, may be useful as an additional metric for typing HAdVs and for determining novel types. Presently, the partial amino acid sequences used for molecular typing derive from loops 1 and 2 of the hexon protein (which are involved in SN) and the fiber knob (which is responsible for hemagglutination). Collectively, these represent ca. 5 to 6% of the genome. The additional region identified here encompasses genes that are encoded contiguously and includes the E3-encoded genes (which may vary in number) and the fiber gene. The E3 and fiber sequences may be extracted from the genome as a single sequence fragment, ca. 4,000 to 5,000 nt in length, and used as a metric for typing. The advantage is that it spans ca. 13% of the genome, represents the 3′ end of the genome, and contains variability that is useful for parsing types. The hexon, E3, and fiber regions could be amplified in two PCR amplicons. These amplicons could be sequenced with 8 to 10 Sanger sequencing reactions to serve as a cost-effective and preliminary alternative to whole-genome data, as requested by some researchers who do not have access to whole-genome sequencing. In all, the aforementioned scheme would provide, in one glance, information on both the two regions desired for molecular typing (hexon and fiber) and the sequence of the most variable part of the genome, which may carry the most phylogenetic information. It is important to note that such a preliminary study should be confirmed eventually with a whole-genome determination for a thorough description, since possible recombination events would not be surveyed.
Genome recombination requires coinfection, which is observed in HAdV infections (8, 26, 39). Putative recombinants, based on the neutralization epsilon and hemagglutination gamma determinants, are recognized as different prototypes by the community, with additional field strains described based on these two markers (SN and hemagglutination inhibition assays) as case studies in the literature (9, 11, 12). Recently, these have been thoroughly characterized using high-resolution genomics-based and bioinformatics-based methods in great detail (31, 40, 41). It has been suggested that recombination is common in species HAdV-C and, probably, in other HAdV species (23). The newly completed genome sequences of HAdV-C6 and -C57 allow for a more detailed analysis of recombination in species HAdV-C.
Both putative hexon recombination events in these two genomes are unique because, unlike other reported HAdV hexon recombination events (40, 41), they do not involve the variable loops of the hexon. Instead, these events occur in the C3 (conserved) region of the hexon. It should be noted that the recombinant areas do not have identical lengths and positions. This area of the hexon gene is highly conserved among HAdVs and usually interferes with the recombination scans of the region. However, the C3 region of HAdV-C species shows a relatively high degree of variability relative to one another. It is possible that recombination events in this area are common but can only be observed in species HAdV-C owing to sufficient sequence variability.
The zPicture and genome percent identity data show that the HAdV-C2, -C6, and -C57 sequences were similar throughout most of their genomes. These genomes differed significantly in only two regions: the hexon and fiber genes. This pattern suggests that HAdV-C2, -C6, and -C57 share an ancestor relative to the other HAdV-C types. This pattern also reveals the possibility that the evolution of these three viruses occurred through a gradual path of divergence. On the contrary, the Bootscan data presented here suggests that recombination was commonly involved in the evolution of HAdV-C2, -C6, and -C57. In this recombinant history scenario, HAdV-C6 could result from a HAdV-C2-like hexon recombination, and HAdV-C57 is the result of HAdV-C6 fiber recombination.
HAdV-C57 is a new type based on phylogenomics and computational analysis of the hexon loop 2 motif. The novel but proven algorithm for HAdV typing calls for the use of genomics-based analysis and genome metrics to identify, characterize, and establish novel HAdVs, along with differences in the biology and/or pathogenicity of the virus (14, 31, 40, 41). One key component is phylogenomics: examining several genes spanning the genome and including the genome regions associated with serological properties and other key virus features. These landmarks include DNA sequences representing the SN epitopes (hexon loops 1 and 2) and the HA epitope (fiber knob) (24, 25). In the past, differences in SN were used to establish a new serotype. Currently, the hexon epitopes are sequenced and commonly substituted for SN. However, this is not identical to SN and should be referred to as, more appropriately, “imputed serum neutralization” (24, 27, 35, 42). The genomic and computational data presented here for HAdV-C6 and HAdV-C57 may be correlated with published serology data (13, 23) and allow a deeper understanding of how these diverse data and research approaches complement and potentially conflict with each other.
Recently, laboratories are sequencing the loops 1 and 2 motifs of the hexon gene and using qualitative phylogenetic approaches to type a particular HAdV rather than the serological methods. There is no clarity as to what degree of sequence divergence could distinguish a new serotype from a previously known one, if based solely on the qualitative interpretation of the phylogenetic data. Quantitatively, Madisch et al. (24) explored the relationships between the hexon loops 1 and 2 regions from all of the prototypes. These authors calculated an amino acid sequence percent identity difference of ≥1.2% as defining a new type. This was based on the difference between the two most closely related HAdV-D39 and -D43 hexon loop 2 motifs. For this report, the amino acid percent identity differences of 10.9 and 13.3%, respectively, are calculated for the loop 2 motif of HAdV-C2 and -C6 (each against HAdV-C57), and these values clearly establish HAdV-C57 as a new type. The corresponding minimal nucleotide sequence identity difference is 2.5% for HAdV-D39 and -D43 (reported by Madisch et al. [24]), and the same analysis yields 16.7 and 19.4% for HAdV-C2 and -C6, respectively (against HAdV-C57).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by U.S. Public Health Service National Institutes of Health (NIH) grants EY013124 (D.S., M.P.W., M.S.J. and J.C.) and P30EY014104 (J.C.). M.S.J. was also supported in part by the U.S. Air Force Surgeon General (Clinical Investigation no. FDG20040024E). J.C. was also funded by an unrestricted grant to the Department of Ophthalmology, Harvard Medical School, from Research to Prevent Blindness, Inc. The sequencing of the genome from HAdV-C6 was undertaken with support from the Department of Defense in which one of the authors (D.S.; 2002 to 2004) was affiliated with the USAF Surgeon General Office, Directorate of Modernization (SGR), and the Epidemic Outbreak Surveillance (EOS) Program, Falls Church, VA. Portions from the HAdV-C6 work were funded specifically, during these time periods, by a grant from the U.S. Army Medical Research and Material Command (USAMRMC; DAMD17-03-2-0089), and additional support was provided through the EOS Project, funded by HQ USAF Surgeon General Office, Directorate of Modernization (SGR), and the Defense Threat Reduction Agency (DTRA).
D.S. thanks Clark Tibbetts (EOS; 2001 to 2005) for initial discussions and for providing the impetus and opportunity to pursue this line of research in adenovirus genomics. We thank David Schnurr for critical readings and thoughtful discussions on the serotyping.
The views expressed in this material are those of the authors and do not reflect the official policy or position of the U.S. Government, the Department of Defense, or the Department of the Air Force.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 17 August 2011.
REFERENCES
- 1. Abd-Jamil J., Teoh B. T., Hassan E. H., Roslan N., Abubakar S. 2010. Molecular identification of adenovirus causing respiratory tract infection in pediatric patients at the University of Malaya Medical Center. BMC Pediatr. 10:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agol V. I., Gmyl A. P. 2010. Viral security proteins: counteracting host defences. Nat. Rev. 8:867–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burgert H. G., Blusch J. H. 2000. Immunomodulatory functions encoded by the E3 transcription unit of adenoviruses. Virus Genes 21:13–25 [PubMed] [Google Scholar]
- 4. Capone S., et al. 2006. A novel adenovirus type 6 (Ad6)-based hepatitis C virus vector that overcomes preexisting anti-Ad5 immunity and induces potent and broad cellular immune responses in rhesus macaques. J. Virol. 80:1688–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Casas I., et al. 2005. Molecular identification of adenoviruses in clinical samples by analyzing a partial hexon genomic region. J. Clin. Microbiol. 43:6176–6182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chroboczek J., Bieber F., Jacrot B. 1992. The sequence of the genome of adenovirus type 5 and its comparison with the genome of adenovirus type 2. Virology 186:280–285 [DOI] [PubMed] [Google Scholar]
- 7. Echavarria M. 2009. Adenovirus, p. 463–488 In Zuckerman A. J., et al. (ed.), Principles and practice of clinical virology, 6th ed John Wiley & Sons, Inc., San Diego, CA [Google Scholar]
- 8. Echavarria M., et al. 2006. Use of PCR to demonstrate presence of adenovirus species B, C, or F as well as coinfection with two adenovirus species in children with flu-like symptoms. J. Clin. Microbiol. 44:625–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Engelmann I., Madisch I., Pommer H., Heim A. 2006. An outbreak of epidemic keratoconjunctivitis caused by a new intermediate adenovirus 22/H8 identified by molecular typing. Clin. Infect. Dis. 43:e64–e66 [DOI] [PubMed] [Google Scholar]
- 10. Garnett C. T., et al. 2009. Latent species C adenoviruses in human tonsil tissues. J. Virol. 83:2417–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Henquell C., et al. 2009. Fatal adenovirus infection in a neonate and transmission to health-care workers. J. Clin. Virol. 45:345–348 [DOI] [PubMed] [Google Scholar]
- 12. Hierholzer J. C., Pumarola A., Rodriguez-Torres A., Beltran M. 1974. Occurrence of respiratory illness due to an atypical strain of adenovirus type 11 during a large outbreak in Spanish military recruits. Am. J. Epidemiol. 99:434–442 [DOI] [PubMed] [Google Scholar]
- 13. Hierholzer J. C., Stone Y. O., Broderson J. R. 1991. Antigenic relationships among the 47 human adenoviruses determined in reference horse antisera. Arch. Virol. 121:179–197 [DOI] [PubMed] [Google Scholar]
- 14. Jones M. S., II, et al. 2007. New adenovirus species found in a patient presenting with gastroenteritis. J. Virol. 81:5978–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kajon A. E., et al. 1996. Molecular epidemiology of adenovirus acute lower respiratory infections of children in the south cone of South America (1991–1994). J. Med. Virol. 48:151–156 [DOI] [PubMed] [Google Scholar]
- 16. Kajon A. E., Portes S. A., de Mello W. A., Nascimento J. P., Siqueira M. M. 1999. Genome type analysis of Brazilian adenovirus strains of serotypes 1, 2, 3, 5, and 7 collected between 1976 and 1995. J. Med. Virol. 58:408–412 [DOI] [PubMed] [Google Scholar]
- 17. Kajon A. E., Suarez M. V., Avendano L. F., Hortal M., Wadell G. 1993. Genome type analysis of South American adenoviruses of subgenus C collected over a 7-year period. Arch. Virol. 132:29–35 [DOI] [PubMed] [Google Scholar]
- 18. Kalyuzhniy O., et al. 2008. Adenovirus serotype 5 hexon is critical for virus infection of hepatocytes in vivo. Proc. Natl. Acad. Sci. U. S. A. 105:5483–5488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Katoh K., Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9:286–298 [DOI] [PubMed] [Google Scholar]
- 20. Lion T., et al. 2003. Molecular monitoring of adenovirus in peripheral blood after allogeneic bone marrow transplantation permits early diagnosis of disseminated disease. Blood 102:1114–1120 [DOI] [PubMed] [Google Scholar]
- 21. Lion T., et al. 2010. Monitoring of adenovirus load in stool by real-time PCR permits early detection of impending invasive infection in patients after allogeneic stem cell transplantation. Leukemia 24:706–714 [DOI] [PubMed] [Google Scholar]
- 22. Lole K. S., et al. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lukashev A. N., Ivanova O. E., Eremeeva T. P., Iggo R. D. 2008. Evidence of frequent recombination among human adenoviruses. J. Gen. Virol. 89:380–388 [DOI] [PubMed] [Google Scholar]
- 24. Madisch I., Harste G., Pommer H., Heim A. 2005. Phylogenetic analysis of the main neutralization and hemagglutination determinants of all human adenovirus prototypes as a basis for molecular classification and taxonomy. J. Virol. 79:15265–15276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Madisch I., Wolfel R., Harste G., Pommer H., Heim A. 2006. Molecular identification of adenovirus sequences: a rapid scheme for early typing of human adenoviruses in diagnostic samples of immunocompetent and immunodeficient patients. J. Med. Virol. 78:1210–1217 [DOI] [PubMed] [Google Scholar]
- 26. Metzgar D., et al. 2005. PCR analysis of egyptian respiratory adenovirus isolates, including identification of species, serotypes, and coinfections. J. Clin. Microbiol. 43:5743–5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miura-Ochiai R., et al. 2007. Quantitative detection and rapid identification of human adenoviruses. J. Clin. Microbiol. 45:958–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Olson S. A. 2002. EMBOSS opens up sequence analysis: European Molecular Biology Open Software Suite. Brief. Bioinform. 3:87–91 [DOI] [PubMed] [Google Scholar]
- 29. Purkayastha A., et al. 2005. Genomic and bioinformatics analysis of HAdV-4, a human adenovirus causing acute respiratory disease: implications for gene therapy and vaccine vector development. J. Virol. 79:2559–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roberts R. J., et al. 1986. A consensus sequence for the adenovirus-2 genome, p. 1–51 In Doerfler W. (ed.), Adenovirus DNA. Martinus Nijhoff, Boston, MA [Google Scholar]
- 31. Robinson C. M., et al. 2011. Computational analysis and identification of an emergent human adenovirus pathogen implicated in a respiratory fatality. Virology 409:141–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rosete D. P., Manjarrez M. E., Barron B. L. 2008. Adenoviruses C in non-hospitalized Mexican children older than five years of age with acute respiratory infection. Mem. Inst. Oswaldo Cruz 103:195–200 [DOI] [PubMed] [Google Scholar]
- 33. Rowe W. P., Hartley J. W., Huebner R. J. 1956. Additional serotypes of the APC virus group. Proc. Soc. Exp. Biol. Med. 91:260–262 [DOI] [PubMed] [Google Scholar]
- 34. Rowe W. P., Huebner R. J., Hartley J. W., Ward T. G., Parrott R. H. 1955. Studies of the adenoidal-pharyngeal-conjunctival (APC) group of viruses. Am. J. Hyg. 61:197–218 [Google Scholar]
- 35. Shimada Y., et al. 2004. Molecular diagnosis of human adenoviruses d and e by a phylogeny-based classification method using a partial hexon sequence. J. Clin. Microbiol. 42:1577–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stone D., et al. 2007. Comparison of adenoviruses from species B, C, E, and F after intravenous delivery. Mol. Ther. 15:2146–2153 [DOI] [PubMed] [Google Scholar]
- 37. Sugarman B. J., Hutchins B. M., McAllister D. L., Lu F., Thomas B. K. 2003. The complete nucleotide acid sequence of the adenovirus type 5 reference material (ARM) genome. Bioproc. J. Sep-Oct:27–32 [Google Scholar]
- 38. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 39. Vora G. J., et al. 2006. Co-infections of adenovirus species in previously vaccinated patients. Emerg. Infect. Dis. 12:921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Walsh M. P., et al. 2009. Evidence of molecular evolution driven by recombination events influencing tropism in a novel human adenovirus that causes epidemic keratoconjunctivitis. PLoS One 4:e5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Walsh M. P., et al. 2010. Computational analysis identifies human adenovirus type 55 as a re-emergent acute respiratory disease pathogen. J. Clin. Microbiol. 48:991–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu W., McDonough M. C., Erdman D. D. 2000. Species-specific identification of human adenoviruses by a multiplex PCR assay. J. Clin. Microbiol 38:4114–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.