Abstract
Homologous recombination repairs DNA double-strand breaks by searching for, invading, and copying information from a homologous template, typically the homologous chromosome or sister chromatid. Tight wrapping of DNA around histone octamers, however, impedes access of repair proteins to DNA damage. To facilitate DNA repair, modifications of histones and energy-dependent remodeling of chromatin are required, but the precise mechanisms by which chromatin modification and remodeling enzymes contribute to homologous DNA repair are unknown. Here we have systematically assessed the role of budding yeast RSC (remodel structure of chromatin), an abundant, ATP-dependent chromatin-remodeling complex, in the cellular response to spontaneous and induced DNA damage. RSC physically interacts with the recombination protein Rad59 and functions in homologous recombination. Multiple recombination assays revealed that RSC is uniquely required for recombination between sister chromatids by virtue of its ability to recruit cohesin at DNA breaks and thereby promoting sister chromatid cohesion. This study provides molecular insights into how chromatin remodeling contributes to DNA repair and maintenance of chromatin fidelity in the face of DNA damage.
INTRODUCTION
Radiation, environmental chemicals, and metabolic by-products produce DNA double-strand breaks (DSBs), lesions that can lead to cell death or chromosome aberrations if not properly repaired. Mechanisms that sense and repair DSBs play important roles in maintaining genome integrity. In organisms from yeast to humans, DSBs trigger a signaling cascade through multiple kinases and adaptor proteins that coordinate DSB repair with other cellular events (17). Cells then employ one of the two mechanisms to repair DSBs: homologous recombination (HR) and nonhomologous end joining (NHEJ).
In NHEJ, the DNA end binding Ku and Mre11-Rad50-Nbs1 (Xrs2) complexes protect DNA ends from general nuclease attack and facilitate synapsis (24, 29, 67, 72). The Dnl4-Lif1-Nej1 complex ligates DNA ends to restore chromosome integrity after processing by nucleases and DNA polymerases according to the DNA end configuration (42).
In HR, the 5′ ends of the DSB are subjected to extensive nucleolytic degradation to yield long single-stranded DNA (ssDNA) (37, 73). The HR machinery engages the ssDNA and then searches for and invades a homologous donor sequence to prime repair DNA synthesis for gene conversion (GC) (46). Alternatively, annealing of complementary strands of homologous regions flanking a DSB will result in deletion of the intervening DNA sequence in a specialized type of recombination termed single-strand annealing (SSA) (30, 35). Moreover, a one-ended invasion of a homologous donor can trigger extensive DNA synthesis by establishing a replication fork-like intermediate, which can copy an entire chromosome arm in a process called break-induced replication (BIR) (32, 36). All of these repair reactions depend on proteins encoded by the RAD52 group genes (46).
Rad51, a key member of the RAD52 group and an orthologue of the Escherichia coli RecA recombinase, catalyzes the formation of DNA joints via a filamentous intermediate assembled on ssDNA (16, 18, 39, 50, 58). All mitotic GC processes therefore depend on Rad51, whereas SSA and BIR still occur in the absence of Rad51 but require the Rad59 protein (2, 14). Rad59 is homologous to the N terminus of Rad52, and it binds and anneals complementary single-strand DNA. This biochemical activity is likely relevant for recombination by SSA and synthesis-dependent strand annealing (14, 22, 44, 57, 71).
DNA transactions during recombination processes occur within the context of chromatin. The tight wrapping of DNA around the histone octamers within the nucleosome in chromatin impedes the access of recombination proteins to the target DNA sequence. To overcome this access constraint, specific histone modifications and ATP-dependent remodeling of chromatin should precede or accompany the repair reaction (12, 65). Indeed, nucleosomes at or near DNA breaks are heavily modified by phosphorylation, acetylation, and ubiquitination. At least four ATP-dependent chromatin-remodeling complexes, namely, INO80, RSC (remodel structure of chromatin), SWR1, and SWI/SNF, have been implicated in distinct steps of recombination and DSB repair by NHEJ (40, 64). Mutations in any of these complexes confer hypersensitivity to DNA break-causative agents. Much remains to be learned about the role of chromatin modifications and remodeling in DSB repair.
RSC is an essential and abundant chromatin-remodeling complex that belongs to the Swi/Snf2 superfamily (4). RSC is comprised of 15 subunits with a central cavity that binds nucleosomes (6). At the expense of ATP hydrolysis, RSC translocates along DNA and can position/slide nucleosomes to promote the access of transcription factors to their target DNA (33). Besides its involvement in gene expression regulation, RSC also participates in loading cohesin onto chromosome arms (20). RSC is rapidly recruited to the DSB to facilitate both NHEJ and HR (7, 48, 49). Yeast RSC physically and functionally interacts with the Ku80 and Mre11 proteins, and these interactions are critical for survival upon genotoxic stress (49). Mutations in RSC reduce the enrichment of γH2AX and Mec1 (the ATR [ataxia telangiectasia-related] orthologue) near DSBs and attenuate the establishment of DNA damage-induced checkpoints at S/G2 (31, 48). Existing evidence suggests that RSC remodels chromatin at DSBs such that DNA repair proteins can gain access to the breaks (25, 48). Indeed, RSC has been implicated in early and late steps of yeast mating type switching that is triggered by a site-specific DSB (7).
Here we further define the function of RSC in recombination and genome maintenance by assessing the functional and physical interactions between RSC and the RAD52 group proteins and by evaluating the role of RSC in distinct recombination pathways. We provide evidence that RSC promotes the DNA damage-induced loading of cohesin at DNA breaks to enhance recombination between sister chromatids.
MATERIALS AND METHODS
Yeast strains.
The strains used in this study and their genotypes are listed in Table 1. To test viability under various stress or DNA-damaging conditions, logarithmic-phase cultures of the wild-type and rsc mutant strains were serially diluted and spotted onto a yeast extract-peptone (YEP)-glucose plate containing the indicated amount of genotoxic chemicals. The plates were photographed after 3 to 4 days of growth at 30°C. Because of their thermosensitivity, rsc2 and rsc7 deletions were generated and maintained at 25°C, except when indicated otherwise.
Table 1.
Genotypes of the strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| JKM161 | hoΔ MATaHMLα hmrΔ::ADE1 ade1-100 leu2-3,112 trp1::hisG′ lys5 ura3-52 ade3::GAL::HO | 29a |
| SLY2890 | JKM161 rsc2Δ::KANR | This work |
| SLY2894 | JKM161 rsc7Δ::KANR | This work |
| SLY2892 | JKM161 rad59Δ::KANR | This work |
| tNS1379 | hoΔ HML MatΔ::leu2::hisG hmrΔ3 leu2-3 ura3-52 trp1 thr4 (ura3[205 bp]-cut site-URA3)/pFH800 | 57 |
| SLY3028 | tNS1379 rsc2Δ::KANR | This work |
| SLY1937 | tNS1379 rsc7Δ::KANR | This work |
| SLY3014 | tNS1379 rad59Δ::KANR | This work |
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3 | Invitrogen |
| JKM179 | hoΔ MATα hmlΔ::ADE1 hmrΔ::ADE1 ade1-100 leu2-3,112 trp1::hisG′ lys5 ura3-52 ade3::GAL::HO | 29a |
| SLY1948 | BY4741 rsc1Δ::KANR | Invitrogen |
| SLY1949 | BY4741 rsc2Δ::KANR | This work |
| SLY1982 | BY4741 rsc7Δ::KANR | This work |
| SLY1951 | BY4741 rsc14Δ::KANR | Invitrogen |
| SLY1912 | BY4741 htl1Δ::KANR | Invitrogen |
| SLY1913 | BY4741 rtt102Δ::KANR | Invitrogen |
| SLY1914 | BY4741 rsc30Δ::KANR | This work |
| SLY2919 | BY4741 rsc8505-557Δ | This work |
| SLY2082 | JKM179 rsc7Δ::KANR | This work |
| SLY2908 | JKM179 rsc2Δ::KANR | This work |
| YNN301 | ura3-52 trp1-Δ1 ade2-101 lys2-801 his3Δ3′ his3Δ5′ | 13 |
| SLY1933 | YNN301 rsc7Δ::KANR | This work |
| SLY3058 | YNN301 rsc2Δ::KANR | This work |
| SLY1940 | YNN301 rsc2Δ::KANR | This work |
| SLY3835 | YNN301 rad59Δ::KANR | This work |
| SLY3846 | YNN301 rad51Δ::KANR | This work |
| SLY3883 | YNN301 rsc7Δ::HYGRrad59Δ::KANR | This work |
| SLY3887 | YNN301 rsc7Δ::HYGRrad51Δ::KANR | This work |
| JKM179-101A | hoΔ MATα hmlΔ::ADE1 hmrΔ::ADE1 ade1-100 leu2-3,112 trp1::hisG′ lys5 ura3-52 ade3::GAL::HO Mcd1-6HA | 62 |
| EU1239 | hoΔ MATα hmlΔ::ADE1 hmrΔ::ADE1 ade1-100 leu2-3,112 trp1::hisG′ lys5 ura3-52 ade3::GAL::HO Smc1-6HA | 62 |
| SLY1944 | EU1239 rsc7Δ::KANR | This study |
| SLY1942 | JKM179-101A rsc7Δ::KANR | This study |
| SLY3041 | EU1239 rsc2Δ::HYGR | This study |
| SLY3042 | JKM179-101A rsc2Δ::HYGR | This study |
| SLY1161 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Mre11-GFP:HIS3 | Invitrogen |
| SLY2862 | SLY1161 rsc2Δ::HYGR | This study |
| SLY2863 | SLY1161 rsc7Δ::HYGR | This study |
| SLY2875 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Ddc2-GFP:HIS3 | Invitrogen |
| SLY2865 | SLY2875 rsc2Δ::HYGR | This study |
| SLY2867 | SLY2875 rsc7Δ::HYGR | This study |
| SLY2876 | BY4743 α1α2::HIS3 | This study |
| SLY2883 | BY4743 α1α2::HIS3 rsc7Δ::KANRrsc7Δ::HYGR | This study |
| SLY2726 | BY4743 α1α2::HIS3 rad52Δ::LEU2 | This study |
| W4138-17D | MATaade2-a leu2ΔBstEII TRP1 lys2Δ | 10a |
| W4091-4C | MATα ade2-n leu2ΔBstEII trp1-1 | 10a |
| SLY3819 | W4138-17D rsc7Δ::HYGR | This study |
| SLY3822 | W4091-4C rsc7Δ::HYGR | This study |
| G354 | MATα FLO1::URA3 in S288C | Gerald Fink |
| SLY3851 | G354 rsc7Δ::HYGR | This study |
| BY4743 | MATα/ahis3Δ1/his3Δ1 leu2Δ/leu2Δ lys2 leu2Δ/LYS2 met15Δ/MET15 ura3Δ/ura3Δ | Invitrogen |
| SLY3861 | BY4743 rsc7Δ::HYGR/rsc7Δ::HYGR | This study |
| SLY3863 | BY4743 rad59Δ::KANR/rad59Δ::KANR | This study |
| SLY3865 | BY4743 rad51Δ::KANR/rad51Δ::KANR | This study |
| SLY3867 | BY4743 rsc7Δ::HYGR/rsc7Δ::HYGRrad59Δ::KANR/rad59Δ::KANR | This study |
| SLY3869 | BY4743 rsc7Δ::HYGR/rsc7Δ::HYGRrad51Δ::KANR/rad51Δ::KANR | This study |
Cell cycle checkpoint assay.
Cell synchronization at G1 by α-factor treatment or at G2 by nocodazole treatment was done as described previously (72). Hyperphosphorylated Rad53 was detected as described in reference 41 using anti-Rad53 polyclonal antibody (Santa Cruz Biotechnology). Flow cytometry analysis was performed on a Becton Dickinson FACScalibur system as described previously (72).
Purification of proteins.
The Rad51 and Rad52 proteins were purified as described previously (47, 59). The RSC complex was purified from yeast cells that chromosomally harbor tandem affinity purification (TAP)-tagged Rsc2 as described previously (69).
Rad59 was purified from E. coli Rosetta cells (Novagen) carrying pET32-RAD59. When the A600 of the culture reached 0.6 to 0.8, Rad59 protein expression was induced by 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 16°C for 16 h. The E. coli cell pellet (50 g) was suspended in 200 ml of cell breakage buffer (50 mM Tris-HCl, pH 7.5; 200 mM KCl; 1 mM 2-mercaptoethanol; 10% sucrose; protease inhibitors aprotinin, chymostatin, leupeptin, and pepstatin A at 2 μg/ml each; 1 mM phenylmethylsulfonyl fluoride) and sonicated. The lysate was clarified by centrifugation (at 100,000 × g for 90 min) and loaded onto a 60-ml Q-Sepharose column. The flowthrough fraction was subjected to precipitation with (NH4)SO4 at a concentration of 0.21 g/ml. The precipitate was harvested by centrifugation (20,000 × g for 30 min), resuspended in 80 ml K buffer (20 mM KH2PO4, pH 7.4; 10% glycerol; 0.5 mM EDTA; 0.5 mM 2-mercaptoethanol; 0.01% Igepal [octylphenoxypolyethoxyethanol]; protease inhibitors), and mixed with 6 ml of Ni-nitrilotriacetic acid agarose beads (Qiagen) for 4 h. The beads were washed with 120 ml of buffer K containing 300 mM KCl and 30 mM imidazole, and Rad59 was eluted with 15 ml of buffer K containing 150 mM KCl and 450 mM imidazole. The Rad59 protein pool was loaded onto a 1-ml Macro hydroxyapatite (Bio-Rad) column and developed with 40 ml of 0 to 300 mM KH2PO4 in buffer K, collecting 1-ml fractions. Peak fractions were pooled (6 ml) and mixed with 1 ml of anti-S-agarose beads (Novagen) for 3 h. The beads were washed with 20 ml of buffer T (50 mM Tris-HCl, pH 7.5; 10% glycerol; 0.5 mM EDTA; 1 mM dithiothreitol [DTT]) containing 500 mM KCl, and Rad59 was eluted with a combination of 4 M MgCl2, 25 mM Tris HCl (pH 7.5), 1 mM DTT, and 0.01% Igepal. The eluate was dialyzed against buffer T containing 1 M KCl, concentrated, divided into small aliquots, and stored at −80°C.
The glutathione S-transferase (GST)-Rsc1 and GST-Rsc2 proteins were overexpressed in Rosetta cells transformed with pGEX-4T-Rsc1 or pGEX-4T-Rsc2. Cells harboring these plasmids were grown at 37°C to an optical density at 600 nm of 0.6 to 0.8, at which time 0.2 mM IPTG was added. The culture was incubated for 16 h at 16°C, and cells were harvested by centrifugation. For protein purification, cell lysate from 10 g of cells was prepared by sonicating the cell suspension in 50 ml cell breakage buffer (50 mM Tris-HCl, pH 7.4; 600 mM KCl; 10 mM EDTA; 1 mM DTT; 10% sucrose; 0.01% Igepal; protease inhibitors) and ultracentrifugation (100,000 × g for 90 min). The clarified lysate was mixed with 1 ml glutathione Sepharose beads (GE Healthcare) for 3.5 h at 4°C. The beads were washed with 50 ml of buffer K containing 500 mM KCl and eluted with the same buffer supplemented with 10 mM glutathione. GST-Rsc1- or GST-Rsc2-containing fractions were pooled, concentrated, divided into small aliquots, and stored at −80°C.
In vitro pulldown assay.
For the pulldown assay with the GST-Rsc1 or GST-Rsc2 protein, glutathione Sepharose beads (5 μl) coated with GST-Rsc1 or GST-Rsc2 (10 μg) were mixed with 2 μg of Rad51, Rad52, or Rad59 at 4°C for 1 h in 30 μl of phosphate-buffered saline. The resin was washed three times with 30 μl of the same buffer and then treated with 30 μl of 2% SDS to elute the bound proteins. The supernatant, the third wash, and the SDS eluate (2 μl of each) were immunoblotted for Rad51, Rad52, or Rad59 protein as indicated.
For the pulldown assay with the RSC complex, the TAP-tagged RSC complex (5 μg) was immobilized on 10 μl of calmodulin resin (Qiagen) and mixed with 200 ng of Rad51, Rad52, or Rad59 in 30 μl of binding buffer containing 50 mM Tris HCl (pH 7.5), 150 mM KCl, 5% glycerol, 1 mM DTT, 0.01% Igepal, and 100 ng/μl bovine serum albumin (BSA). The resin was washed three times with 30 μl of the same buffer and then treated with 30 μl of 2% SDS to elute the bound proteins. The supernatant, the third wash, and the SDS eluate (6 μl of each) were analyzed by immunoblotting as described above.
ATPase assay.
ATP hydrolysis by the RSC complex was examined as described previously (51). Plasmid DNA (pBluescript) assembled into chromatin (40 μM nucleotides) was incubated with the purified RSC complex (15 nM) and 0.1 mM ATP (0.1 μCi/μl [γ-32P]ATP) in 10 μl buffer (50 mM HEPES-KOH, pH 7.3; 2 mM MgCl2; 1 mM DTT; 50 mM KCl; 100 μg/ml BSA) at 30°C. To check the effect of Rad59 (200 nM), it was preincubated with the RSC complex for 45 min on ice before the addition of radioactive ATP and a shift to incubation at 30°C. The level of ATP hydrolysis was determined by thin-layer chromatography as described previously (51).
Yeast two-hybrid assay.
Yeast two-hybrid analysis was carried out as previously described (49). Briefly, RSC1 or RSC2 fused to the GAL4 activation sequence of pGAD-T7 was mated with yeast PJ69α harboring RAD51, RAD52, or RAD59 fused to the GAL4 DNA binding sequence in vector pGBT9. Interactions were determined based on the ability to express the HIS3 reporter gene by spotting cells on synthetic complete (SC) media in the presence or absence of histidine.
Chromatin-remodeling reaction.
The chromatin-remodeling activity of the RSC complex was assayed as described previously (69). Briefly, the reconstituted 201-11 nucleosomal array (3 μM base pairs) was mixed with the restriction enzyme SalI (1 U/μl) in 10 μl buffer A (10 mM HEPES, pH 7.3; 2 mM MgCl2; 1 mM DTT; 100 mM KCl; 1 mM ATP; 25 mM creatine phosphate; 25 μg/ml creatine kinase) on ice. RSC (2.5, 5, or 10 nM) was preincubated with or without Rad52 or Rad59 (50, 100, or 200 nM) in 10 μl buffer T (25 mM Tris HCl, pH 7.5; 0.5 mM EDTA; 5% glycerol; 1 mM DTT; 0.01% Igepal) containing 300 mM KCl for 45 min on ice. These were added to the assembled chromatin with SalI, and the resulting reaction mixtures (20-μl final volume) were incubated at 30°C for 30 min. The reaction was stopped by adding an equal volume of 1% SDS with proteinase K (1 mg/ml). Following a 5-min incubation at 37°C, the deproteinized reaction mixtures were analyzed in a 1% agarose gel. The gel was dried on a sheet of DEAE paper (Whatman) and subjected to phosphorimaging analysis.
HO expression and recombination assays.
HO endonuclease was induced by adding 2% galactose (wt/vol) to logarithmic-phase cultures (1 × 107 cells ml−1) grown in YEP-glycerol or SC medium minus leucine at 30°C (56). The extent of cleavage at the HO site was determined by quantitative PCR as previously described (48), and the colonies plated onto YEP-dextrose (YEPD) or YEP-galactose plates were counted after 3 to 4 days at 30°C following 1 to 3 h of incubation in the galactose-containing medium. The survival frequency was calculated by normalization with the number of colonies obtained without galactose addition. For detection of SSA products, logarithmic-phase cultures were induced to express HO and genomic DNA from each indicated time point was digested with BglII restriction enzyme, separated on a 1.0% agarose gel, and probed with a 32P-labeled HindIII-BamHI fragment distal to URA3 (55) and a BglII fragment of RAD10. We determined the amount of DNA in each band in a PhosphorImager by using the RAD10 band as an internal loading control. To detect mating type GC products, JKM161 and its mutant derivatives were induced for HO cleavage and the genomic DNA was digested with EcoRV, separated by an agarose gel (1%), and transferred to a nylon membrane (Hybond N+). The blot was probed with a 1-kb 32P-labeled MAT distal probe that recognizes the 3.4-kb (MATα) or the 5.0-kb (MATa) fragment containing the HO cleavage site on yeast chromosome III. The levels of recombination products were determined after normalization to a HIS4 sequence that served as an internal loading control. Recombination between leu2 heteroalleles, intrachromosomal recombination between repeated sequences at the FLO1 gene locus, ectopic GC, and BIR assays were performed as described previously (10, 21, 28, 52, 66).
Pulsed-field gel electrophoresis (PFGE).
Cells (0.5 × 107/ml) arrested at G1 or G2 were incubated with 0.2% (vol/vol) methyl methanesulfonate (MMS) for 20 min at 30°C. Cells were washed 3 times with YEPD with 10 μg/ml nocodazole, resuspended in fresh YEPD containing 10 μg/ml nocodazole, and incubated at 30°C for an additional 8 h. Plug formation and PFGE were carried out as described in the CHEF Yeast Genomic DNA Plug Kit manual (Bio-Rad). Southern blot analysis was performed for quantitative measurement of chromosome recovery after DNA damage using a 32P-labeled PEX10 probe that is specific for chromosome IV.
Measurement of unequal sister recombination rates.
To measure unequal sister recombination rates, 11 colonies of strain YNN301 (13) and its mutant derivatives were cultured in YEPD and plated onto an SC plate lacking histidine to score for the recombination events that lead to histidine auxotrophy after 4 days of plating. Recombination rates were estimated using the method of Lea and Coulson (28). Error bars represent three or four independent experiments.
Chromatin immunoprecipitation (ChIP).
Asynchronous or nocodazole (10 μg/ml)-arrested cells were induced for HO break formation for the indicated lengths of time. For ChIP, immunoprecipitation of Smc1-hemagglutinin (HA), Scc1-HA, and Rad59; PCR amplification; and additional analysis were performed as previously described (48).
RESULTS
Inactivation of RSC induces a DNA damage response.
All but seven of the RSC subunits are essential for viability in haploid yeast cells (5, 49). As an initial measurement of the role of RSC in repair and signaling of DNA damage, we surveyed the DNA damage sensitivity of haploid yeast strains with RSC1, RSC2, RSC7, RSC14, RSC30, HTL1, and RTT102 deleted. According to previous results, deletion of RSC7 or HTL1 likely affects the function of all RSC complexes, while RSC1 or RSC2 deletion is thought to affect distinct subclasses (6). The C-terminal truncation of 54 amino acids in RSC8 was also included in the analysis because it is viable at 30°C (61). In parallel, we examined the sensitivity of diploid strains with each nonessential RSC gene deleted to test whether RSC functions differently according to ploidy.
Three strains, the rsc2Δ, rsc7Δ, and htl1Δ mutants, exhibit moderate-to-extreme thermosensitivity at 30 to 37°C (Fig. 1 A). Indeed, the results are consistent with the finding reported by various groups that deletion of RSC genes leads to temperature-sensitive growth and hypersensitivity to genotoxic agents (5, 7, 15, 27, 31, 34, 45, 61, 68). The apparent temperature sensitivity of these mutants must be the consequence of the corresponding RSC gene deletions because thermosensitivity can be rescued by ectopic expression of each RSC gene (data not shown). Our results showed that neither ploidy nor the strain background influenced the thermosensitivity engendered by RSC gene deletions (Fig. 1A and data not shown).
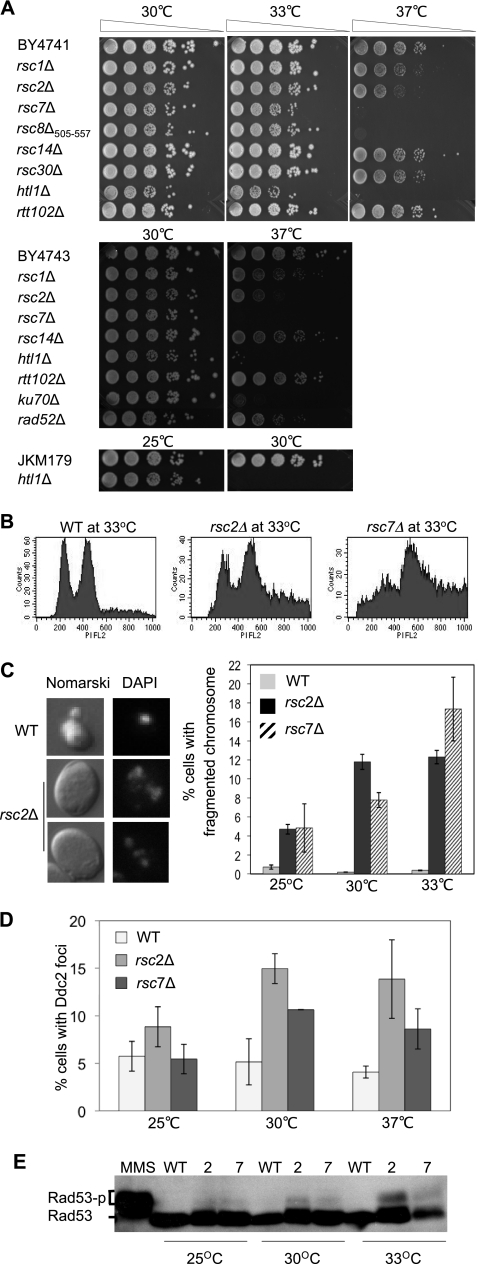
Fig. 1.
rsc mutations induce a spontaneous DNA damage response. (A) Mutants lacking nonessential RSC subunits in the BY4741, BY4743 diploid, and JKM179 backgrounds were tested for growth at 30°C, 33°C, and 37°C. Fivefold serial dilutions were plated and photographed after 3 to 4 days of growth. (B) Cell cycle distribution was assessed by flow cytometry as described in Materials and Methods. Wild-type and rsc2 and rsc7 mutant cells grown overnight in YEPD at 33°C were analyzed by FACS. (C) The nuclear morphology of BY4741 and its rsc mutant derivatives grown at 33°C was examined by DeltaVision fluorescence microscopy (left). The images shown were obtained with Nomarski optics after 4′,6-diamidino-2-phenylindole (DAPI) staining. Percentages of cells containing fragmented nuclei were scored from at least 300 cells in each mutant and plotted as a function of increasing growth temperature (right). The results are the average of at least three independent experiments ± the standard deviation. (D) Percentages of wild-type (WT), rsc2Δ mutant, and rsc7Δ mutant cells showing Ddc2-GFP foci at the indicated growth temperatures. More than 300 cells were analyzed in each case. (E) Phosphorylation of Rad53 in wild-type cells and the rsc2 and rsc7 mutants at different growth temperatures was analyzed by Western blotting using anti-Rad53 antibody. For comparison, hyperphosphorylation of Rad53 was induced by treating logarithmically growing wild-type cells with 0.2% MMS for 20 min (lane 1).
To elucidate the basis of the growth defect of the rsc mutants at the nonpermissive temperature, we examined the cell cycle profiles of the rsc2Δ and rsc7Δ mutants in the BY4741 strain background at the semipermissive temperature of 33°C. Analysis at the semipermissive temperature of 33°C instead of 37°C was done to avoid complications from cells undergoing necrosis. We found that deletion of RSC2 or RSC7 moderately but reproducibly enriched for G2 cells, whereas G1- and S-phase cell populations were decreased (Table 2 and Fig. 1B). Furthermore, the rsc2Δ and rsc7Δ mutants accumulated (4- to 8-fold, respectively) cells with aberrant morphology and fragmented nuclei (Fig. 1C). To test if the aberrant morphology reflects induction of DNA damage-induced cell cycle checkpoints, we monitored the formation of spontaneous Ddc2-green fluorescent protein (GFP) foci and Rad53 hyperphosphorylation in strains grown at the permissive, semipermissive, or nonpermissive temperature. We found that the absence of RSC2 or RSC7 increases spontaneous Ddc2-GFP foci 2- to 3-fold and accumulates phosphorylated Rad53 at the semi- or nonpermissive temperature (Fig. 1D and E). Together, the results revealed that the deletion of RSC2 or RSC7 induces the DNA damage response.
Table 2.
Altered cell cycle distribution and morphology in rsc mutantsa
| Strain | % of cells |
|||
|---|---|---|---|---|
| Single | Small budded | Large budded | Aberrantb | |
| Wild type | 36.95 | 21.37 | 35.86 | 5.82 |
| rsc2Δ mutant | 18.44 | 12.13 | 43.20 | 26.23 |
| rsc7Δ mutant | 12.29 | 5.88 | 39.00 | 42.83 |
The distributions of single, small-budded, large-budded, and aberrant rsc mutant and wild-type cells grown at 33°C in YEPD are compared.
Aberrant morphology represents enlarged, elongated, or multibudded single cells. We analyzed 500 to 600 cells of each strain.
Rsc2 and Rsc7 participate in the Rad51-independent but Rad59-dependent homologous DNA repair pathway.
We found that deletion of RSC7 or HTL1 causes severe sensitivity to MMS, ionizing radiation (IR), phleomycin, hydroxyurea, and camptothecin, whereas deletion of RSC2 or the C-terminal truncation of RSC8 engenders moderate but clear sensitivity to these agents (Fig. 2 A). The variations in DNA damage sensitivity among the rsc mutants suggest that the RSC complex may exist in multiple isoforms (5) that may play distinct roles in conferring tolerance to specific types of DNA damage.
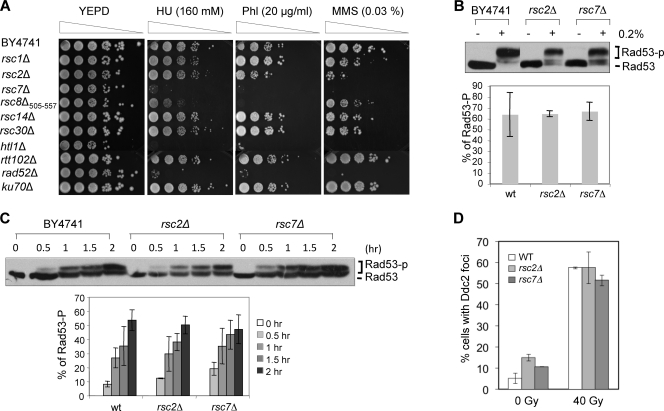
Fig. 2.
Integrity of cell cycle checkpoint activation in rsc mutants. (A) Viability of mutants after DNA-damaging treatment was determined by spotting cells (5-fold serial dilutions) onto YEPD plates containing hydroxyurea (HU), phleomycin (Phl), or MMS. The plates were photographed after 3 to 4 days of growth at 30°C. (B) The integrity of damage-induced cell cycle checkpoint activation was assessed in cells treated with 0.2% MMS for 30 min and examined for the level of Rad53 phosphorylation by Western blotting using anti-Rad53 antibody. Percent Rad53 phosphorylation was calculated as intensity of phosphorylated Rad53/intensity of total Rad53 using the ImageJ program. The averages of three independent experiments ± the standard deviations are shown. (C) Kinetics of Rad53 phosphorylation in cells that were arrested at S/G2 and exposed to 0.033% MMS for the indicated times. Percent Rad53 phosphorylation was calculated as described for panel B. The averages of three independent experiments ± the standard deviations are shown. (D) Yeast cells expressing Ddc2-GFP were gamma irradiated and fixed with paraformaldehyde, and the number of cells showing Ddc2-GFP foci was scored using a DeltaVision microscope. Over 300 cells of each strain were analyzed. wt, wild type.
Reduced survival upon DNA damage can be attributed to either a DNA damage checkpoint or a repair defect. We thus interrogated checkpoint functions in the rsc2 and rsc7 mutants by assessing the level and kinetics of Rad53 hyperphosphorylation in 0.033 and 0.2% MMS. A severe growth deficiency, even at 30°C (Fig. 1A), kept us from analyzing the htl1Δ mutant further. We focused most of our analysis on the rsc2 and rsc7 mutants. We found that both the rsc2 and rsc7 mutations efficiently induce Rad53 hyperphosphorylation within 30 min of MMS treatment (Fig. 2B and C). Importantly, our results showed that gamma irradiation-induced Ddc2-GFP focus formation occurs normally in the rsc mutants (Fig. 2D). Thus, the hypersensitivity of the rsc2 and rsc7 mutants to genotoxic agents is not due to an inability to mount the checkpoint response.
We next tested whether RSC is required for the repair of DSBs by HR, as the elimination of genotoxin-induced damage in yeast is mediated primarily by this repair mechanism. HR is either Rad51 dependent or Rad51 independent (2, 9). Rad51-independent HR requires the Rad59 protein and includes subsets of GC, BIR, and SSA (57). Therefore, we assessed the epistatic relationship between RSC genes and RAD51 or RAD59. We found that the rsc2 rad51 or the rsc7 rad51 double mutant exhibits synergistic hypersensitivity to DNA-damaging agents compared to single gene deletion mutants (Fig. 3 A and B). In contrast, the rsc2 rad59 or rsc7 rad59 mutant strain is no more sensitive to DNA-damaging treatment than the rad59 strain (Fig. 3A and B). The severe hypersensitivity seen in the rsc2 rad51 or rsc7 rad51 strain is indistinguishable from that of the rad51 rad59 strain (Fig. 3B). The results provide evidence that RSC operates in Rad51-independent but Rad59-dependent DSB repair.
Fig. 3.
RSC participates in Rad59-dependent DNA repair. (A) The viability of single and double rsc mutant combinations and the indicated HR genes was determined by spotting cells onto culture plates containing the indicated genotoxic chemicals. Fivefold serial dilutions were spotted, and the plates were photographed after 3 to 4 days of growth at 30°C. (B) Percent survival after gamma irradiation or MMS treatment. Logarithmic-phase cultures of yeast cells were harvested, resuspended in phosphate-buffered saline, irradiated with the indicated dose of gamma rays or treated with the indicated concentration of MMS for 20 min, and plated onto YEPD plates. After 3 to 4 days, surviving colonies were counted and the survival rate was calculated by comparison to the number of colonies of mock-treated cells. Each experimental point represents the average of three independent experiments ± the standard deviation. HU, hydroxyurea.
Rad59 interacts with Rsc1 and Rsc2.
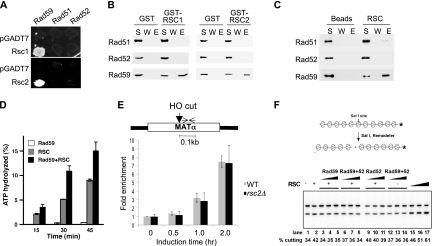
Given the role of Rsc2 in Rad59-dependent HR, we examined whether a physical interaction between Rad59 and RSC exists (Fig. 4). Using the yeast two-hybrid assay, we found robust interaction between Rsc1 or Rsc2 with Rad59, but not with Rad51 or Rad52 (Fig. 4A). The interaction between Rad59 and Rsc1 or Rsc2 is further confirmed by affinity pulldown assays using purified GST-tagged Rsc1 or Rsc2 and Rad51, Rad52, or Rad59 proteins. We found specific association of Rad59 with GST-Rsc1 or GST-Rsc2 and, as expected, Rad59 did not interact with GST alone (Fig. 4B). Additionally, RSC complex immobilized on resin was tested for association with Rad51, Rad52, and Rad59. Rad59, but not Rad51 or Rad52, was specifically retained on the RSC resin (Fig. 4C). These results suggest that RSC physically interacts with Rad59.
Fig. 4.
Rad59 interacts with and enhances the ATPase activity of the RSC complex. (A) Interactions of Rsc1 and Rsc2 with Rad51, Rad52, and Rad59 were determined by yeast two-hybrid assays. Interactions were discerned based on expression of the HIS3 reporter gene by spotting cells on SC in the absence of histidine. (B) Pulldown experiment was performed using purified GST-RSC1 and -RSC2 and purified Rad51, Rad52, and Rad59 proteins. Proteins bound to glutathione Sepharose resin were eluted with 2% SDS. The supernatant (S), wash (W), and SDS eluate (E) fractions were analyzed by immunoblot assay with anti-Rad51, anti-Rad52, or anti-Rad59 antibodies. (C) The purified RSC complex with TAP-tagged Rsc2 was incubated with Rad51, Rad52, or Rad59 protein, and the RSC-associated proteins were pulled down with calmodulin resin. The different fractions from the affinity pulldown reactions were probed for Rad51, Rad52, and Rad59 as described for panel B. (D) ATP hydrolysis as a function of time was measured for RSC, Rad59, and a mixture of the two in the presence of chromatinized dsDNA. (E) ChIP assays measured the level of Rad59 using anti-Rad59 antibody as described in Materials and Methods. Fold enrichment represents the ratio of the anti-Rad59 immunoprecipitate PCR signal before and after HO induction, normalized against the PCR signal of the PRE1 control and the amount of input DNA. The location of the primers used for PCR at the MAT locus is shown by the opposing arrows. (F) Chromatin-remodeling assay using 201-11 nucleosomal arrays. The 201-11 nucleosomal substrate was incubated with RSC (lanes 2 to 11 and 15, 2.5 nM; lane 16, 5 nM; lane 17, 10 nM), Rad59 (50, 100, or 200 nM), Rad52 (50, 100, or 200 nM), or Rad52 and Rad59 (50, 100, or 200 nM each) at 30°C for 30 min. WT, wild type.
The physical and functional interaction between RSC and Rad59 prompted us to test if RSC plays a role in recruiting Rad59 to the DSB by the ChIP assay. Rad59 was specifically recruited to the HO-induced DNA break at the MAT locus (Fig. 4E). Importantly, deletion of RSC2 had no influence on the level and timing of Rad59 recruitment to the DSB. Since RSC remodels chromatin at the expense of ATP hydrolysis, we explored if Rad59 stimulates the ATPase activity of RSC. Indeed, Rad59 enhanced RSC-dependent ATP hydrolysis 2-fold, whereas Rad59 alone did not hydrolyze ATP (Fig. 4D). Nevertheless, enhanced ATP hydrolysis by Rad59 did not result in more efficient RSC-dependent chromatin remodeling (Fig. 4F) (23). The results suggest that RSC participates in Rad59-dependent HR in a step other than DSB recruitment of Rad59.
RSC is dispensable for SSA, GC, and BIR.
The SSA type of recombination is dependent on Rad59 (60). We thus examined whether Rsc2 or Rsc7 is required for SSA. To this end, the HO endonuclease was induced in the tNS1379 strain, in which a HO cut site is situated between two homologous ura3 sequences of 205 bp on chromosome V (Fig. 5 A). SSA between the ura3 repeats was monitored by scoring cell viability after HO induction and the formation of the SSA product by Southern blot hybridization (Fig. 5B, C, and D) (55). We found that deletion of RSC2 or RSC7 impairs neither cell survival nor SSA product formation subsequent to HO induction (Fig. 5B and D). The slow disappearance of the HO cleavage product (labeled C in Fig. 5C) in rsc2 and rsc7 cells may be attributed to the attenuated DSB resection in these mutants owing to a reduced DSB recruitment of MRX (48).
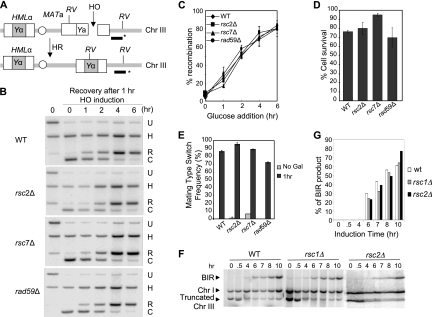
Fig. 5.
RSC is dispensable for SSA recombination. (A) Diagram showing SSA in tNS1379. The 205-bp ura3 direct repeats, the HO recognition site, the locations of BglII sites (arrows), and the probe (black bar with asterisk) used to detect SSA products in the Southern blot assay are shown. (B) Percent survival was determined by dividing the number of colonies on a YEP-galactose plate with that in YEPD. Each experimental point represents the average of three independent experiments ± the standard deviation. (C) SSA products in rsc mutants were detected by Southern blot hybridization. Genomic DNA digested with BglII was separated by agarose gel electrophoresis and subjected to Southern blot hybridization with 32P-labeled probes. One anneals to the 3′ region of the ura3 sequences (as indicated in panel A), and the other anneals to the RAD1 gene at chromosome XVI. U, signal represents uncut fragment; C, signal resulting from the HO cleavage; R, signal from recombination; H, signal from the RAD1 locus used as a control. (D) Percent SSA repair was determined by normalizing the amount of SSA products with that of the control RAD1 signal and plotted as a function of time of HO expression. Results obtained with the wild-type RSC strain (WT; tNS1379) and the rsc2Δ, rsc7Δ, and rad59Δ mutants are shown. Each point represents the average of three independent experiments ± the standard deviation.
We examined the influence of RSC on mating type switch GC between the HMLα and MATa loci on chromosome III and also on ectopic GC between the MAT and MATa-inc sequences located on chromosomes V and III, respectively, upon induction of the HO gene integrated at the ADE3 gene locus. We found that both the recombination frequency and the level of repair product in the rsc2 and rsc7 mutants were comparable to those in the wild-type strain, suggesting that RSC is dispensable for GC (Fig. 6 A to E; data not shown).
Fig. 6.
RSC is dispensable for mating type GC and BIR. (A) Schematic of mating type switch GC between MATa and HMLα. The HO recognition site, the locations of EcoRV sites (RV), and the DNA probe (black bar with asterisk) used to detect GC products by Southern blot assay are shown. (B) Effects of rsc2Δ and rsc7Δ on the mating type GC reaction initiated by induction of HO endonuclease expression. H, signal from the HIS3 locus used as a control; U, signal from the MAT locus that represents the uncut fragment; C, signal resulting from HO cleavage; R, signal from the recombination reaction; WT, wild type. (C) Summary of Southern blot analysis results. Percent repair was determined as the ratio of the recombination signal (R in panel B) to the signal from the HIS3 control (H in panel B) and is plotted as a function of recovery time after glucose addition to turn off HO expression. Data represent the mean ± the standard deviation of three or more independent experiments. (D) Percent survival was determined by dividing the number of surviving colonies growing on a YEPD plate after 1 h of HO expression, normalized by the number of colonies growing after mock HO expression. Each experimental point represents the average of three independent experiments ± the standard deviation. (E) Percent mating type switching was determined by normalizing the number of colonies becoming MATα type among survivors. Each point represents the average of three independent experiments ± the standard deviation. Gal, galactose. (F) DSB repair in AM1003 and rsc mutants was analyzed using PFGE at intervals after induction of DSB at MATa. Southern blots were probed with ADE1, which hybridized to the truncated chromosome III (Chr III) BIR product and its native position on chromosome I. The chromosome I signal was used as a loading control. (G) Quantitation of BIR repair product.
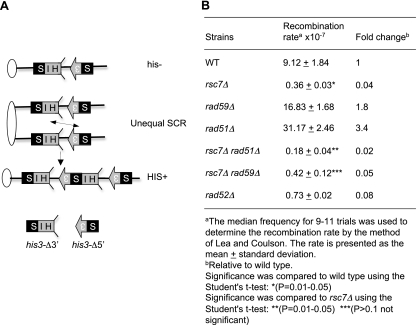
We next examined whether RSC7 gene deletion affects recombination between leu2 heteroalleles in diploid cells or SSA between repeated sequences at the FLO1 gene locus (Table 3). We found that RSC7 gene deletion does not reduce heteroallelic recombination and that, interestingly, it dramatically elevates SSA. These results suggest that Rsc7 is not required for heteroallelic recombination but likely suppresses SSA between repeats. We also examined if RSC1 or RSC2 deletion decreases BIR frequency in a disomic strain carrying a truncated chromosome III with a HO cleavage site by recombining with an extra copy of chromosome III that harbors an uncleavable MAT-inc sequence as the template (11). Neither the RSC1 nor the RSC2 deletion reduces the BIR-dependent cell survival or the formation of BIR products in Southern blot analysis (Fig. 6F and G). Collectively, these results suggest that RSC is dispensable for interhomolog or intrachromosomal recombination, regardless of the recombination subtype.
Table 3.
Effects of Δrsc7 on mitotic recombination
| Strain | Mean rate,a 10−7 ± SD (fold change)b |
|
|---|---|---|
| Heteroallelic recombination | Direct-repeat recombination (FLO1) | |
| Wild type | 6.63 ± 0.55 (1) | 1.54 ± 0.4 (1) |
| rsc7Δ mutant | 5.24 ± 1.0c (0.79) | 292 ± 57d (189) |
The median frequency for 9 to 11 trials was used to determine the recombination rate by the method of Lea and Coulson.
Relative to the wild type.
P > 0.1 (not significantly different from the wild type).
P = 0.01 to 0.05 (significantly different from the wild type).
RSC is required for MMS-induced chromosome damage repair in the G2 phase.
We then asked if RSC is involved in recombination between sister chromatids. If so, repairing DSBs in S/G2, when sister chromatids are available, will be dependent on the RSC complex, whereas the repair of a DSB made in G1 should be largely independent of RSC. To test this premise, we monitored repair product formation in haploid (Fig. 7 A and B) and diploid rsc7Δ mutants with MATα deleted (that thus behaving like the MATa type) challenged with the alkylating agent MMS at G1 and G2 (Fig. 7C and D). Briefly, cells were first arrested at G1 or G2 with α-factor or nocodazole, treated with 0.2% MMS for 20 min, and then released to a drug-free medium for various time intervals to allow recovery (Fig. 7A to D). Chromosomes were isolated and examined by PFGE. G1 or G2 arrest was ensured by monitoring cell cycle progression by fluorescence-activated cell sorting (FACS), as well as cell counting under a microscope (Fig. 7C and D and data not shown). The relative repair efficiency was calculated by comparing the integrity of chromosome IV or XV after MMS exposure normalized against the same chromosome in cells that had not been exposed to MMS, setting the wild-type value to 1 (Fig. 7). To normalize different growth rates among mutants (e.g., the rsc7 mutant grows more slowly than the wild type), we used drug-free medium containing nocodazole to keep cells at G2/M upon their release from either G1 or G2 arrest. In wild-type cells, chromosomes fragmented by MMS treatment are partially recovered after growth in drug-free medium for 8 h upon release from G1 or G2 arrest, regardless of ploidy (Fig. 7A to D). At both G1 and G2, repair of MMS damage depends primarily on Rad52, as chromosome recovery after MMS treatment was severely deficient in rad52Δ mutant cells (Fig. 7A to D). Interestingly, when the RSC7 gene deletion strain was challenged with MMS at G2, the repair efficiency was reduced to less than 20% of that of the wild type 8 h after MMS exposure (Fig. 7B and D). In contrast, deletion of RSC7 only moderately reduced the repair of MMS damage induced at G1 (Fig. 7A and C). We also found that deletion of RAD51, but not RAD59, further reduced the repair in diploid rsc7Δ mutant cells, supporting the role of Rsc7 in Rad51-independent but Rad59-dependent repair (Fig. 7F). Deletion of RSC2 in haploid cells also reduces chromosome recovery after MMS treatment at G2, and the repair deficiency is further reduced by the deletion of RAD51 but not by the deletion of RAD59 (Fig. 7E).
Fig. 7.
rsc mutants are defective in DSB repair in G2 but not in G1. (A and B) Haploid BY4741 cells were grown to mid-log phase, arrested at G1 (10 μg/ml α-factor for 3 h) (A) or G2 (10 μg/ml nocodazole) (B), and then incubated with MMS (0.2% for 20 min). Cells were washed and resuspended in YEPD supplemented with 10 μg/ml nocodazole and allowed to recover for up to 8 h with samples for PFGE taken at the indicated time points. DNA separated by PFGE as shown in panels A to F was subjected to Southern blot analysis to determine the percentage of repair using the PEX10 fragment in chromosome IV as a probe. The percentage of the repair of broken chromosome IV was calculated by measuring the amount of chromosome IV after MMS damage at each time point and then normalized to the mock-treated samples at the same time point. The repair efficiency after 8 h of recovery in wild-type (wt) cells was set to 1. The averages of three independent experiments ± the standard deviation are shown. (C and D) Diploid yeast cells with a MATα deletion (and thus of the MATa type instead of the typical MATa/α type) were arrested at G1 (C) or G2 (D), treated with MMS, and allowed to recover in nocodazole-containing but MMS-free medium. Shown are FACS profiles of diploid wild-type and rsc7 and rad52 mutant cells arrested at G1(C) or G2 (D), treated with MMS, and allowed to recover in nocodazole-containing but MMS-free medium. (E and F) Quantification of the repair efficiency of haploid yeast cells with MATa (BY4741) (E) and diploid cells (BY4743) (F) at G2 using Southern blotting with rad59Δ and rad51Δ mutant combinations. Repair efficiency at G2 was determined as described for panel A and plotted. The averages of three independent experiments ± the standard deviation are shown.
We also measured the frequency of unequal sister chromatid recombination (SCR) that occurs spontaneously between his3 repeats, one containing a deletion of the 5′ protein coding sequence and the other containing a deletion of the 3′ coding sequence, in the wild-type, rad52Δ mutant, or rsc7Δ mutant strain (Fig. 8 A) (2, 13). As shown in Fig. 8B, the absence of RSC7 caused a dramatic reduction (as severe as that caused by RAD52 deletion) in the unequal sister recombination rate, judging from the reduction in the number of histidine prototrophs. Deletion of RAD51 further (∼2-fold) reduced sister chromatid exchange in the rsc7Δ mutant, whereas deletion of RAD59 did not (Fig. 8B). The above results support the premise that RSC is required for recombination between sister chromatids.
Fig. 8.
Rsc7 is required for unequal SCR. (A) Schematic representation of the unequal SCR assay. Ovals represent centromeres, and lines represent chromosomes. Arrows and boxes denote HIS3, S3 represents the 5′ deletion, and HIS represents the 3′ deletion. The region of homology is shown in black. (B) Unequal SCR rates were determined by dividing the number of HIS+ colonies in an SC plate without histidine with that in YEPD. The recombination rate was estimated by the method of Lea and Coulson (28).
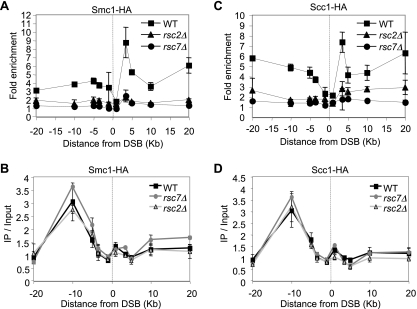
Rsc7 is required for loading of cohesin at DNA breaks.
Previous studies demonstrated that RSC loads cohesin onto chromosome arms (1, 20). Furthermore, recruitment of cohesin to DSBs depends on MRX and phosphorylated H2AX, both of which require a functional RSC complex to associate with the DSB region (26). We hypothesized that RSC promotes cohesin loading at or near DSBs, where the newly loaded cohesin molecules help tether the broken and intact sister chromatids to facilitate recombination between the chromatids (54, 62). To test this model, we carried out ChIP of HA-tagged cohesin subunit Smc1 or Scc1 to monitor the association of cohesin with the HO-induced DSB at MAT in donorless wild-type cells and the rsc2 or rsc7 mutant. We found that cohesin recruitment was dramatically reduced when RSC2 or RSC7 was deleted (Fig. 9 A and C). Deletion of RSC2 or RSC7 did not have a significant impact on the level of cohesin before HO expression (Fig. 9B and D). The results suggest that RSC is involved in the DSB recruitment of cohesin to facilitate homologous repair using the intact sister chromatid.
Fig. 9.
Lack of RSC reduces cohesin recruitment at DNA breaks. ChIP assays measured the levels of Smc1-HA (A) and Scc1-HA (C) using anti-HA antibody as described previously (48). Fold enrichment represents the ratio of the anti-HA immunoprecipitate PCR signal before and after HO induction, normalized against the PCR signal of the PRE1 control and the amount of input DNA. The mean values ± the standard deviations from three independent experiments are shown. The site of the HO break is indicated by the dotted line. The levels of Smc1-HA (B) and Scc1-HA (D) before HO induction are shown as immunoprecipitate (IP)/input, which represents the anti-HA immunoprecipitate PCR signal normalized against the PCR signal of the PRE1 control and the amount of input DNA. WT, wild type.
DISCUSSION
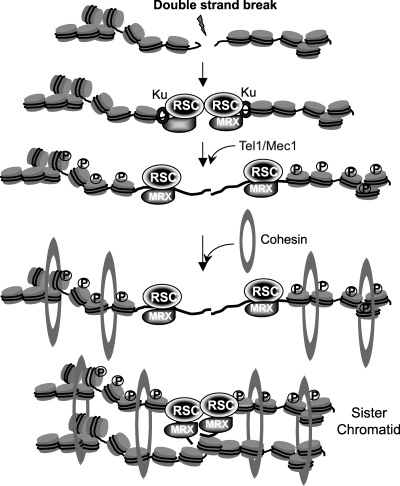
Repair of chromosomal breaks relies on distinct histone modifications and chromatin-remodeling complexes (3, 12, 40, 64, 70). Here we have evaluated the role of RSC in the repair and signaling of DNA damage incurred by exposure to genotoxic chemicals and radiation and found that this chromatin remodeler plays a key role in enforcing interactions between sister chromatids upon DNA damage and thereby promoting sister chromatid recombination (SCR) (Fig. 10). We also found that the DNA damage response and nuclear deformation are induced in certain rsc gene mutants in a temperature-dependent fashion. These new results shed light on the role of RSC in DSB repair/signaling to help maintain chromosome integrity.
Fig. 10.
Possible mechanisms of RSC-mediated SCR.
Inactivation of RSC leads to DNA damage.
We found that deletion of three nonessential subunits of the RSC complex leads to a moderate-to-severe growth deficiency at temperatures over 30°C (Fig. 1A). Growing these mutants at a semi-or nonpermissive temperature leads to accumulation of Ddc2 foci and the hyperphosphorylated form of Rad53 (Fig. 1D and E), suggesting that the DNA damage response is activated. The mutants also accumulate at the G2/M stage of the cell cycle and exhibit abnormal nuclear morphology, likely representing the Cut (cells untimely torn) phenotype (Fig. 1C and Table 2). The precise reason for checkpoint activation and mitotic catastrophe in rsc mutants at non- or semipermissive temperatures is unknown. Several possibilities, such as a lack of proper cohesin deposition in S phase, improper segregation and deposition of nucleosomes during DNA replication, or deficient repair of DNA damage that arises during S phase, may account for these phenotypes. Our results provide evidence of an important role for RSC in repairing DNA damage induced by genotoxic chemicals in the S/G2 phase of the cell cycle. It is possible that the growth deficiency of the rsc mutants stems from pleiotropic deficiencies, as discussed above.
RSC is involved in the Rad59-dependent pathways of HR.
Being able to anneal complementary strands, the Rad59 protein has been implicated in strand annealing for SSA and second-end capture and/or strand annealing for GC (8, 44, 71). Rad59 is also implicated in recombination between sister chromatids, especially for unequal sister chromatid exchange (USCE) induced by DNA damage or unequal SCR involving an HO-cleaved plasmid molecule (11). The results of this study suggest that RSC contributes to Rad51-independent but Rad59-dependent recombination processes (Fig. 3, 7, and 8). Rsc1 and Rsc2 also physically interact with Rad59, and the interaction stimulates ATP hydrolysis by the RSC complex (Fig. 4). We propose that RSC and Rad59 participate in a distinct pathway or step(s) of SCR. However, the precise function of the Rad59-RSC complex is not clear. Our ChIP analysis rules out the possibility that RSC promotes the association of Rad59 at the HO-induced DSB (Fig. 4E). However, it remains possible that RSC catalyzes the access of Rad59 to a unique SCR intermediate.
We found that RSC is not defective in yeast mating type switch GC, nonallelic or heteroallelic recombination, SSA, and BIR (Fig. 5 and 6 and Table 3). It is not clear why we did not detect mating type switch deficiency, as has been reported by Chai et al. (7). However, during the course of these studies, we realized that rsc2 deletion mutants grow substantially more slowly than the wild type. This slow growth leads to a delay in the formation of recombination products, even though recombinants eventually arise at the wild-type frequency (Fig. 5 and 6). We thus postulate that the reported deficiency in mating type switch recombination may reflect the delayed product formation in this mutant.
RSC is required for recombination between sister chromatids.
We have presented evidence that RSC participates in recombination between sister chromatids. First, deletion of RSC2 or RSC7 primarily reduces repair of MMS damage induced at G2 when sister chromatids are available for efficient recombination (Fig. 7) (43). Second, RSC promotes the phosphorylation of H2AX and the DSB association of Mre11, which is a prerequisite for cohesin binding at the break (62). Third, inactivation of RSC does not reduce recombination events between substrates situated ectopically or intrachromosomally even though they are delayed substantially (Fig. 5 and 6 and Table 3). Fourth, deletion of RSC7 dramatically reduces exchange between his3Δ-5′ and his3Δ-3′ situated on sister chromatids (Fig. 8). Together, these results argue that RSC functions in recombination between sister chromatids.
Previously, it was shown that cohesin is recruited to the site of DNA breaks with a dependence on Mre11 and γH2AX, and sister chromatid cohesion is established at a postreplication stage in a DNA damage- and Chk1/EcoI-dependent fashion (19, 53, 62, 63). Using ChIP, we have found that RSC is essential for maximum accumulation of cohesin at DNA breaks. Along with the finding that RSC loads cohesin at chromosome arms during replication (31), our results highlight the role of RSC in cohesin loading both in the normal cell cycle and upon DNA damage.
We showed previously that Rsc1 and Rsc2 physically and functionally interact with the Mre11 protein (49). Deletion of RSC2 or depletion of Sth1 compromises DNA damage-induced nucleosome repositioning and reduces the binding of the MRX complex and Ku at DNA ends (48). The reduced MRX level will then attenuate Tel1 recruitment and yield a suboptimal level of γH2AX at the DSB (48), resulting in reduced deposition of cohesin and delaying DNA end resection (Fig. 10). Indeed, many of the rsc mutant phenotypes overlap those of hypomorphic MRX complex mutants, i.e., HR and NHEJ deficiencies, telomere shortening, and compromised sister chromatid cohesion (7, 49). The model further predicts that telomere shortening in rsc mutants can be attributed to reduced MRX levels at telomeres. It is possible that involvement of RSC in cohesin loading onto chromosome arms in the normal cell cycle may be tied to its role in modulating the access of MRX to the newly replicated DNA for cohesion establishment (26, 62).
Owing to the higher fidelity of SCR than of allelic or ectopic recombination, it is widely believed that SCR is a preferred HR pathway and that a deficiency in this mechanism predisposes cells to the gross chromosomal rearrangements seen in cancer. In addition, the unfaithful chromosome segregation and mitotic catastrophe in the rsc mutants represent yet further key attributes of cancer cells. These observations are consistent with the role of SNF5 in tumor suppression (38). It will be of considerable importance to further define the involvement of RSC in the maintenance of chromosome stability.
ACKNOWLEDGMENTS
We thank the members of the S.E.L., P.S., A.M., and G.I. laboratories for helpful comments; K. J. Myung for sharing strains and discussions; and B. J. Daniel for FACS support.
This work was funded by grants from the NIH to S.E.L. (GM071011, 3R01 GM071011), P.S. (R01GM57814), A.M. (GM084242-01), and G.I. (GM080600 and 3R01GM080600). S.E.L. is a scholar of the Leukemia and Lymphoma Society.
Footnotes
Published ahead of print on 1 August 2011.
REFERENCES
- 1. Baetz K. K., Krogan N. J., Emili A., Greenblatt J., Hieter P. 2004. The ctf13-30/CTF13 genomic haploinsufficiency modifier screen identifies the yeast chromatin remodeling complex RSC, which is required for the establishment of sister chromatid cohesion. Mol. Cell. Biol. 24:1232–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bai Y., Symington L. S. 1996. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 10:2025–2037 [DOI] [PubMed] [Google Scholar]
- 3. Bao Y., Shen X. 2007. Chromatin remodeling in DNA double-strand break repair. Curr. Opin. Genet. Dev. 17:126–131 [DOI] [PubMed] [Google Scholar]
- 4. Cairns B. R., et al. 1996. RSC, an essential, abundant chromatin-remodeling complex. Cell 87:1249–1260 [DOI] [PubMed] [Google Scholar]
- 5. Cairns B. R., et al. 1999. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol. Cell 4:715–723 [DOI] [PubMed] [Google Scholar]
- 6. Chaban Y., et al. 2008. Structure of a RSC-nucleosome complex and insights into chromatin remodeling. Nat. Struct. Mol. Biol. 15:1272–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chai B., Huang J., Cairns B. R., Laurent B. C. 2005. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 19:1656–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Davis A. P., Symington L. S. 2003. The Rad52-Rad59 complex interacts with Rad51 and replication protein A. DNA Repair (Amst.) 2:1127–1134 [DOI] [PubMed] [Google Scholar]
- 9. Davis A. P., Symington L. S. 2001. The yeast recombinational repair protein Rad59 interacts with Rad52 and stimulates single-strand annealing. Genetics 159:515–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deem A., et al. 2008. Defective break-induced replication leads to half-crossovers in Saccharomyces cerevisiae. Genetics 179:1845–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a. de Mayolo A. A., et al. 2010. The rad52-y66A allele alters the choice of donor template during spontaneous chromosomal recombination. DNA Repair (Amst.) 9:23–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dong Z., Fasullo M. 2003. Multiple recombination pathways for sister chromatid exchange in Saccharomyces cerevisiae: role of RAD1 and the RAD52 epistasis group genes. Nucleic Acids Res. 31:2576–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Downs J. A., Nussenzweig M. C., Nussenzweig A. 2007. Chromatin dynamics and the preservation of genetic information. Nature 447:951–958 [DOI] [PubMed] [Google Scholar]
- 13. Fasullo M. T., Davis R. W. 1987. Recombinational substrates designed to study recombination between unique and repetitive sequences in vivo. Proc. Natl. Acad. Sci. U. S. A. 84:6215–6219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feng Q., et al. 2007. Rad52 and Rad59 exhibit both overlapping and distinct functions. DNA Repair (Amst.) 6:27–37 [DOI] [PubMed] [Google Scholar]
- 15. Florio C., et al. 2007. A study of biochemical and functional interactions of Htl1p, a putative component of the Saccharomyces cerevisiae, Rsc chromatin-remodeling complex. Gene 395:72–85 [DOI] [PubMed] [Google Scholar]
- 16. Gasior S. L., Wong A. K., Kora Y., Shinohara A., Bishop D. K. 1998. Rad52 associates with RPA and functions with rad55 and rad57 to assemble meiotic recombination complexes. Genes Dev. 12:2208–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harrison J. C., Haber J. E. 2006. Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 40:209–235 [DOI] [PubMed] [Google Scholar]
- 18. Hays S. L., Firmenich A. A., Massey P., Banerjee R., Berg P. 1998. Studies of the interaction between Rad52 protein and the yeast single-stranded DNA binding protein RPA. Mol. Cell. Biol. 18:4400–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heidinger-Pauli J. M., Unal E., Guacci V., Koshland D. 2008. The kleisin subunit of cohesin dictates damage-induced cohesion. Mol. Cell 31:47–56 [DOI] [PubMed] [Google Scholar]
- 20. Huang J., Hsu J. M., Laurent B. C. 2004. The RSC nucleosome-remodeling complex is required for cohesin's association with chromosome arms. Mol. Cell 13:739–750 [DOI] [PubMed] [Google Scholar]
- 21. Ira G., Satory D., Haber J. E. 2006. Conservative inheritance of newly synthesized DNA in double-strand break-induced gene conversion. Mol. Cell. Biol. 26:9424–9429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jablonovich Z., Liefshitz B., Steinlauf R., Kupiec M. 1999. Characterization of the role played by the RAD59 gene of Saccharomyces cerevisiae in ectopic recombination. Curr. Genet. 36:13–20 [DOI] [PubMed] [Google Scholar]
- 23. Jaskelioff M., Van Komen S., Krebs J. E., Sung P., Peterson C. L. 2003. Rad54p is a chromatin remodeling enzyme required for heteroduplex DNA joint formation with chromatin. J. Biol. Chem. 278:9212–9218 [DOI] [PubMed] [Google Scholar]
- 24. Kaye J. A., et al. 2004. DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr. Biol. 14:2096–2106 [DOI] [PubMed] [Google Scholar]
- 25. Kent N. A., Chambers A. L., Downs J. A. 2007. Dual chromatin remodeling roles for RSC during DNA double-strand break induction and repair at the yeast MAT locus. J. Biol. Chem. 282:27693–27701 [DOI] [PubMed] [Google Scholar]
- 26. Kim J. S., Krasieva T. B., LaMorte V., Taylor A. M., Yokomori K. 2002. Specific recruitment of human cohesin to laser-induced DNA damage. J. Biol. Chem. 277:45149–45153 [DOI] [PubMed] [Google Scholar]
- 27. Lanzuolo C., et al. 2001. The HTL1 gene (YCR020W-b) of Saccharomyces cerevisiae is necessary for growth at 37 degrees C, and for the conservation of chromosome stability and fertility. Yeast 18:1317–1330 [DOI] [PubMed] [Google Scholar]
- 28. Lea D. E., Coulson C. A. 1949. The distribution in the numbers of mutants in bacterial populations. J. Genet. 49:264–285 [DOI] [PubMed] [Google Scholar]
- 29. Lee K., Zhang Y., Lee S. E. 2008. Saccharomyces cerevisiae ATM orthologue suppresses break-induced chromosome translocations. Nature 454:543–546 [DOI] [PubMed] [Google Scholar]
- 29a. Lee S. E., et al. 1998. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94:399–409 [DOI] [PubMed] [Google Scholar]
- 30. Li F., et al. 2008. Microarray-based genetic screen defines SAW1, a gene required for Rad1/Rad10-dependent processing of recombination intermediates. Mol. Cell 30:325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liang B., Qiu J., Ratnakumar K., Laurent B. C. 2007. RSC functions as an early double-strand-break sensor in the cell's response to DNA damage. Curr. Biol. 17:1432–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Llorente B., Smith C. E., Symington L. S. 2008. Break-induced replication: what is it and what is it for? Cell Cycle 7:859–864 [DOI] [PubMed] [Google Scholar]
- 33. Lorch Y., Zhang M., Kornberg R. D. 1999. Histone octamer transfer by a chromatin-remodeling complex. Cell 96:389–392 [DOI] [PubMed] [Google Scholar]
- 34. Lu Y. M., et al. 2003. Dissecting the pet18 mutation in Saccharomyces cerevisiae: HTL1 encodes a 7-kDa polypeptide that interacts with components of the RSC complex. Mol. Genet. Genomics 269:321–330 [DOI] [PubMed] [Google Scholar]
- 35. Lyndaker A. M., Alani E. 2009. A tale of tails: insights into the coordination of 3′ end processing during homologous recombination. Bioessays 31:315–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McEachern M. J., Haber J. E. 2006. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 75:111–135 [DOI] [PubMed] [Google Scholar]
- 37. Mimitou E. P., Symington L. S. 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455:770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Neely K. E., Workman J. L. 2002. The complexity of chromatin remodeling and its links to cancer. Biochim. Biophys. Acta 1603:19–29 [DOI] [PubMed] [Google Scholar]
- 39. New J. H., Sugiyama T., Zaitseva E., Kowalczykowski S. C. 1998. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391:407–410 [DOI] [PubMed] [Google Scholar]
- 40. Osley M. A., Tsukuda T., Nickoloff J. A. 2007. ATP-dependent chromatin remodeling factors and DNA damage repair. Mutat. Res. 618:65–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pabla R., Pawar V., Zhang H., Siede W. 2006. Characterization of checkpoint responses to DNA damage in Saccharomyces cerevisiae: basic protocols. Methods Enzymol. 409:101–117 [DOI] [PubMed] [Google Scholar]
- 42. Palmbos P. L., Daley J. M., Wilson T. E. 2005. Mutations of the Yku80 C terminus and Xrs2 FHA domain specifically block yeast nonhomologous end joining. Mol. Cell. Biol. 25:10782–10790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pâques F., Haber J. E. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Petukhova G., Stratton S. A., Sung P. 1999. Single strand DNA binding and annealing activities in the yeast recombination factor Rad59. J. Biol. Chem. 274:33839–33842 [DOI] [PubMed] [Google Scholar]
- 45. Romeo M. J., et al. 2002. HTL1 encodes a novel factor that interacts with the RSC chromatin remodeling complex in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:8165–8174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. San Filippo J., Sung P., Klein H. 2008. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77:229–257 [DOI] [PubMed] [Google Scholar]
- 47. Seong C., et al. 2008. Molecular anatomy of the recombination mediator function of Saccharomyces cerevisiae Rad52. J. Biol. Chem. 283:12166–12174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shim E. Y., et al. 2007. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol. Cell. Biol. 27:1602–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shim E. Y., Ma J. L., Oum J. H., Yanez Y., Lee S. E. 2005. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol. Cell. Biol. 25:3934–3944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shinohara A., Ogawa T. 1998. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature 391:404–407 [DOI] [PubMed] [Google Scholar]
- 51. Smith C. L., Peterson C. L. 2005. A conserved Swi2/Snf2 ATPase motif couples ATP hydrolysis to chromatin remodeling. Mol. Cell. Biol. 25:5880–5892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Smith J., Rothstein R. 1999. An allele of RFA1 suppresses RAD52-dependent double-strand break repair in Saccharomyces cerevisiae. Genetics 151:447–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ström L., et al. 2007. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science 317:242–245 [DOI] [PubMed] [Google Scholar]
- 54. Ström L., Lindroos H. B., Shirahige K., Sjogren C. 2004. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell 16:1003–1015 [DOI] [PubMed] [Google Scholar]
- 55. Sugawara N., Goldfarb T., Studamire B., Alani E., Haber J. E. 2004. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc. Natl. Acad. Sci. U. S. A. 101:9315–9320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sugawara N., Haber J. E. 1992. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol. Cell. Biol. 12:563–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sugawara N., Ira G., Haber J. E. 2000. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 20:5300–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sung P. 1997. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 272:28194–28197 [DOI] [PubMed] [Google Scholar]
- 59. Sung P., Stratton S. A. 1996. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J. Biol. Chem. 271:27983–27986 [DOI] [PubMed] [Google Scholar]
- 60. Symington L. S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66:630–670, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Treich I., Ho L., Carlson M. 1998. Direct interaction between Rsc6 and Rsc8/Swh3, two proteins that are conserved in SWI/SNF-related complexes. Nucleic Acids Res. 26:3739–3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Unal E., et al. 2004. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16:991–1002 [DOI] [PubMed] [Google Scholar]
- 63. Unal E., Heidinger-Pauli J. M., Koshland D. 2007. DNA double-strand breaks trigger genome-wide sister chromatid cohesion through EcoI (Ctf7). Science 317:245–248 [DOI] [PubMed] [Google Scholar]
- 64. van Attikum H., Gasser S. M. 2005. ATP-dependent chromatin remodeling and DNA double-strand break repair. Cell Cycle 4:1011–1014 [DOI] [PubMed] [Google Scholar]
- 65. van Attikum H., Gasser S. M. 2005. The histone code at DNA breaks: a guide to repair? Nat. Rev. Mol. Cell Biol. 6:757–765 [DOI] [PubMed] [Google Scholar]
- 66. Verstrepen K. J., Jansen A., Lewitter F., Fink G. R. 2005. Intragenic tandem repeats generate functional variability. Nat. Genet. 37:986–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Williams R. S., et al. 2009. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell 139:87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wilson B., Erdjument-Bromage H., Tempst P., Cairns B. R. 2006. The RSC chromatin remodeling complex bears an essential fungal-specific protein module with broad functional roles. Genetics 172:795–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wittmeyer J., Saha A., Cairns B. 2004. DNA translocation and nucleosome remodeling assays by the RSC chromatin remodeling complex. Methods Enzymol. 377:322–343 [DOI] [PubMed] [Google Scholar]
- 70. Wong L. Y., Recht J., Laurent B. C. 2006. Chromatin remodeling and repair of DNA double-strand breaks. J. Mol. Histol. 37:261–269 [DOI] [PubMed] [Google Scholar]
- 71. Wu Y., Sugiyama T., Kowalczykowski S. C. 2006. DNA annealing mediated by Rad52 and Rad59 proteins. J. Biol. Chem. 281:15441–15449 [DOI] [PubMed] [Google Scholar]
- 72. Zhang Y., et al. 2007. Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat. Struct. Mol. Biol. 14:639–646 [DOI] [PubMed] [Google Scholar]
- 73. Zhu Z., Chung W. H., Shim E. Y., Lee S. E., Ira G. 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134:981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]