Abstract
The assembly of the cytochrome bc1 complex in Saccharomyces cerevisiae is shown to be conditionally dependent on a novel factor, Mzm1. Cells lacking Mzm1 exhibit a modest bc1 defect at 30°C, but the defect is exacerbated at elevated temperatures. Formation of bc1 is stalled in mzm1Δ cells at a late assembly intermediate lacking the Rieske iron-sulfur protein Rip1. Rip1 levels are markedly attenuated in mzm1Δ cells at elevated temperatures. Respiratory growth can be restored in the mutant cells by the overexpression of the Rip1 subunit. Elevated levels of Mzm1 enhance the stabilization of Rip1 through physical interaction, suggesting that Mzm1 may be an important Rip1 chaperone especially under heat stress. Mzm1 may function primarily to stabilize Rip1 prior to inner membrane (IM) insertion or alternatively to aid in the presentation of Rip1 to the inner membrane translocation complex for extrusion of the folded domain containing the iron-sulfur center.
INTRODUCTION
The ubiquinol-cytochrome c reductase (bc1 complex, complex III) is a key component of the mitochondrial OXPHOS respiratory chain linking ubiquinol oxidation to cytochrome c reduction for terminal oxidation by cytochrome oxidase (CcO, complex IV). The eukaryotic bc1 complex contains three redox subunits involved in electron transfer and 7 or 8 supernumerary subunits (18, 35). In contrast, the bacterial complex contains the core redox subunits and a single supernumerary subunit, if any (8). Of the core redox subunits, only the cytochrome b (Cob) subunit is encoded by the mitochondrial genome, whereas the other subunits are nuclear DNA encoded and imported into mitochondria from the cytoplasm. The Cob subunit anchors the core of the bc1 complex that exists as a homodimeric unit within the inner mitochondrial membrane. The interface within the homodimeric unit consists of interactions of the Cob subunits and the matrix-facing Cor2 subunit (18, 35). In addition, the Rieske Fe/S protein Rip1 is intertwined between adjacent monomers with its globular domain in one unit and its transmembrane helix in the adjacent unit. The bc1 complex is organized into higher-order supercomplexes, associating with CcO in Saccharomyces cerevisiae and both CcO and the complex I NADH oxidase in metazoans (30, 31). The bc1 subunit Qcr9 is important for formation of this supercomplex and is likely to reside at the interaction interface (16).
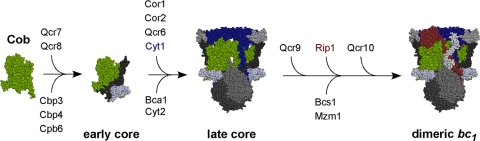
The biogenesis of the bc1 complex occurs as a modular assembly pathway with the mitochondrially encoded Cob seeding the complex assembly (37, 38). The nucleation by cytochrome b may explain why it is mitochondrially encoded in all known eukaryotes. An early core assembly complex containing Cob associated with two nucleus-encoded subunits, Qcr7 and Qcr8, is documented in yeast (Fig. 1) (3, 38). A second subassembly intermediate consists of the early core complex along with Cyt1, Cor1, Cor2, and Qcr6 (3, 37), lacking only Qcr9, Qcr10, and Rip1. This second intermediate of ∼500 kDa, herein referred to as the late core intermediate, stably accumulates in yeast lacking either Rip1 or Qcr9. Since Rip1 contains the essential Fe/S center that facilitates electron transfer to the cytochrome c1 subunit Cyt1, the assembly intermediate is devoid of function. Qcr10 is a late addition with Rip1 (5, 37) and appears to stabilize the complex, as Rip1 is labile within the bc1 complex in the absence of Qcr10 (2). Consistent with its late addition, Rip1 can be extracted from the purified complex and reconstituted back to restore function (7).
Fig. 1.
Schematic for assembly of the bc1 complex in yeast mitochondria. The Cob translational activator Cbp6 and assembly factor Cbp3 associate with the nascent polypeptide as it exits the mitochondrial ribosome. Cbp3 and Cbp6 remain bound to the newly synthesized Cob, and Cbp4 is recruited to this complex. The supernumerary subunits Qcr7 and Qcr8 associate with Cob (green) to form the early core complex. The core protein subunits, Cor1 and Cor2, along with Qcr6 subunits and the catalytic subunits of Cyt1 (blue), are added to form the late core complex. Bca1 is a recently identified assembly factor for bc1 that acts prior to formation of the late core, and Cyt2 is the heme lyase. The assembly factors Bcs1 and Mzm1 assist in formation of the functional homodimeric bc1 complex by assisting in the insertion of Rip1 (shown in ruby). Qcr9 and Qcr10 subunits are also added at a late step. Protein subunits of the bc1 enzyme complex are listed above the arrows; nonsubunit proteins are listed below.
The late core intermediate containing Cyt1 is likely a dimeric complex, but it fails to associate with CcO (4, 37). While the predicted mass for a dimeric complex lacking Qcr9, Qcr10, and Rip1 would be 400 kDa, the size of the second assembly intermediate is approximately 500 kDa. An additional molecule associated with the late core intermediate is the assembly factor Bcs1 (5). Bcs1 is an AAA ATPase that is critical for the biogenesis of the bc1 complex. In the absence of Bcs1, yeast accumulates the late core intermediate and steady-state levels of Rip1 are diminished. Bcs1 was proposed to be an ATP-dependent chaperone that maintains the assembly intermediate in a competent state for subsequent insertion of Rip1 and Qcr10 (5).
A series of other assembly factors in addition to Bcs1 participate in biogenesis of the bc1 complex in yeast (Fig. 1). Cbp3, Cbp4, and Cbp6 function at an early step in Cob translation and in addition associate with the newly synthesized Cob polypeptide (14, 19). Cyt2 is the CC1HL heme lyase that covalently attaches heme to apo-Cyt1 within the intermembrane space (IMS) (39, 40). Recently, Bca1 was shown to function in bc1 assembly in fungi (22). Bca1 is postulated to act at an undefined step prior to insertion of Rip1 (22). A novel metazoan bc1 assembly factor, TTC19, was also recently reported (9). Patients with mutations in TTC19 show a marked bc1 deficiency, with assembly intermediates being devoid of Rip1.
We identified a mitochondrial protein, Mzm1 (mitochondrial zinc maintenance), that when depleted in yeast results in a diminution in a mitochondrial labile, cationic zinc pool (1). In addition to the reduced labile zinc pool, mzm1Δ cells exhibit a destabilized bc1 complex without perturbation in cytochrome oxidase or ATP synthase. The defect in bc1 biogenesis in mzm1Δ cells is especially marked at 37°C. We show presently that Mzm1 is a bc1 assembly factor functioning at the late assembly step of Rip1 insertion into the late core assembly intermediate. Mzm1 cooperates with Bcs1 in the Rip1 insertion step.
MATERIALS AND METHODS
Yeast strains and vectors.
Strains were BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0). The Cor2::TAP strain was purchased from Open Biosystems. The Bcs1::Myc strain was constructed through insertion of a PCR cassette (32) to append a 3′ tag with 13 repeats of the Myc epitope (EQKLISEEDL) and the HIS3MX6 coding sequence for selection. To generate the mzm1Δ deletion in these tagged strains, MZM1 was replaced with a Candida albicans URA3 coding sequence. Strains were grown either in 1% dextrose rich medium (yeast peptone dextrose) or in 2% galactose synthetic complete medium lacking relevant amino acids to maintain plasmid selection. For growth assays on solid medium, synthetic complete medium was supplemented with either 2% glucose or glycerol-lactate and relevant amino acids. Cultures for growth tests were grown in 2% glucose synthetic complete medium overnight and then normalized to absorbance at 600 nm and applied to solid medium in 1:10 serial dilutions.
Plasmids containing MZM1, RIP1, and BCS1 were constructed as follows. MZM1 was 3′ tagged with Myc and six histidine repeats and placed under the control of the MET25 promoter and the CYC1 terminator on a pRS413 vector. This Mzm1-MycHis plasmid was created by cloning a 5′ BamHI-to-3′ SalI fragment created by PCR into a vector containing the MET25 promoter and CYC1 terminator (24). To make point mutations in this plasmid-borne MZM1, Tyr11 and Arg23 were replaced with alanine by overlap extension PCR utilizing primers encoding the relevant substitution together with vector-based primers. The final PCR product was cloned into a pRS413 vector containing the MET25 promoter and CYC1 terminator by in vivo ligation. To target the C-terminal portion of Mzm1 beginning at glutamate 27 to the mitochondrial matrix, the Sod2 mitochondrial targeting sequence corresponding to amino acids 1 to 26 was cloned into a pRS413 vector as an XbaI/BamHI fragment, and the MZM1 sequence (from either the Mzm1-HisMyc clone or the point mutants) was appended following the BamHI site using overlapping oligonucleotides and in vivo gap repair. RIP1 was cloned by PCR using primers anchored in the RIP1 promoter and terminator appended with vector sequence to facilitate in vivo gap repair into BamHI- and NotI-digested pRS vector systems. This strategy created both pRS415 and pRS425 vectors bearing RIP1 under the control of its endogenous promoter and terminator. Using these RIP1 plasmids as a template, a plasmid containing the S183C point mutation in RIP1 was generated by overlap extension PCR utilizing primers encoding the substitution together with vector-based primers. BCS1 was 3′ tagged with Myc and cloned into either pRS416 or pRS426 vector under the control of the MET25 promoter and the CYC1 terminator (24). The tag was added by PCR, using primers appended with vector sequence to facilitate in vivo gap repair of the PCR product into the vectors digested with either 5′ BamHI/3′ PstI (pRS416) or 5′ EcoRI/3′ XhoI (pRS426). All vectors were confirmed by sequencing.
Suppression of mzm1Δ cells.
The mzm1Δ strain was transformed with a YEp library derived from the coa2Δ strain (29). Approximately 52,000 transformants were recovered on glucose-containing synthetic complete medium lacking uracil. Colonies were then replica plated onto glycerol-lactate synthetic complete plates lacking uracil and grown at 37°C. Plasmids from recovered colonies were isolated and sequenced with vector-based primers.
Preparation of mitochondria.
Mitochondria were isolated as described previously (6). Briefly, lyticase was used to create spheroplasts that were subsequently ruptured by vortexing with glass beads or Dounce homogenizing, and mitochondria were isolated by differential centrifugation. For some experiments, mitochondria were further purified using ultracentrifugation through a discontinuous Histodenz (Sigma-Aldrich) gradient (14% and 22%). The total mitochondrial protein concentration was estimated using the Bradford method.
Immunoblotting and BN-PAGE analysis.
For SDS-PAGE, 10 to 30 μg of protein samples or mitochondria was separated on 12 or 15% polyacrylamide gels and transferred to nitrocellulose membranes. Membranes were blocked prior to detection using 5 to 10% nonfat dry milk in phosphate-buffered saline (PBS) with or without 0.01% Tween 20. Blue native PAGE (BN-PAGE) was performed essentially as described previously (34) with either 1% digitonin or 1% dodecylmaltoside. Either chemiluminescent reagents with horseradish peroxidase-conjugated secondary antibodies or infrared (IR) dye-conjugated secondary antibodies (Li-Cor) were used to visualize proteins. Antibodies either were purchased or were generous gifts: anti-Myc (Roche Diagnostics), anti-Sdh2 (21st Century Biochemicals), antiporin (Molecular Probes), anti-Cox2 (Mitosciences), anti-TAP (Open Biosystems), anti-Rip1 and anti-Cor1/Cor2 (B. Trumpower), anti-Cyt1 (B. Meunier), anti-Yah1 and anti-Aco1 (R. Lill), and anti-F1 ATPase beta subunit Atp2 (A. Tzagoloff).
Depletion of Yah1.
Strains were a kind gift from R. Lill and are described in reference 20. Gal-YAH1 or wild-type (WT) cells were precultivated in galactose-containing minimal medium for 24 h at 30°C. Cells were washed and transferred to minimal medium containing 2% lactate and 0.1% glucose. Cells were diluted 20-fold into fresh lactate medium containing 0.1% glucose every 20 h. After 85 h, mitochondria from WT and Gal-YAH1 cells were isolated as described above. Depletion of Yah1 was verified by immunoblotting.
Complex III purification.
Mitochondria from both the Cor2::TAP and Cor2::TAP mzm1Δ strains (200 μg) were lysed in 450 μl of IPP150 buffer (10 mM Tris, pH 7.4, 150 mM NaCl2, 1 mM phenylmethylsulfonyl fluoride [PMSF], with 1% digitonin) on ice for 10 min. Lysates were clarified by centrifugation at 10,000 × g for 10 min at 4°C. IgG magnetic beads (50 μl; NEB S1431S) were rinsed three times in IPP150 buffer (with 0.5% digitonin). Beads were added to each lysate and incubated overnight at 4°C with rotation. Beads were precipitated magnetically and rinsed two times in tobacco etch virus (TEV) protease buffer (10 mM Tris, pH 7.4, 150 mM NaCl2, 0.5 mM EDTA, 1 mM PMSF, 0.1 M dithiothreitol [DTT], and 0.5% digitonin). TEV protease (100 units; Invitrogen) was added to 1 ml TEV buffer, and 500 μl was added to beads and incubated for 1 h at room temperature. As the Cor2::TAP strain remained associated with the IgG beads after TEV treatment, IgG beads were precipitated and rinsed three times in 0.5 ml calmodulin binding buffer (10 mM Tris, pH 7.4, 150 mM NaCl2, 4 mM CaCl2, 1 mM magnesium acetate, 1 mM imidazole, 10 mM 2-mercaptoethanol, and 0.5% digitonin). Calmodulin beads (100 μl) were rinsed in 0.5 ml binding buffer three times, resuspended in an 0.5-ml final volume, and added directly to precipitated IgG beads. The mixed beads were incubated at 4°C with rotation for 4 h. IgG beads were removed from the calmodulin bead bed magnetically. Calmodulin beads were rinsed three times in 0.5 ml buffer, with precipitation between rinses with brief spins. Precipitated beads were boiled in 0.1 ml Laemmli buffer and loaded into duplicate lanes of Bis-Tris 12% NuPAGE gels (Invitrogen) and run in the manufacturer's recommended morpholineethanesulfonic acid (MES) SDS running buffer. Gels were divided for parallel immunoblot analysis and Sypro ruby staining (Invitrogen) as recommended by the manufacturer. For mass spectrometry, indicated Sypro ruby-stained bands were excised from the gel for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis at the University of Utah Mass Spectrometry and Proteomics Core.
Immunoprecipitation of Mzm1.
mzm1Δ cells expressing Sod2-Mzm1-MycHis and WT cells were grown on synthetic galactose medium with selection, and mitochondria were isolated as described above. Mitochondria (1 mg protein/ml) were lysed at 4°C in lysis buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 1 mM PMSF, 1% digitonin, 0.1 mM EDTA, and 10% glycerol) and clarified by centrifugation at 20,000 × g for 15 min. Solubilized mitochondrial proteins were applied to Myc-agarose beads (20-μl settled volume) at 4°C and then washed with 60 column volumes of wash buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 1 mM PMSF, 0.3% digitonin, 0.1 mM EDTA, and 10% glycerol). Bound proteins were eluted either by boiling or with 10% SDS.
Mitochondrial enzymatic activity assays.
The bc1 activity of isolated mitochondria was measured spectrophotometrically by supplying decylubiquinol and cytochrome c and following the rate of reduction of cytochrome c (while inhibiting CcO with 2 mM KCN). Decylubiquinol was prepared as described previously (1). The activity assay was performed with 30 μM decylubiquinol, 0.4 mg/ml cytochrome c (from horse heart; Sigma-Aldrich), and 10 to 30 μg total mitochondrial protein in 40 mM potassium phosphate, pH 6.8, with 0.5% Tween 80. The initial rate of cytochrome c reduction at 550 nm was measured by an Agilent 8453 spectrophotometer. Succinate dehydrogenase (SDH) activity of 15 μg of isolated mitochondria in potassium phosphate (pH 6.8) buffer with 0.5% Tween 80 (TMPD) was measured with 10 mM succinate, 90 μM decylubiquinol, and 80 μM dichlorophenolindophenol (DCPIP) (Sigma-Aldrich) spectrophotometrically at 600 nm. Aconitase activity of 15 μg of isolated mitochondria in TMPD was measured spectrophotometrically with cis-aconitate at 240 nm.
Metal analysis.
Total metal levels were determined by inductively coupled plasma-optical emission spectroscopy (ICP-OES) as described previously (1). The zinc-specific fluorophore RhodZin-3 dipotassium salt (Molecular Probes) was used to assess labile zinc concentrations as described previously (1).
Absorbance spectroscopy of heme.
For optical absorption spectra, 1.5 mg of purified mitochondria grown in galactose medium was solubilized in PBS buffer containing 1% dodecylmaltoside. Each spectrum represents the calculated difference spectrum of the reduced (dithionite) minus oxidized (ferricyanide) cytochromes and was recorded by an Agilent 8453 spectrophotometer. Absorption maxima at 550, 560, and 603 nm correspond to cytochromes b, c, and a,a3, respectively.
RESULTS
Labile zinc pool in mitochondria is dependent on functional bc1 complex.
We reported previously that mzm1Δ cells have reduced mitochondrial zinc, especially in a labile matrix zinc pool accessible to the Zn(II)-chelating fluorophore Rhodzin-3 (1). The mutant cells are hypersensitive to further attenuation of this labile zinc pool. In addition, mzm1Δ cells are impaired in respiration with a specific defect in the bc1 complex, especially at 37°C. We addressed the link between bc1 complex function and mitochondrial labile zinc by quantifying zinc levels in yeast mutants impaired in bc1 assembly. Qcr7 is an essential bc1 complex subunit that stabilizes the first assembly intermediate containing cytochrome b (Fig. 1) (3, 38). In contrast, Qcr9 is one of the last subunits added during the assembly process and, much like Mzm1, is critical only for bc1 function at 37°C (12, 28). Mitochondria isolated from yeast lacking either Qcr7 or Qcr9 were used for quantitation of total zinc by ICP-OES and of labile zinc with the RhodZin-3 fluorophore. The two mutants exhibit similar diminutions in labile zinc (data not shown). Likewise, the labile mitochondrial zinc pool was also attenuated in cells lacking the bc1 assembly factor Bcs1. Thus, the zinc impairment in mzm1Δ cells likely arises from the deficiency of the bc1 complex and not from a specific effect of Mzm1 on mitochondrial zinc.
Rip1 is deficient in mzm1Δ cells.
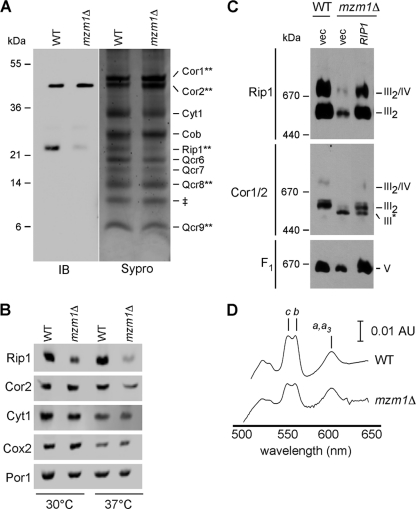
We sought to deduce the basis for the defect in bc1 function in mzm1Δ cells. Using a yeast strain with a chromosomal TAP purification tag on the Cor2 bc1 subunit, we purified the complex from wild-type (WT) and mzm1Δ cells. After a two-step affinity purification, visualization of the final eluate by Sypro staining after SDS-PAGE revealed an array of bc1 complex subunits (Fig. 2A). Mass spectrometry was used to validate the identity of several of the bands in the WT cells, including Cor1, Cor2, Rip1, Qcr8, and Qcr9. The sample recovered from mzm1Δ cells showed similar levels of most subunits, with the exception of Rip1. To confirm the attenuation in Rip1, immunoblotting was performed (Fig. 2A, IB). Rip1 was markedly low in mzm1Δ cells, despite equal loading of Cor2. Qcr9 and Qcr10 are normally added late in assembly along with Rip1 (37), but no obvious defect was observed in the level of Qcr9 in the complex isolated from mzm1Δ cells. Steady-state levels of Rip1 were attenuated in the mutant cells, especially at 37°C (Fig. 2B).
Fig. 2.
Characterization of bc1 complex components and supercomplex formation in mzm1Δ cells. (A) bc1 complex purification via tandem affinity purification of Cor2::TAP in strains with and without Mzm1 from mitochondria solubilized by 1% digitonin. SDS-PAGE gels of the purification products were stained with Sypro ruby, and the bands were identified by mass spectrometry (where indicated by **) along with immunoblotting (IB) to identify Cor2 (anti-TAP) and Rip1 (anti-Rip1). ‡, a band with a mass similar to that of Qcr10 was observed by Sypro staining; we failed to confirm its identity as Qcr10. (B) SDS-PAGE immunoblot of mitochondria isolated from cultures grown at 30 and 37°C. (C) BN-PAGE immunoblot of mitochondria solubilized by 1% digitonin. The late core subassembly intermediate is designated by III*. (D) Optical absorbance spectra were recorded of reduced minus oxidized cytochromes in WT and mzm1Δ mitochondrial detergent lysates. AU, absorbance unit.
Previous studies on bc1 biogenesis revealed that a stable assembly intermediate lacking Rip1 is apparent in cells lacking Bcs1 or Qcr9 (5). This late core assembly intermediate is visualized by BN-PAGE using antisera to subunits preceding Rip1 insertion. Digitonin extracts of mitochondria isolated from mzm1Δ cells separated on BN-PAGE show a bc1 assembly of lower mass than the dimeric complex (Fig. 2C, III*). Since the bc1 assembly intermediate is deficient in Rip1, the lower-mass complex is not apparent using antisera to Rip1. The bc1 intermediate as purified above contains the mitochondrion-encoded Cob subunit and its two cytochrome b moieties. The presence of hemylated Cob was shown by difference spectroscopy of membranes from mzm1Δ cells (Fig. 2D). Heme b levels were not markedly attenuated in mzm1Δ cells relative to WT. This result is consistent with the previous observation that heme insertion into Cob occurs upon formation of the early core assembly intermediate (27).
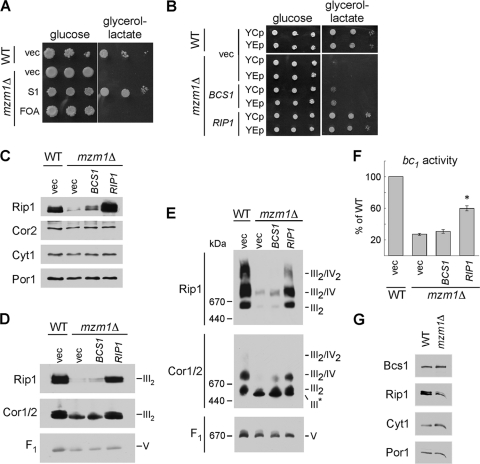
Since residual bc1 activity exists in mzm1Δ cells, we attempted a suppressor screen of mzm1Δ cells to isolate extragenic suppressors. The mutant cells exhibit a marked growth defect at 37°C on glycerol-lactate medium (Fig. 3A, “vec”). Although no spontaneous suppressor mutants were recovered, respiration-competent clones were obtained after transformation of mzm1Δ cells with a high-copy-number yeast DNA library. Transformants were screened by replica plating at 30 and 37°C. In addition to the expected MZM1 clones, we recovered two clones with inserts spanning the RIP1 locus (Fig. 3A, S1). The respiratory growth of the clones was abrogated by pregrowth in 5-fluoroorotic acid used to shed the vector. Two new vectors were engineered containing the RIP1 locus as low-copy-number (YCp) and high-copy-number (YEp) plasmids. Transformation of mzm1Δ cells with these vectors revealed that 37°C respiratory growth was restored in the mutant cells with either RIP1-containing vector (Fig. 3B). The overexpression of RIP1 restored steady-state Rip1 levels in mzm1Δ cells (Fig. 3C), along with restoring the intact bc1 complex visualized by BN-PAGE after dodecylmaltoside solubilization of mitochondria (Fig. 3D). Immunoblotting for the F1/Fo ATP synthase (complex V) was used to show equivalent protein loads per lane. BN-PAGE with digitonin solubilization revealed the restoration of CcO/bc1 supercomplexes upon RIP1 overexpression (Fig. 3E). Consistent with the BN-PAGE results, elevated levels of Rip1 increased bc1 enzymatic activity to approximately 60% of WT levels (Fig. 3F).
Fig. 3.
Suppression of the 37°C growth defect and rescue of the reduced Rip1 levels, supercomplex formation, and bc1 complex activity of mzm1Δ. (A) Growth test at 37°C on glucose or glycerol-lactate medium of mzm1Δ cells transformed with vector control (vec) or a plasmid isolated from the suppression screen (S1), which was identified by sequencing to contain RIP1. Loss of this RIP1-containing plasmid was induced by 5-fluoroorotic acid treatment (FOA). (B) Growth test at 37°C on glucose or glycerol-lactate medium of mzm1Δ cells transformed with low-copy-number (YCp) and high-copy-number (YEp) vector control or plasmids containing BCS1 or RIP1. (C to E) SDS-PAGE (C) and BN-PAGE immunoblots with either 1% dodecylmaltoside solubilization (D) or 1% digitonin solubilization (E) of mitochondria from mzm1Δ cells transformed with YEp vector control or plasmids containing BCS1 or RIP1. The late core subassembly intermediate is designated by III*. (F) bc1 complex activity of mitochondria from mzm1Δ cells transformed with high-copy-number vector control or plasmids containing BCS1 or RIP1, shown as a percentage of WT activity, with the asterisk indicating significantly increased activity relative to mzm1Δ with vector control (n = 6 ± standard deviation). Statistical significance was determined by analysis of variance with Bonferroni's post hoc test in Kaleidagraph. (G) SDS-PAGE immunoblot of mitochondria from Bcs1::Myc cultures with and without Mzm1. Bcs1 protein levels were identified using anti-Myc antibody.
As evidenced above, cells lacking Mzm1 have impaired bc1 activity due to attenuated levels of Rip1 insertion, similar to the phenotype reported for bcs1Δ cells (5). To assess the relationship between Mzm1 and Bcs1, we tested whether overexpression of BCS1 would restore respiratory growth to mzm1Δ cells (Fig. 3B). Overexpression of BCS1 gave only a modest improvement in glycerol-lactate growth and bc1 activity of the mutant cells, which was clearly not to the same extent as overexpression of RIP1 (Fig. 3B and F). Bcs1 steady-state protein levels are not attenuated in mzm1Δ cells, unlike Rip1 levels (Fig. 3G). Thus, the defect in mzm1Δ cells correlates with impaired Rip1 insertion into the bc1 complex.
Late step in Rip1 maturation is impaired in mzm1Δ cells.
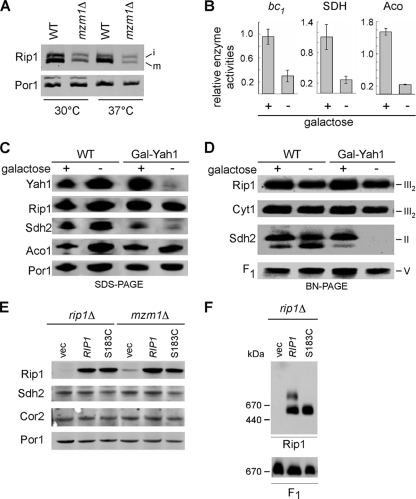
The maturation of Rip1 into the bc1 complex involves a series of steps. Rip1 is synthesized in the cytoplasm with an N-terminal mitochondrial targeting sequence that is proteolytically cleaved by the matrix processing protease (MPP) during import through the TIM23 complex (15). The protein is fully translocated into the matrix compartment for subsequent insertion into the inner membrane (IM). In yeast, a second proteolytic cleavage by Oct1 removes an additional 8 residues. The remaining steps in Rip1 maturation involve insertion of the 2Fe-2S cluster within the matrix, translocation of the Fe/S domain across the IM, and finally the formation of a disulfide bond (11, 23, 25, 26). Processing of the residual Rip1 by MPP and Oct1 appears normal in mzm1Δ cells (Fig. 4A). No precursor polypeptide consistent with impaired MPP cleavage was evident in mzm1Δ cells, although incomplete cleavage of the intermediate precursor was apparent in both WT and mutant cells. This cleavage of the intermediate Rip1 precursor by Oct1 is not required for Rip1 insertion into the bc1 complex (26).
Fig. 4.
Rip1 processing and Fe/S cluster insertion are not impaired in mzm1Δ cells. (A) SDS-PAGE immunoblot of mitochondria isolated from WT and mzm1Δ cultures grown at 30 and 37°C. Intermediate (i) and mature (m) forms of Rip1 are indicated. (B) Analysis of cytochrome c reductase (bc1), succinate dehydrogenase (SDH), and aconitase (Aco) activities of mitochondria from WT or Gal-YAH1 cells grown in either galactose-replete (+) or -deficient (−) medium, where the depletion of Yah1 by lactate replacement of galactose results in a lack of Fe/S cluster biogenesis. The results are presented relative to the activities measured for the mitochondrial enzymes of either WT or Gal-YAH1 cells grown in the presence of galactose (+). Error bars indicate standard deviations; n = 3. (C and D) SDS-PAGE (C) and BN-PAGE (D) with 1% dodecylmaltoside solubilization of mitochondria from WT or Gal-YAH1 cells grown in either galactose or lactate-replete medium. (E) SDS-PAGE and immunoblotting of mitochondria from rip1Δ or mzm1Δ cells transformed with high-copy-number vector control or plasmids containing WT or S183C Rip1. (F) BN-PAGE with 1% digitonin solubilization of mitochondria isolated from rip1Δ cells expressing either WT or S183C Rip1 (mutant lacking a Fe/S cluster).
The Rip1 defect in mzm1Δ cells is unlikely to be due to impaired Fe/S cluster formation based on previous studies. Neither formation of the Fe/S center nor the disulfide bond was found to be important for the IM insertion of Rip1 (13, 23). To confirm this observation on the formation of the Fe/S cluster, we repressed the expression of the essential mitochondrial matrix ferredoxin, Yah1, required for Fe/S cluster biogenesis using a GAL10-YAH1 strain (20). Propagation of these cells on lactate-containing medium results in the repression of YAH1 expression, resulting in an impairment in Fe/S cluster biogenesis within the mitochondrial matrix (21). Depletion of Yah1 results in a marked attenuation in the activity of mitochondrial Fe/S cluster proteins aconitase, succinate dehydrogenase, and bc1 (Fig. 4B), as was shown previously (20). Whereas steady-state levels of Sdh2 (Fig. 4C) and the SDH complex on BN-PAGE (Fig. 4D) were markedly attenuated, Rip1 levels were not significantly perturbed and Rip1 was associated with the bc1 complex by BN-PAGE (Fig. 4C and D). Likewise, the S183C Rip1 mutant that was reported to lack a Fe/S cluster is stably expressed in both mzm1Δ and rip1Δ cells (Fig. 4E) and is inserted into the bc1 complex (Fig. 4F). These studies confirm the previous observations showing that Fe/S cluster insertion is not a prerequisite for Rip1 IM insertion (5, 13). Thus, the impaired insertion of Rip1 into the bc1 complex in mzm1Δ cells is not a result of a failure to insert Fe/S into Rip1. The remaining step in Rip1 maturation is its insertion in the IM with the globular domain containing the Fe/S cluster facing the IMS side of the membrane.
Rip1 is stabilized by Mzm1.
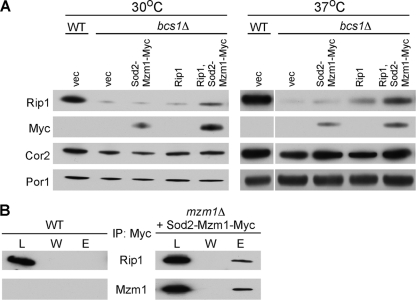
As mentioned, mzm1Δ cells exhibit attenuated levels of Rip1. To assess whether Mzm1 contributes to the stabilization of Rip1 prior to membrane insertion, we took advantage of the low, albeit unincorporated, levels of Rip1 in bcs1Δ cells (27). Overexpression of Mzm1 in bcs1Δ cells gave variable results on Rip1 stabilization, but coexpression of Rip1 and Mzm1 gave a highly reproducible stabilization of Rip1 levels compared to expression of Rip1 alone in bcs1Δ cells (Fig. 5A). The Mzm1-mediated stabilization of Rip1 was apparent in cells isolated at both 30 and 37°C. The observed stabilization of Rip1 by Mzm1 suggested that the two proteins may transiently interact. Immunoprecipitation of a Myc-tagged Mzm1 allele in WT cells with anti-Myc antibody-conjugated beads resulted in the coprecipitation of Rip1 (Fig. 5B). No Rip1 was precipitated in WT cells lacking a Myc-tagged Mzm1 allele.
Fig. 5.
Mzm1 stabilizes and interacts with Rip1. (A) SDS-PAGE immunoblot of mitochondria from WT and bcs1Δ cells cultured at either 30 or 37°C expressing indicated plasmids. Mzm1 levels were identified with anti-Myc antibody. (B) Immunoprecipitation of WT or mzm1Δ cells overexpressing Sod2-Mzm1-Myc with anti-Myc-agarose beads. Three percent of the total extracts (L), the entire fraction of the last wash (W), and 20% of the bead eluate (E) were analyzed by SDS-PAGE and immunoblotting with anti-Rip1 or anti-Myc (Mzm1) antibodies.
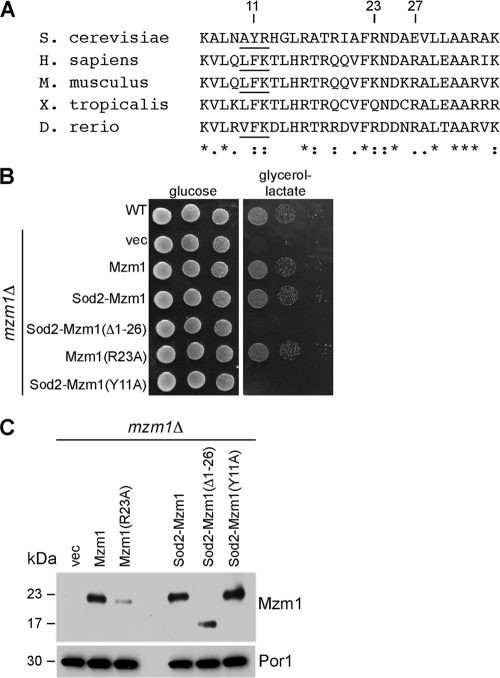
The LYR motif of Mzm1 is important for its function.
Mzm1 is a 14-kDa polypeptide that contains only one conserved motif, a poorly defined variant of the LYR motif that is seen in Isd11 and Sdh6 (10, 33) (Fig. 6 A). To address whether the LYR motif is functionally important, a Tyr11Ala (Y11A) mutant was generated for in vivo testing. The mutations were introduced in the gene expressed from its own promoter as well as full- and partial-length fusion genes containing the MET25 promoter and Sod2 mitochondrial target sequence (MTS).
Fig. 6.
The LYR motif of Mzm1 is important for its function. (A) Mzm1 protein sequences are shown with conserved segments in the N-terminal region. The Tyr in the LYR (AYR) motif is residue 11. H. sapiens, Homo sapiens; M. musculus, Mus musculus; X. tropicalis, Xenopus tropicalis; D. rerio, Danio rerio. (B) WT cells and mzm1Δ cells expressing vector, high-copy-number Mzm1, low-copy-number Sod2-Mzm1, low-copy-number Sod2-Mzm1 (Δ1-26), high-copy-number Mzm1 (R23A), and low-copy-number Sod2-Mzm1 (Y11A) were grown in synthetic selective medium, serially diluted, and spotted on yeast extract-peptone medium containing glucose or glycerol-lactate carbon sources. Cells were incubated at 37°C. (C) Mitochondria from the respective strains in panel B were analyzed by SDS-PAGE and immunoblotted with anti-Myc antibody to visualize Mzm1.
The LYR motif in Mzm1 occurs in the N-terminal segment of the protein that is predicted by the MitoProt II algorithm to be a cleaved mitochondrial target sequence consisting of 26 residues. Removal of the candidate MTS from Mzm1 abolished function (data not shown). Growth of mzm1Δ cells at 37°C was supported by fusion of the Sod2 MTS to full-length Mzm1; however, Sod2 MTS fused to residues 27 to 124 of Mzm1 failed to support respiratory growth in mzm1Δ cells at 37°C (Fig. 6B). Lastly, in either the native or the Sod2-Mzm1 fusion protein, replacement of Tyr11 in the LYR motif with alanine (Y11A) failed to support growth on glycerol-lactate (Fig. 6B and data not shown).
To assess whether Mzm1 is processed by the MPP protease, we utilized the Sod2 fusion proteins together with mutating the candidate Arg (Arg23) that may serve as the MPP protease docking site to an alanine (R23A). Mitochondria isolated from mzm1Δ cells harboring either WT, R23A, or Sod2-Mzm1 fusions were used for steady-state PAGE protein analysis (Fig. 6C). The size of the Mzm1 polypeptide was equivalent in comparing the WT Mzm1 protein to either R23A or the Sod2 fusion with full-length Mzm1. However, the Sod2 MTS fusion with the truncated Mzm1 lacking its putative MTS exhibited a markedly smaller polypeptide. These results show that Mzm1 lacks a cleavable MTS and that Tyr11 within the LYR motif of the mature protein is functionally important.
DISCUSSION
Mzm1 is presently shown to be an assembly factor for the bc1 complex in yeast. The conservation of Mzm1 in metazoans suggests that it may have a conserved role in higher eukaryotes. Mzm1 functions at a late step in bc1 complex assembly during the insertion of the Rip1 Rieske Fe/S protein. Cells lacking Mzm1 exhibit a modest bc1 defect at 30°C, but the defect is exacerbated at elevated temperatures. Rip1 levels are markedly attenuated at elevated temperatures in mzm1Δ cells. Previously, we demonstrated that cells lacking Mzm1 are compromised in a mitochondrial zinc pool accessible to the zinc-responsive RhodZin-3 fluorophore (1). Further attenuation in the mitochondrial zinc pool by targeting a heterologous Zn-binding protein to the mitochondrial matrix leads to a respiratory deficiency. We show presently that the mitochondrial zinc deficiency is not a unique phenotype of mzm1Δ cells. Rather, cells lacking the bc1 complex show the same attenuated labile zinc pool. Yeasts lacking either of two structural subunits of the bc1 complex, Qcr7 and Qcr9, or lacking an essential bc1 assembly factor Bcs1 are compromised in the zinc pool. The basis for the attenuation in labile matrix zinc in bc1-deficient cells as well as cells deficient in the citric acid cycle enzymes Cit1 and Mdh1 shown previously (1) remains unclear.
The biogenesis of the bc1 complex occurs in a modular assembly pathway (3, 36, 38) (Fig. 1). The early central core complex consisting of Cob, Qcr7, and Qcr8 is evident in cells stalled at downstream assembly steps (38). The early core complex is converted to the late core subassembly intermediate with the addition of Cyt1, Cor1, Cor2, and Qcr6 subunits and occasionally Qcr9 (3, 37). This late core intermediate lacks only Qcr10 and Rip1 (5, 37). Cob and Cyt1 contain their heme b and heme c cofactors, respectively (3, 37). This late core complex is of greater stability than the early core complex and can be isolated after solubilization in either digitonin or Triton X-100. An unresolved issue concerns the step of bc1 dimerization (37). The late core intermediate is either a dimeric unit with an associated Bcs1 assembly factor or a monomeric unit with additional assembly subunits such as an oligomerized Bcs1. However, since the dimer interface consists mainly of cytochrome b helices, dimerization is believed to occur early in the assembly pathway (37, 38).
Cells lacking Mzm1 are stalled at the late core assembly intermediate, which in this case contains Qcr9. Respiratory growth at 37°C can be restored in the mzm1Δ cells by the overexpression of the Rip1 subunit. The attenuated level of Rip1 in mzm1Δ cells is consistent with a role of Mzm1 in Rip1 maturation.
Rip1 maturation has previously been shown to involve the complete import of Rip1 into the matrix, without a stop transfer within the IM. In support of a complete import of Rip1 into the mitochondrial matrix, expression of Rip1 from the mitochondrial genome is able to complement a rip1Δ strain, suggesting that Rip1 maturation occurs within the matrix (26). In yeast, two proteolytic processing steps occur, one being the removal of the mitochondrial target sequence by MPP and the second the removal of an octapeptide sequence by Oct1. The 2Fe-2S center is likely formed in Rip1 within the matrix by the resident iron-sulfur cluster (ISC) Fe/S cluster system. Subsequent to Fe/S cluster formation, membrane insertion of Rip1 occurs along with the extrusion of the C-terminal Fe/S domain to the IMS side of the IM. An important disulfide bond forms in the Fe/S domain, and this may occur after extrusion of the folded domain to the IMS side of the IM. The redox potential of the IMS is more oxidizing than the matrix or cytoplasm, and numerous proteins within this space contain disulfides (17).
The block in Rip1 insertion into the bc1 complex in mzm1Δ cells likely occurs at a later step in Rip1 maturation, as evidenced by the lack of impairment in either MPP or Oct1 processing in mzm1Δ cells. Although the mutant cells have somewhat higher levels of the i-Rip1 species without the Oct1 cleavage, the Oct1 processing step is not essential for IM insertion or catalytic activity (26). Formation of the Fe/S center is not a prerequisite for IM insertion of Rip1, so that step is not likely impaired in mzm1Δ cells. IM insertion and extrusion of the Fe/S domain are steps that may be impaired in the mutant cells. However, Mzm1 is not essential to these steps since limited bc1 activity is apparent at 30°C and a modest increase in Rip1 expression can suppress the respiratory defect in mzm1Δ cells. One revealing defect in mzm1Δ cells is the impaired stabilization of Rip1 prior to IM insertion. Rip1 levels are markedly attenuated at elevated temperatures in mzm1Δ cells. Rip1 is also destabilized in bcs1Δ cells in which Rip1 IM insertion is likely blocked. In the absence of IM insertion, Rip1 has limited stability, especially at elevated temperatures. Under these conditions, Mzm1 has a greater role in Rip1 maturation. Elevated levels of Mzm1 enhance the stabilization of Rip1, suggesting that Mzm1 may be an important Rip1 chaperone under cell stress.
Mzm1 may function primarily as a Rip1 chaperone to stabilize Rip1 prior to IM insertion or, alternatively, as a chaperone that aids in the presentation of Rip1 to the maturing bc1 complex. This maturation step appears to necessitate translocation of the folded, metallated Rip1 across the IM. Bcs1 may mediate the extrusion of the Rip1 C-terminal globular domain with the Fe/S center across a bilayer. Mzm1 may stabilize a conformation of Rip1 that creates an optimal recognition interface for Bcs1. Studies are under way to assess whether Mzm1 mediates presentation of Rip1 to Bcs1.
ACKNOWLEDGMENTS
This work was supported by grant GM083292 to D.R.W. A.A. and P.S. were supported by training grant T32 DK007115. J.L.F. was supported by training grant T32 HL007576-25. O.K. was supported by American Heart Association fellowship 10POST4300044.
Footnotes
Published ahead of print on 1 August 2011.
REFERENCES
- 1. Atkinson A., et al. 2010. Mzm1 influences a labile pool of mitochondrial zinc important for respiratory function. J. Biol. Chem. 285:19450–19459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brandt U., Uribe S., Schagger H., Trumpower B. L. 1994. Isolation and characterization of QCR10, the nuclear gene encoding the 8.5-kDa subunit 10 of the Saccharomyces cerevisiae cytochrome bc1 complex. J. Biol. Chem. 269:12947–12953 [PubMed] [Google Scholar]
- 3. Crivellone M. D., Wu M. A., Tzagoloff A. 1988. Assembly of the mitochondrial membrane system. Analysis of structural mutants of the yeast coenzyme QH2-cytochrome c reductase complex. J. Biol. Chem. 263:14323–14333 [PubMed] [Google Scholar]
- 4. Cruciat C. M., Brunner S., Baumann F., Neupert W., Stuart R. A. 2000. The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J. Biol. Chem. 275:18093–18098 [DOI] [PubMed] [Google Scholar]
- 5. Cruciat C. M., Hell K., Folsch H., Neupert W., Stuart R. A. 1999. Bcs1, an AAA-family member, is a chaperone for the assembly of the cytochrome bc1 complex. EMBO J. 18:5226–5233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Diekert K., De Kroon A. I., Kispal G., Lill R. 2001. Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 65:37–51 [DOI] [PubMed] [Google Scholar]
- 7. Edwards C. A., Trumpower B. L. 1986. Resolution and reconstitution of the iron-sulfur protein of the cytochrome bc1 segment of the mitochondrial respiratory chain. Methods Enzymol. 126:211–224 [DOI] [PubMed] [Google Scholar]
- 8. Esser L., et al. 2008. Inhibitor-complexed structures of the cytochrome bc1 from the photosynthetic bacterium Rhodobacter sphaeroides. J. Biol. Chem. 283:2846–2857 [DOI] [PubMed] [Google Scholar]
- 9. Ghezzi D., et al. 2011. Mutations in TTC19 cause mitochondrial complex III deficiency and neurological impairment in humans and flies. Nat. Genet. 43:259–263 [DOI] [PubMed] [Google Scholar]
- 10. Ghezzi D., et al. 2011. SDHAF1, encoding a LYR complex-II specific assembly factor, is mutated in SDH-defective infantile leukoencephalopathy. Nat. Genet. 43:259–265 [DOI] [PubMed] [Google Scholar]
- 11. Graham L. A., Brandt U., Trumpower B. L. 1994. Protease maturation of the Rieske iron-sulphur protein after its insertion into the mitochondrial cytochrome bc1 complex of Saccharomyces cerevisiae. Biochem. Soc. Trans. 22:188–191 [DOI] [PubMed] [Google Scholar]
- 12. Graham L. A., Phillips J. D., Trumpower B. L. 1992. Deletion of subunit 9 of the Saccharomyces cerevisiae cytochrome bc1 complex specifically impairs electron transfer at the ubiquinol oxidase site (center P) in the bc1 complex. FEBS Lett. 313:251–254 [DOI] [PubMed] [Google Scholar]
- 13. Graham L. A., Trumpower B. L. 1991. Mutational analysis of the mitochondrial Rieske iron-sulfur protein of Saccharomyces cerevisiae. III. Import, protease processing, and assembly into the cytochrome bc1 complex of iron-sulfur protein lacking the iron-sulfur cluster. J. Biol. Chem. 266:22485–22492 [PubMed] [Google Scholar]
- 14. Gruschke S., et al. 2011. Cbp3-Cbp6 interacts with the yeast mitochondrial ribosomal tunnel exit and promotes cytochrome b synthesis and assembly. J. Cell Biol. 193:1101–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hartl F. U., Schmidt B., Wachter E., Weiss H., Neupert W. 1986. Transport into mitochondria and intramitochondrial sorting of the Fe/S protein of ubiquinol-cytochrome c reductase. Cell 47:939–951 [DOI] [PubMed] [Google Scholar]
- 16. Heinemeyer J., Braun H. P., Boekema E. J., Kouril R. 2007. A structural model of the cytochrome c reductase/oxidase supercomplex from yeast mitochondria. J. Biol. Chem. 282:12240–12248 [DOI] [PubMed] [Google Scholar]
- 17. Hu J., Dong L., Outten C. E. 2008. The redox environment in the mitochondrial intermembrane space is maintained separately from the cytosol and matrix. J. Biol. Chem. 283:29126–29134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Iwata S., et al. 1998. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science 281:64–71 [DOI] [PubMed] [Google Scholar]
- 19. Kronekova Z., Rodel G. 2005. Organization of assembly factors Cbp3 and Cbp4 and their effect on bc1 complex assembly in Saccharomyces cerevisiae. Curr. Genet. 47:203–212 [DOI] [PubMed] [Google Scholar]
- 20. Lange H., Kaut A., Kispal G., Lill R. 2000. A mitochondrial ferredoxin is essential for biogenesis of cellular iron-sulfur proteins. Proc. Natl. Acad. Sci. U. S. A. 97:1050–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lill R., Muhlenhoff U. 2008. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 77:669–700 [DOI] [PubMed] [Google Scholar]
- 22. Mathieu L., Marsy S., Saint-Georges Y., Jacq C., Dujardin G. 2011. A transcriptome screen in yeast identifies a novel assembly factor for the mitochondrial complex III. Mitochondrion 11:391–396 [DOI] [PubMed] [Google Scholar]
- 23. Merbitz-Zahradnik T., Zwicker K., Nett J. H., Link T. A., Trumpower B. L. 2003. Elimination of the disulfide bridge in the Rieske iron-sulfur protein allows assembly of the [2Fe-2S] cluster into the Rieske protein but damages the ubiquinol oxidation site in the cytochrome bc1 complex. Biochemistry 42:13637–13645 [DOI] [PubMed] [Google Scholar]
- 24. Mumberg D., Muller R., Funk M. 1994. Regulatable promoters of Saccharomyces cerevisiae: Comparison of transcriptional activity and their use of heterologous expression. Nucleic Acids Res. 22:5767–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nett J. H., Trumpower B. L. 1996. Dissociation of import of the Rieske iron-sulfur protein into Saccharomyces cerevisiae mitochondria from proteolytic processing of the presequence. J. Biol. Chem. 271:26713–26716 [DOI] [PubMed] [Google Scholar]
- 26. Nett J. H., Trumpower B. L. 1999. Intermediate length Rieske iron-sulfur protein is present and functionally active in the cytochrome bc1 complex of Saccharomyces cerevisiae. J. Biol. Chem. 274:9253–9257 [DOI] [PubMed] [Google Scholar]
- 27. Nobrega F. G., Nobrega M. P., Tzagoloff A. 1992. BCS1, a novel gene required for the expression of functional Rieske iron-sulfur protein in Saccharomyces cerevisiae. EMBO J. 11:3821–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Phillips J. D., Graham L. A., Trumpower B. L. 1993. Subunit 9 of the Saccharomyces cerevisiae cytochrome bc1 complex is required for insertion of EPR-detectable iron-sulfur cluster into the Rieske iron-sulfur protein. J. Biol. Chem. 268:11727–11736 [PubMed] [Google Scholar]
- 29. Pierrel F., Khalimonchuk O., Cobine P. A., Bestwick M., Winge D. R. 2008. Coa2 is an assembly factor for yeast cytochrome c oxidase biogenesis facilitating the maturation of Cox1. Mol. Cell. Biol. 28:4927–4939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stuart R. A. 2008. Supercomplex organization of the oxidative phosphorylation enzymes in yeast mitochondria. J. Bioenerg. Biomembr. 40:411–417 [DOI] [PubMed] [Google Scholar]
- 31. Vonck J., Schafer E. 2009. Supramolecular organization of protein complexes in the mitochondrial inner membrane. Biochim. Biophys. Acta 1793:117–124 [DOI] [PubMed] [Google Scholar]
- 32. Wach A., Brachat A., Alberti-Segui C., Rebischung C., Philippsen P. 1997. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast 13:1065–1075 [DOI] [PubMed] [Google Scholar]
- 33. Wiedemann N., et al. 2006. Essential role of Isd11 in mitochondrial iron-sulfur cluster synthesis on Isu scaffold proteins. EMBO J. 25:184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wittig I., Braun H. P., Schagger H. 2006. Blue native PAGE. Nat. Protoc. 1:418–428 [DOI] [PubMed] [Google Scholar]
- 35. Xia D., et al. 1997. Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science 277:60–66 [DOI] [PubMed] [Google Scholar]
- 36. Zara V., Conte L., Trumpower B. L. 2009. Biogenesis of the yeast cytochrome bc1 complex. Biochim. Biophys. Acta 1793:89–96 [DOI] [PubMed] [Google Scholar]
- 37. Zara V., Conte L., Trumpower B. L. 2009. Evidence that the assembly of the yeast cytochrome bc1 complex involves the formation of a large core structure in the inner mitochondrial membrane. FEBS J. 276:1900–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zara V., Conte L., Trumpower B. L. 2007. Identification and characterization of cytochrome bc1 subcomplexes in mitochondria from yeast with single and double deletions of genes encoding cytochrome bc1 subunits. FEBS J. 274:4526–4539 [DOI] [PubMed] [Google Scholar]
- 39. Zollner A., Rodel G., Haid A. 1994. Expression of the Saccharomyces cerevisiae CYT2 gene, encoding cytochrome c1 heme lyase. Curr. Genet. 25:291–298 [DOI] [PubMed] [Google Scholar]
- 40. Zollner A., Rodel G., Haid A. 1992. Molecular cloning and characterization of the Saccharomyces cerevisiae CYT2 gene encoding cytochrome-c1-heme lyase. Eur. J. Biochem. 207:1093–1100 [DOI] [PubMed] [Google Scholar]