Abstract
The hypoxia-inducible factor (HIF) is a key regulator of the transcriptional response to hypoxia. While the mechanism underpinning HIF activation is well understood, little is known about its resolution. Both the protein and the mRNA levels of HIF-1α (but not HIF-2α) were decreased in intestinal epithelial cells exposed to prolonged hypoxia. Coincident with this, microRNA (miRNA) array analysis revealed multiple hypoxia-inducible miRNAs. Among these was miRNA-155 (miR-155), which is predicted to target HIF-1α mRNA. We confirmed the hypoxic upregulation of miR-155 in cultured cells and intestinal tissue from mice exposed to hypoxia. Furthermore, a role for HIF-1α in the induction of miR-155 in hypoxia was suggested by the identification of hypoxia response elements in the miR-155 promoter and confirmed experimentally. Application of miR-155 decreased the HIF-1α mRNA, protein, and transcriptional activity in hypoxia, and neutralization of endogenous miR-155 reversed the resolution of HIF-1α stabilization and activity. Based on these data and a mathematical model of HIF-1α suppression by miR-155, we propose that miR-155 induction contributes to an isoform-specific negative-feedback loop for the resolution of HIF-1α activity in cells exposed to prolonged hypoxia, leading to oscillatory behavior of HIF-1α-dependent transcription.
INTRODUCTION
Tissue hypoxia is a common feature in a range of physiologic and pathophysiologic states, including exercise, development, cancer, and chronic inflammation. The hypoxia-inducible factor (HIF) is a ubiquitous hypoxia-responsive transcription factor that regulates the expression of a range of genes that promote adaptation to hypoxia (32, 57). The mechanism by which HIF is stabilized in hypoxia is well understood and is due to reduced activity of a family of oxygen-dependent HIF-hydroxylases that target HIFα subunits for degradation and block transactivation in normoxia (5). Several studies (including the present one) have shown that the upregulation of HIF-1α that occurs in response to hypoxia is transient and involves a resolution phase even while the cells are maintained in hypoxia (23, 26, 59). However, the mechanism(s) underpinning the resolution of HIF-1α during prolonged hypoxia remains incompletely understood. Negative-feedback mechanisms involving HIF-dependent upregulation of PHD2 and PHD3 have been identified (5, 26, 47, 59). In the present study, we aimed to expand our understanding of how the HIF response is resolved in prolonged hypoxia by investigating a possible role for hypoxia-induced microRNAs (miRNAs).
miRNAs are endogenous small RNA molecules of approximately 22 nucleotides that regulate gene expression by destabilizing mRNA or repressing translation (4, 25). Approximately one-third of all genes in mammals have been predicted to be regulated by miRNAs (43, 71), and the development of our understanding of the role of miRNAs in various species is ongoing (39, 40). Individual miRNAs may target hundreds of distinct mRNAs. miRNA target prediction algorithms, including miRanda, TargetScan, and PicTar, can be used to predict theoretical targets for specific miRNAs; however, biological confirmation of these targets is required to confirm bona fide targets (24, 36, 44). The regulation of protein expression by miRNAs impacts upon all physiological processes examined thus far, including hematopoiesis, development, cell proliferation, apoptosis, immunity, and metabolism (3, 69). Furthermore, altered expression of specific miRNAs is often associated with the interrelated pathologies of chronic inflammation and tumor development (18, 31, 46, 72). Importantly, such pathophysiologic events often feature microenvironmental hypoxia due to decreased tissue perfusion and/or increased oxygen consumption (14).
Recent work has demonstrated altered global miRNA expression in response to hypoxia (22, 38). While a limited number of individual miRNAs (e.g., miRNA-210 [miR-210]) are regulated by hypoxia in most models tested, many other miRNAs appear to be regulated by hypoxia in a cell type/tissue-specific manner (29, 37). The functions of hypoxia-induced miRNAs such as miR-210 include regulation of apoptosis, proliferation, cell cycle, DNA repair, cell migration, and mitochondrial function (7, 13, 15, 17, 19, 29, 34, 37, 52). However, our understanding of the function of alternative hypoxia-induced miRNAs at the cellular level remains limited. In the present study, we provide evidence that HIF-1α is a direct target of miR-155, a hypoxia-inducible miRNA, in intestinal epithelial cells. HIF-1α-dependent signaling is reduced by miR-155. Furthermore, HIF-1α is the transcription factor primarily responsible for miR-155 induction in hypoxia. We hypothesize, based on these data and a mathematical model of miR-155 regulation of HIF-1α mRNA, that miR-155 is a component of the network of negative-feedback loops responsible for the resolution of HIF-1α-dependent transcription in prolonged hypoxia.
MATERIALS AND METHODS
Cell culture and hypoxia.
Caco-2 cells and murine embryonic fibroblasts (MEFs) were maintained in Dulbecco modified Eagle medium containing 4.5 g of glucose/liter, 10% fetal calf serum (FCS), 2 mM l-glutamine, and 10 U/ml of penicillin-streptomycin. HeLa and HepG2 cells were maintained in minimum essential medium containing 10% FCS, 2 mM l-glutamine, nonessential amino acids, and 10 U of penicillin-streptomycin/ml. Medium for stable transfected HepG2 cells additionally contained 2 μg of puromycin dihydrochloride/ml. Caco-2, MEF, HepG2, and HeLa cells were placed in a hypoxia chamber (Coy Laboratories, Grass Lake, MI) with a normobaric atmosphere of humidified, ambient hypoxia of 1%O2, 5%CO2, and a balance of N2. Temperature was maintained at 37°C, and instantaneous hypoxia was achieved by feeding cells in the hypoxia chamber immediately with preconditioned medium.
Microarray analysis.
Caco-2 cells were exposed to 1%O2 for 0, 8, 24, or 48 h. Total RNA was purified using an miRNeasy minikit (Qiagen) according to the manufacturer's protocol. The integrity of total RNA was analyzed by agarose gel electrophoresis and by RNA 6000 Nano assay using a 2100 Bioanalyzer (Agilent Technologies). miRNA array analysis was carried out on 20 μg of each sample (LC Sciences). The μParaflo microfluidics chip allowed the analysis of 856 known human miRNAs in each sample using RNA labeled with cyanin 3 (Cy3) and Cy5 dyes prior to hybridization. Significant changes in miRNA expression were identified using an analysis of variance (ANOVA) t test (P < 0.01).
Immunoblot analysis.
Cells were washed with phosphate-buffered saline and lysed on ice in whole-cell lysis buffer (150 mM sodium chloride, 25 mM Tris [pH 8.0], 1 mM EDTA, 1% [vol/vol] NP-40) containing protease inhibitor cocktail (Sigma, St. Louis, MO). Cytosolic and nuclear extracts were prepared as described previously (16). After normalization (DC protein assay; Bio-Rad), samples (15 μg/lane) were separated by SDS-PAGE and transferred to nitrocellulose. For immunoblotting, blots were incubated with anti-human HIF-1α (BD Biosciences), anti-HIF-2α (Novus Biosciences), TATA box binding protein (TBP; Abcam), α-tubulin (Cedarlane), and anti-β-actin (Sigma) for 8 h at 4°C on a rocking platform. Blots were then washed and species matched secondary horseradish peroxidase-conjugated antibodies were added for 1 h. Bands were detected by using ECL reagent (Santa Cruz Biotechnology).
Transient transfections and luciferase reporter assay.
Caco-2 cells were transfected with 20 to 80 nM mature miRNAs, anti-miRs (Ambion), HIF-1α siRNA (30 nM), HIF-2α siRNA (100 nM), or RelA/p65 siRNA (40 nM) (ON-TARGETplus SMARTpool; Dharmacon) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. An anti-miR negative control and nontarget small interfering (siRNA) were used as controls where appropriate. At 24 h after transfection, the medium was replaced, and hypoxia exposure was started 48 h after transfection. For luciferase experiments 24 h after siRNA/miRNA transfection, the cells were transfected with either an HRE-luciferase reporter construct (Promega) or pmiR-report construct (Applied Biosystems) (cloning was performed according the manufacturer's protocol) using FuGene 6 transfection reagent (Roche Applied Science). A pSV-β-Gal vector (Promega) was cotransfected for normalization. For transient overexpression of PHD2 or PHD3, 0.5 μg of a V5-tag construct was transfected into Caco-2 cells using FuGene 6, and pcDNA 3.1 empty vector was used as negative control. At 24 h after transfection, the medium was replaced, and the cells were exposed to hypoxia for the indicated experimental conditions. The cells were lysed in 1× reporter lysis buffer (Promega). Luciferin/ATP substrate (Promega) was added to cell lysate, and the luciferase activity was detected in a luminometer (Berthold Technologies). All samples were run in duplicate and normalized to cotransfected β-galactosidase (β-Gal). A secreted luciferase assay was carried out according to the manufacturer's protocol (Biolux Gaussia luciferase flex assay kit; NEB) and normalized to the cell number.
miRNA real-time PCR.
Total RNA was purified from cells or tissue by using a miRNeasy minikit (Qiagen) according to the manufacturer's protocol. The RNA concentration was measured by using a NanoDrop apparatus (NanoDrop Technology, Inc.) and normalized, and the RNA integrity was determined by agarose gel electrophoresis. Reverse transcription was carried out using a TaqMan MicroRNA reverse transcription kit (Applied Biosystems) according to the manufacturer's protocol. Hsa-miR-155, mmu-miR-155, pri-miR-155, and hsa-miR-215 were detected using TaqMan MicroRNA assays. Real-time quantification of cDNA was carried out on an ABI Prism 7900HT sequence detection system (Applied Biosystems) and normalized to 18S rRNA.
mRNA real-time PCR.
Total RNA was purified using the TRIzol (Invitrogen) method, and reverse transcription was carried out using Superscript II (Invitrogen). Real time quantification of cDNA was carried out on the ABI Prism 7900HT sequence detection system and normalized to 18S rRNA. For HIF-1α, HIF-2α, and PHD-2, the following primers were used: HIF-1α fwd 5′-ACCATGCCCCAGATTCAGG-3′; HIF-1α rev, 5′-AGTGCTTCCATCGGAAGGACT-3′; HIF-2α fwd, 5′-CTACGCCACCCAGTACCAGG-3′; HIF-2α rev, 5′-GACACCTTGTGGGCTGACG-3′; PHD-2 fwd, 5′-ACATAACCCGTTCCATTGCC-3′; and PHD-2 rev, 5′-ATCAATGGCCGGACGAAAG-3′.
Animal studies.
C57BL/6 mice were exposed to 10% O2 for the indicated times, and the tissue was snap-frozen and stored at −80°C. All experiments were carried out with prior ethical approval from the University College Dublin Animal Research Ethics Committee.
Bioinformatic analysis.
The miRanda algorithm (www.microRNA.org) was used to identify predicted potential miRNA targets. For promoter analysis studies, MatInspector software (Genomatix) was used.
Mathematical model of the HIF pathway.
We developed a computational model, based on differential equations, to study the role of miR-155 and PHD2/3 in downregulating HIF-1α mRNA and, consequently, the HIF-1α protein levels. Numerical simulations and parameter estimation have been performed using Matlab.
Statistical analysis.
All data are presented as mean ± the SEM of the indicated n independent experiments. Statistical significance was assessed using an ANOVA t test for the array data and the unpaired two-tailed Student t test for all other experiments. The significance is indicated in the figures (*, P < 0.05; **, P < 0.01.
RESULTS
Transient activation of HIF-1α in hypoxia.
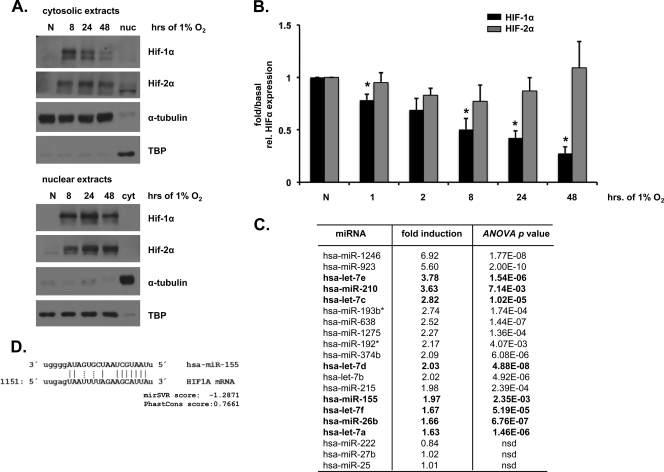
Caco-2 cells exposed to hypoxia (0 to 48 h) demonstrated differential HIFα subunit expression patterns. Although both nuclear and cytoplasmic HIF-1α protein levels were transiently increased, HIF-2α protein levels were increased in a sustained manner (Fig. 1A). The HIF-1α mRNA levels were also reduced at later time points of hypoxic exposure, whereas the HIF-2α mRNA levels were unchanged (Fig. 1B). These data led us to investigate the presence of a specific resolving mechanism for HIF-1α in prolonged hypoxia mediated through decreased expression of HIF-1α mRNA.
Fig. 1.
Differential expression of HIFα subunits in hypoxia and potential role for hypoxia-induced miRNAs. (A) Caco-2 colonic epithelial cells exposed to 0 to 48 h of hypoxia (1% O2) demonstrate a transient increase in cytosolic and nuclear HIF-1α and a sustained increase in HIF-2α protein levels. α-Tubulin and TATA-box binding protein (TBP) were used as loading controls. (B) Real-time PCR analysis normalized to the corresponding normoxic control was used to determine HIF-1α and HIF-2 α mRNA levels in Caco-2 cells exposed to hypoxia. (C) miRNA array analysis identified a cohort of hypoxia-induced miRNAs in Caco-2 cells. Those miRNAs previously identified as hypoxia-inducible are outlined in bold. The last three miRNAs are included as sample controls that were not induced in hypoxia. (D) HIF-1α is a predicted target of miR-155. Watson-Crick base pairing is indicated by solid lines, while non-Watson-Crick base pairing is indicated by dashed lines. In all cases, n = 3 to 4 independent experiments. The data are shown as representative blots or means ± the standard errors of the mean (SEM) (*, P < 0.05; nsd, no significant difference).
Hypoxia induces the expression of miR-155.
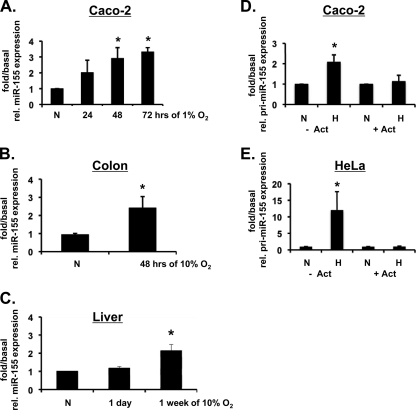
miRNA array analysis of Caco-2 cells exposed to hypoxia revealed ca. 12% of miRNAs studied to demonstrate significant and consistent altered expression levels in response to hypoxia. In Fig. 1C, we list the cohort of the most significantly upregulated miRNAs, which included the previously characterized hypoxia-dependent miRNA, miR-210 (38). miRNAs previously reported to be hypoxia inducible are indicated in Fig. 1C in boldface. Three control miRNAs with unchanged expression levels in hypoxia are also included (miR-222, miR-27b, and miR-25). We next used the bioinformatic algorithms miRanda and PicTar (6, 12, 36) to identify the predicted targets of the hypoxia-upregulated miRNAs. HIF-1α was identified as a potential target for just one of the hypoxia-induced miRNAs, miR-155 (Fig. 1D). The alignment scores represent a weighted summary of matches and mismatches for individual base pairs and gaps. miR-155 was not predicted to target HIF-2α mRNA since it does not contain a miR-155 binding site in its 3′ untranslated region (3′UTR). miR-215, which was increased in hypoxia to approximately the same degree as miR-155 but was not predicted to target HIF-1α mRNA, was selected for use as a negative control miRNA for subsequent experiments. Real-time PCR was used to validate the array data and confirm the upregulation of mature miR-155 in Caco-2 cells (Fig. 2A). Hypoxia also induced miR-155 in HeLa cells, another transformed epithelial cell line (data not shown), but not in primary MEFs (data not shown). In order to determine whether hypoxic induction of miR-155 occurs in vivo, we determined mature miR-155 levels in tissues from mice exposed to atmospheric hypoxia. Hypoxia upregulated miR-155 expression levels in the colon (Fig. 2B) and liver (Fig. 2C) but not in the small intestine (data not shown), indicating a tissue-specific response. To determine whether the miR-155 upregulation in hypoxia is due to increased transcription, the expression of the primary transcript pri-miR-155 was measured in Caco-2 and HeLa cells. The pri-miR-155 transcript was induced in response to hypoxia, and this effect was diminished when cells were treated with the inhibitor of transcription actinomycin D (Fig. 2D and E). Together, these results implicate miR-155 to be a cell type-specific and tissue-specific hypoxia-induced miRNA and that the induction of miR-155 occurs at least in part at the level of transcriptional upregulation.
Fig. 2.
Transcriptional upregulation of miR-155 in response to hypoxia. (A) Real-time PCR analysis demonstrates mature miR-155 expression in Caco-2 cells exposed to hypoxia. (B and C) Mir-155 expression levels from colon and liver tissue, respectively, harvested from mice exposed to 10% atmospheric O2 for the indicated periods. (D and E) Caco-2 and HeLa cells were treated with (or without) actinomycin D and exposed to 48 h of hypoxia. Real-time PCR analysis was used to measure primary transcript pri-miR-155 expression in response to hypoxia. All data are shown as the fold over basal miR-155 or pri-miR-155 expression, with 18S rRNA being used as an endogenous control. n = 3 independent experiments for in vitro experiments. n = 6 independent mice were used for in vivo measurements. Values are presented as means ± the SEM (*, P < 0.05).
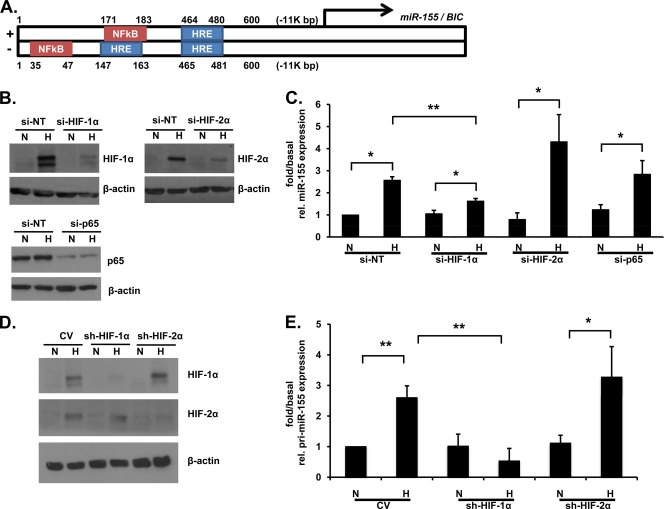
HIF-1α contributes to hypoxic miR-155 induction.
We next investigated the potential molecular regulators of miRNA-155 expression in hypoxia. Using bioinformatic analysis (MatInspector), three HIF response elements (HREs) and two NF-κB consensus motifs (NREs) were identified in the 600-bp promoter region of miR-155 that is ∼11 kbp upstream from the actual miR-155 gene BIC on chromosome 21 (Fig. 3A). Protein knockdown with all siRNA treatments was confirmed by immunoblotting (Fig. 3B). Caco-2 cells transfected with siRNA against HIF-1α but not HIF-2α or the RelA/p65 subunit of NF-κB demonstrate significantly decreased induction of miR-155 expression in response to hypoxia compared to cells pretreated with nontarget siRNA (Fig. 3C). The incomplete knockdown of HIF-1α with siRNA likely explains the residual miR-155 induction in hypoxia seen in Fig. 3C. In the hepatocellular carcinoma cell line HepG2 stably expressing short hairpin RNA (shRNA) targeted against HIF-1α or HIF-2α, HIF-1α dependence for hypoxic pri-miR-155 induction was also observed (Fig. 3D and E). These data implicate HIF-1α as the transcription factor primarily responsible for the hypoxic induction of miR-155.
Fig. 3.
HIF-1-dependent miR-155 expression in hypoxia. (A) Bioinformatic analysis using the Genomatix software tool MatInspector reveals the presence of three HRE consensus motifs and two NRE consensus motifs in the promoter of the miR-155 gene. (B) Effective siRNA-dependent knockdown of HIF-1α, HIF-2α, and p65 was confirmed by immunoblot analysis. (C) Caco-2 cells were transfected with HIF-1α, HIF-2α, or p65-siRNA prior to hypoxic exposure (48 h), and mature miR-155 expression was analyzed by real-time PCR. (D) Protein levels of HIFα subunits in HIF-1α or HIF-2α knockdown HepG2 cells were analyzed by immunoblotting. (E) HepG2 cells stably expressing shRNA directed against HIF-1α or HIF-2α were exposed to hypoxia, and pri-miR-155 expression was measured by using real-time PCR. n = four independent experiments throughout. Values are presented as means ± the SEM (*, P < 0.05; **, P < 0.01).
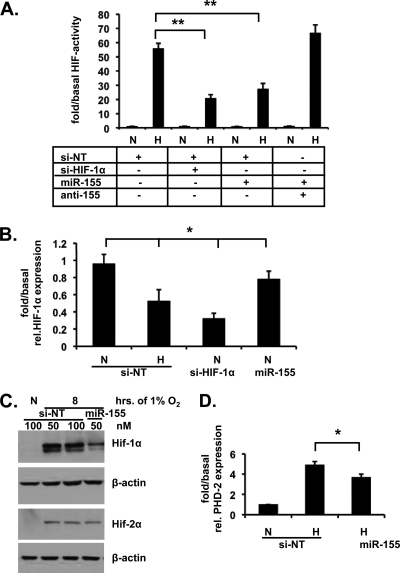
miR-155 reduces HIF signaling.
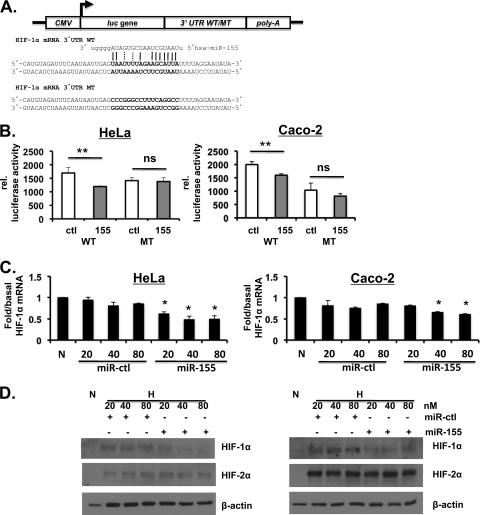
We next investigated whether HIF-1α was a bona fide functional target for miR-155. Using a HRE-luciferase reporter assay, we found that exogenously applied mature miR-155 reduced hypoxia-induced HIF activity in a manner that could be reversed by cotreatment with neutralizing anti-miR-155 (Fig. 4A). In contrast, miR-215, also induced in Caco-2 cells exposed to hypoxia, was without effect on basal or hypoxia-induced HIF activity (data not shown). Transfection with exogenous miRNA-155 induced a mild but significant reduction in HIF-1α mRNA levels in normoxic cells (Fig. 4B). Transfection of miR-155 also resulted in an inhibition of hypoxia-induced HIF-1α (but not HIF-2α) protein expression (Fig. 4C). Furthermore, real-time PCR analysis of hypoxia-induced expression of prolyl hydroxylase 2 (PHD2), a downstream target gene of HIF-1α, also showed a significant reduction in cells treated with miR-155 (Fig. 4D). Using a miRNA expression reporter vector system (pmiR-report) with wild-type and mutant versions of the predicted miR-155 binding site from the HIF-1α 3′UTR inserted into a firefly luciferase construct showed HIF-1α to be a direct target of miR-155 (Fig. 5A and B). Furthermore, increasing concentrations of miR-155 reduce HIF-1α mRNA and protein specifically in a dose-response manner (Fig. 5C and D). Taken together, these data indicate that miR-155 specifically and selectively targets HIF-1α over HIF-2α through direct targeting of HIF-1α (but not HIF-2α) mRNA.
Fig. 4.
miR-155 reduces HIF signaling. (A) Caco-2 cells cotransfected with HRE-luciferase and either control siRNA (si-NT), HIF-1α siRNA (si-HIF-1α), or miR-155 with or without anti-miR-155 and exposed to 6 h of 1% O2. A luciferase assay was carried out, and the values were normalized to cotransfected β-Gal. (B) Real-time PCR analysis of HIF-1α mRNA levels in Caco-2 cells in normoxia following transfection with miR-155. HIF-1α siRNA was used as positive control. (C) Immunoblot analysis reveals HIF-1α and HIF-2α protein levels after transfection with miR-155 for 48 h prior to hypoxic exposure for 8 h. (D) Caco-2 cells were transfected with miR-155, and hypoxia-induced PHD2 mRNA expression was determined by real-time PCR analysis. In all cases n = 3 to 4 independent experiments. The data are shown as representative blots or as means ± the SEM (*, P < 0.05; **, P < 0.01).
Fig. 5.
HIF-1α is a direct target of miR-155. (A) Wild-type (WT) and mutant (MT) versions of the predicted miR-155 binding site in the 3′UTR of HIF-1α mRNA were cloned into a luciferase vector. (B) Caco-2 and HeLa cells were transfected with miR-155 (155) or miR-control (ctl) 24 h prior to transfection with the reporter luciferase construct. After 24 h, a luciferase assay was carried out and normalized to the cotransfected β-Gal. (C and D) HeLa and Caco-2 cells were transfected with increasing doses of either miR-155 or miR-control (miR-ctl), and the HIFα protein or mRNA levels were measured. For the HIF-1α mRNA statistical data, comparisons were made to the corresponding concentration of the control RNA. n = 3 to 4 independent experiments throughout. The data shown are representative blots or means ± the SEM (*, P < 0.05; **, P < 0.01).
Inhibition of endogenous miR-155 induces sustained HIF-1α activation in hypoxia.
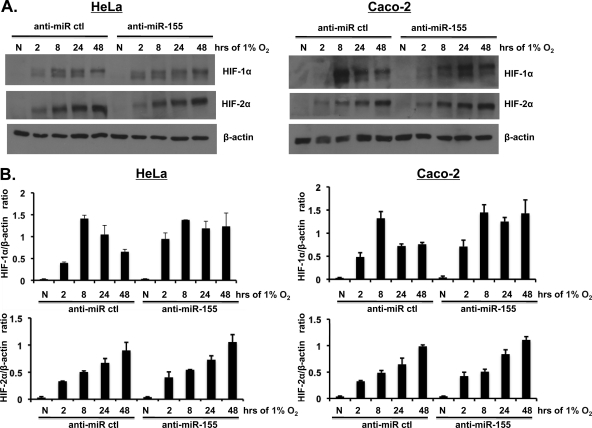
To investigate whether inhibition of miR-155 alters the transient nature of HIF-1α activation in hypoxia, cells were treated with anti-miR-155 prior to hypoxic exposure. As described earlier, Caco-2 cells exposed to hypoxia demonstrated a transient increase in whole-cell HIF-1α protein and a sustained increase of HIF-2α protein levels (Fig. 6) in both Caco-2 and HeLa cells. However, cells pretreated with anti-miR-155 demonstrated sustained increase in HIF-1α protein levels compared to the transient HIF-1α activation in control anti-miR treatment (Fig. 6). Total HIF-2α protein levels over time in hypoxia were unchanged between all treatments (Fig. 6). These data indicate a role for hypoxia-induced miR-155 in the resolution of hypoxia-induced HIF-1α (but not HIF-2α) activation in prolonged periods of hypoxia.
Fig. 6.
Inhibition of miR-155 sustains HIF-1α activation in hypoxia. (A) Caco-2 and HeLa cells were transfected with nonspecific anti-miR control or anti-miR-155 oligonucleotides 24 h prior to hypoxic exposure over time (0 to 48 h). The HIF-1α and HIF-2α levels were analyzed in whole-cell protein extracts by immunoblotting. (B) HIFα/β-actin ratios of densitometric analysis are shown from n = 3 independent experiments in each case.
Mathematical model of HIF-1α regulation by miR-155 and PHD2/3 negative-feedback loops.
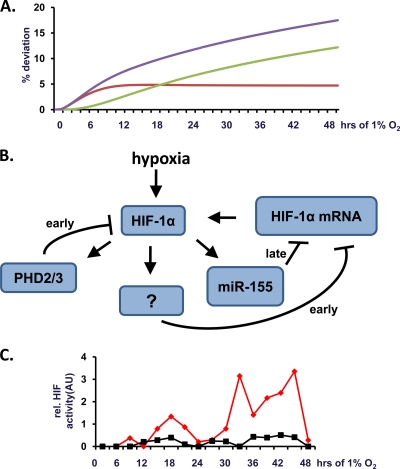
A predictive computational model (based on differential equations) was developed to study the relative roles of miR-155 and PHD2/3 in downregulating HIF-1α mRNA and, consequently, reducing the HIF-1α protein level. In order for this model to accurately predict HIF-1α activity in prolonged hypoxia, it was necessary to include a direct or indirect negative autoregulation of HIF-1α mRNA when the HIF-1α protein increases above a threshold value. Although this type of regulation has not yet been reported for HIF in the literature, such negative autoregulation has been proposed to be a general control motif in gene transcription networks and has a compensatory role allowing for a faster response to changes in transcription factor activity (1). Using this model, in Fig. 7A the difference from the x axis reflects deviation from the normal temporal HIF-1α activation profile. Removal of the PHD-dependent feedback loop from the model leads to predictive deviation in the early but not in the late regulation of HIF-1α response to hypoxia (red). In contrast, removal of the miR-155-dependent negative-feedback loop leads to late but not early deviation from the normal HIF-1α response to hypoxia (green). Removal of both feedback loops leads to both early and late deviation (blue). In summary, using this predictive model, we estimate that PHD2/3 and miR-155 feedback loops play temporally distinct roles in the regulation of HIF-1α activity in prolonged hypoxia (Fig. 7B). Interestingly, on longer timescales (>50 h) the computational model predicts that the establishment of periodic oscillations in HIF-1α protein concentration is a consequence of the negative-feedback loop through miR-155. Such oscillations have been proposed to be important in selective target gene activation for other transcription factors such as NF-κB (2, 51, 60, 63). In order to test the prediction of oscillatory behavior of HIF, we generated a vector containing a secreted form of luciferase (derived from Gaussia princeps) under the control of an HRE promoter. This construct allows high-resolution, temporal measurement of HIF transcriptional activity in response to hypoxia. In a time course experiment, we presented the first evidence that HIF activity is increasing in an oscillatory and not sustained fashion in response to prolonged hypoxia (Fig. 7C).
Fig. 7.
PHD2/3 and miR-155 negative-feedback loops are part of the regulatory network of HIF-1α in prolonged hypoxia. A computational model based on experimental data was generated to estimate the relative contributions of miR-155 and PHD2/3 in determining temporal HIF-1α activity during prolonged periods of hypoxia. (A) Temporal deviation from normal (transient) HIF-1α activity predicted by removal of the PHD2/3 feedback loop, the miR-155 feedback loop, or both feedback loops from the model are shown in red, green, and blue, respectively. (B) Schematic representation of the proposed network of known and predicted negative-feedback loops in the HIF-1a pathway. The “?” indicates currently unidentified direct or indirect negative-feedback mechanism(s). (C) Caco-2 cells were transfected with a vector containing an HRE promoter controlling expression of a secreted form of luciferase derived from Gaussia princeps and cultured under normoxia (21% O2; red) or hypoxia (1% O2; black). The medium was sampled every 3 h, and the HRE-luciferase activity assessed for each time period and normalized to the cell number. Representative traces are shown for HIF activity in arbitrary units (AU).
Multiple negative-feedback loops in the HIF pathway.
A number of previous studies have demonstrated a role for increased expression/activity of PHD2 and PHD3 in prolonged hypoxia in the resolution of HIF signaling. In agreement with this, we found that the overexpression of either PHD2 or PHD3 was sufficient to decrease hypoxia-induced HIF activity (data not shown). However, since PHDs suppress the level of the HIF-1α protein stability (and do not suppress HIF-1α mRNA expression), this pathway does not fully account for the negative feedback we observed in the present study. In summary, miR-155, an miRNA induced in hypoxia in a HIF-1α-dependent manner, represents a component of the negative-feedback loop network (Fig. 7B) responsible for the resolution of selectively HIF-1α activation in prolonged hypoxia, and this negative feedback is effective at the level of the inhibition of translation.
DISCUSSION
HIF-1α is a key transcriptional regulator of the cellular response to hypoxia in all metazoans (57). The stabilization and transactivation of HIF-1α during hypoxia is primarily regulated by altered HIF-1α hydroxylation (32). Less is known about the processes involved in the resolution of HIF-1α activity. A number of studies have demonstrated that the resolution of HIF-1α observed in prolonged hypoxia is due to increased activity and expression of PHD2 and PHD3 (23, 26, 47, 59). Because PHD2 and PHD3 are themselves HIF-dependent genes and retain some activity even under hypoxic conditions, this represents an effective negative-feedback loop for HIF signaling that prevents excessive activation of the HIF pathway that could prove detrimental to cells and tissues. A further negative-feedback loop for HIF signaling in hypoxia that involves components of the E3 ubiquitin ligase has also been proposed (62). In the present study, we have found that intestinal epithelial cells exposed to prolonged hypoxia demonstrate a transient and resolving HIF-1α (but not HIF-2α) response and that the resolution of this response is at least in part due to decreased expression of HIF-1α mRNA. Because this response is specific for HIF-1α and is associated with decreased mRNA levels, it cannot be fully accounted for by altered PHD2/3 expression or HIF-1α protein degradation. Thus, we hypothesized the existence of a further negative-feedback loop in the HIF pathway involving decreased translation of the HIF-1α gene.
miRNAs are key regulators of gene expression that act at the posttranscriptional level (20, 70). Multiple studies have used microarray analysis to investigate a hypoxia-inducible microRNA signature in various model systems. Although it is clear that hypoxia regulates the expression of miRNAs, strong evidence for biological targets and functions remain to be confirmed. To date, miR-210 is the miRNA most convincingly shown to be hypoxia induced in a manner independent of tissue or cell line used (37, 38). Notably, while HIF can increase the expression of miRNAs, it can also be an miRNA target. For example, various reports have shown that HIF is a target of the miR-17-92 cluster, miR-199a, miR-519c, and miR-20b (9, 11, 41, 54, 58, 61). The constitutively expressed HIF-1β subunit has been shown to be a target of p53-induced miR-107 with repressive effects on tumor angiogenesis (72). A very recent study investigated an indirect positive-feedback loop for HIF-1α in hypoxia through miR-210 by targeting glycerol-3-phosphate dehydrogenase 1-like (GPD1L), a negative regulator of PHDs (33). In the present study, we have identified miR-155 as a hypoxia-inducible HIF-1-dependent miRNA in colon cancer cells that specifically targets HIF-1α mRNA and thus forms part of the network of HIF-1α negative-feedback loops responsible for the resolution of HIF-dependent transcription in hypoxia. Interestingly, miR-155 has been shown to be an important regulator of gene translation in multiple physiological and pathophysiological states, including hematopoiesis, inflammation (49, 50), and cancer (42). In one study miR-155 was shown to target HIF-1α in a monocyte cell line (50). We found miR-155 to be induced by hypoxia in a cell type and tissue-specific manner, which indicates a possible specific role in intestinal epithelial cells. In the study of Lei et al. (41) the expression of miR-155 was unchanged in hypoxia. That study used one time point and did not define the degree of hypoxia. In the present study miR-155 expression was upregulated in prolonged hypoxia in multiple cell lines and tissues, a finding that is in agreement with previous studies (28, 48; reviewed in reference 37). We show here for the first time that inhibition of HIF-1α expression through miR-155 results in a reduction of HIF-1-dependent signaling. The HIF-2α isoform is not affected by miR-155. miRNA-155 has been predicted to play a role in tumor development. For example, miR-155 expression contributes to tumor size in a breast cancer model and it has thus been termed an “OncomiR” (30). In colorectal cancer, miR-155 was shown to target key proteins of DNA damage repair (MLH1, MSH2, and MSH6) and therefore induce a mutative phenotype in cancer tissues (67). These results demonstrate that a single miRNA can be sufficient to impact upon disease progression in cancer. The overexpression of miR-155 has been reported in acute myeloid leukemia patients (49), and miR-155 was shown to regulate the survival and proliferation in melanoma cells (42). Overall, in recent studies a high expression of miR-155 in tumors is correlated with aggressive tumor growth in different systems. By targeting the tumor-suppressor SMAD5, miR-155 was demonstrated to contribute to the aggressiveness of lymphomagenesis (53). Two groups showed independently the pro-proliferative effect of miR-155 in breast cancer. Suppressor of cytokine signaling 1 (SOCS1) was identified to be a target of miR-155, which led to an induction of proliferation in breast cancer cells (30). In a similar model miR-155 was shown to promote survival, tumor growth, and chemoresistance by targeting FOXO3a (35). Taken together, the wide variety of miR-155 targets leads to a broad function of miR-155 in physiological processes, such as hematopoiesis, and in pathophysiological processes, such as tumor development. Although miR-155 was reported to be upregulated under hypoxic conditions in a nasopharyngeal cell line (28) and in VHL-deficient renal cancer cells (48), the role of miR-155 in hypoxia remains unclear. Because hypoxia is a key microenvironmental feature of a growing tumor, hypoxic regulation of miR-155 may have profound implications for tumor development. Clinical applications of miRNA technology include potential use of miR-210 as a prognostic marker for breast, pancreatic and head and neck cancer (8, 21, 27). miRNA-155 may also play an important role in regulating the immune response (45, 50, 66) and has been proposed to be a critical player in the cross talk between neoplastic development and inflammation (65). Furthermore, in hematopoiesis, miR-155 is differentially expressed in certain stages of blood cell differentiation, where it plays an important role in B cell development (3). Multiple targets of miR-155 (Meis1, Ets-1, IKKε, IL-1, Pu.1, AID, and c-MAF) have been identified which define its role both in hematopoiesis and in immune response (10, 45, 55, 56, 64, 68).
In summary, while HIF-1α-dependent gene expression is a key determinant of cell, tissue and organism adaptation to hypoxia, excessive or inappropriate activation of this pathway may result in pathological outcomes for the host. As a result, negative-feedback loops that keep tight control of the HIF-1α pathway have evolved. In the present study, we propose that hypoxia-induced miR-155 plays a role as a component of a network of such negative-feedback loops which acts at the level of control of HIF-1α mRNA translation. Such negative-feedback loops are likely crucial to ultimately determining the temporal nature of the cellular transcriptional response to hypoxia.
ACKNOWLEDGMENTS
This study was funded by a principal investigator award from Science Foundation Ireland (to C.T.T.) and by Systems Biology Ireland.
We thank Bernhard Bruene (Frankfurt Am Main, Germany) for the kind gift and permission to use the HepG2 cells with stable knockdown of HIF-1α or HIF-2α.
We declare no conflict of interest.
Footnotes
Published ahead of print on 1 August 2011.
REFERENCES
- 1. Alon U. 2007. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8:450–461 [DOI] [PubMed] [Google Scholar]
- 2. Ashall L., et al. 2009. Pulsatile stimulation determines timing and specificity of NF-κB-dependent transcription. Science 324:242–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baltimore D., Boldin M. P., O'Connell R. M., Rao D. S., Taganov K. D. 2008. MicroRNAs: new regulators of immune cell development and function. Nat. Immunol. 9:839–845 [DOI] [PubMed] [Google Scholar]
- 4. Bartel D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297 [DOI] [PubMed] [Google Scholar]
- 5. Berra E., et al. 2003. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J. 22:4082–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Betel D., Wilson M., Gabow A., Marks D. S., Sander C. 2008. The microRNA.org resource: targets and expression. Nucleic Acids Res. 36:D149–D53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Biswas S. Hypoxia inducible microRNA 210 attenuates keratinocyte proliferation and impairs closure in a murine model of ischemic wounds. Proc. Natl. Acad. Sci. U. S. A. 107:6976–6981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Camps C., et al. 2008. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin. Cancer Res. 14:1340–1348 [DOI] [PubMed] [Google Scholar]
- 9. Cascio S., D'Andrea A., Ferla R., Surmacz E., Gulotta E., Amodeo V., Bazan V., Gebbia N., Russo A. miR-20b modulates VEGF expression by targeting HIF-1α and STAT3 in MCF-7 breast cancer cells. J. Cell Physiol. 224:242–249 [DOI] [PubMed] [Google Scholar]
- 10. Ceppi M., et al. 2009. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc. Natl. Acad. Sci. U. S. A. 106:2735–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cha S. T. MicroRNA-519c suppresses hypoxia-inducible factor-1α expression and tumor angiogenesis. Cancer Res. 70:2675–2685 [DOI] [PubMed] [Google Scholar]
- 12. Chen K., Rajewsky N. 2006. Natural selection on human microRNA binding sites inferred from SNP data. Nat. Genet. 38:1452–1456 [DOI] [PubMed] [Google Scholar]
- 13. Chen Z., Li Y., Zhang H., Huang P., Luthra R. Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene 29:4362–4368 [DOI] [PubMed] [Google Scholar]
- 14. Colgan S. P., Taylor C. T. Hypoxia: an alarm signal during intestinal inflammation. Nat. Rev. Gastroenterol. Hepatol. 7:281–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crosby M. E., Kulshreshtha R., Ivan M., Glazer P. M. 2009. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 69:1221–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cummins E. P. NF-κB links CO2 sensing to innate immunity and inflammation in mammalian cells. J. Immunol. 185:4439–4445 [DOI] [PubMed] [Google Scholar]
- 17. Fasanaro P., et al. 2008. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J. Biol. Chem. 283:15878–15883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fasseu M. Identification of restricted subsets of mature microRNA abnormally expressed in inactive colonic mucosa of patients with inflammatory bowel disease. PLoS One. 5::e13160. doi: 10.1371/journal.pone.0013160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Favaro E. MicroRNA-210 regulates mitochondrial free radical response to hypoxia and Krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PLoS One. 5::e10345. doi: 10.1371/journal.pone.0010345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Filipowicz W., Bhattacharyya S. N., Sonenberg N. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9:102–114 [DOI] [PubMed] [Google Scholar]
- 21. Gee H. E. hsa-miR-210 is a marker of tumor hypoxia and a prognostic factor in head and neck cancer. Cancer 116:2148–2158 [DOI] [PubMed] [Google Scholar]
- 22. Giannakakis A., et al. 2008. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol. Ther. 7:255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ginouves A., Ilc K., Macias N., Pouyssegur J., Berra E. 2008. PHDs overactivation during chronic hypoxia “desensitizes” HIFα and protects cells from necrosis. Proc. Natl. Acad. Sci. U. S. A. 105:4745–4750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Griffiths-Jones S., Grocock R. J., van Dongen S., Bateman A., Enright A. J. 2006. miRBase: microRNA sequences, targets, and gene nomenclature. Nucleic Acids Res. 34:D140–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo H., Ingolia N. T., Weissman J. S., Bartel D. P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466:835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Henze A. T. Prolyl hydroxylases 2 and 3 act in gliomas as protective negative feedback regulators of hypoxia-inducible factors. Cancer Res. 70:357–366 [DOI] [PubMed] [Google Scholar]
- 27. Ho A. S. Circulating miR-210 as a novel hypoxia marker in pancreatic cancer. Transl. Oncol. 3:109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hua Z., et al. 2006. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One 1:e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ivan M., Harris A. L., Martelli F., Kulshreshtha R. 2008. Hypoxia response and microRNAs: no longer two separate worlds. J. Cell Mol. Med. 12:1426–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jiang S. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 70:3119–3127 [DOI] [PubMed] [Google Scholar]
- 31. Johnnidis J. B., et al. 2008. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 451:1125–1129 [DOI] [PubMed] [Google Scholar]
- 32. Kaelin W. G., Jr., Ratcliffe P. J. 2008. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell 30:393–402 [DOI] [PubMed] [Google Scholar]
- 33. Kelly T. J., Souza A. L., Clish C. B., Puigserver P. A hypoxia induced positive feedback loop promotes HIF-1α stability through miR-210 suppression of glycerol-3-phosphate dehydrogenase 1-like. Mol. Cell. Biol. 31:2696–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim H. W., Haider H. K., Jiang S., Ashraf M. 2009. Ischemic preconditioning augments survival of stem cells via miR-210 expression by targeting caspase-8-associated protein 2. J. Biol. Chem. 284:33161–33168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kong W. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J. Biol. Chem. 285:17869–17879 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36. Krek A., et al. 2005. Combinatorial microRNA target predictions. Nat. Genet. 37:495–500 [DOI] [PubMed] [Google Scholar]
- 37. Kulshreshtha R., Davuluri R. V., Calin G. A., Ivan M. 2008. A microRNA component of the hypoxic response. Cell Death Differ. 15:667–671 [DOI] [PubMed] [Google Scholar]
- 38. Kulshreshtha R., et al. 2007. A microRNA signature of hypoxia. Mol. Cell. Biol. 27:1859–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294:853–858 [DOI] [PubMed] [Google Scholar]
- 40. Lagos-Quintana M., Rauhut R., Meyer J., Borkhardt A., Tuschl T. 2003. New microRNAs from mouse and human. RNA 9:175–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lei Z., et al. 2009. Regulation of HIF-1α and VEGF by miR-20b tunes tumor cells to adapt to the alteration of oxygen concentration. PLoS One 4:e7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Levati L., et al. 2009. Altered expression of selected microRNAs in melanoma: antiproliferative and proapoptotic activity of miRNA-155. Int. J. Oncol. 35:393–400 [PubMed] [Google Scholar]
- 43. Lewis B. P., Burge C. B., Bartel D. P. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20 [DOI] [PubMed] [Google Scholar]
- 44. Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. 2003. Prediction of mammalian microRNA targets. Cell 115:787–798 [DOI] [PubMed] [Google Scholar]
- 45. Lu F., et al. 2008. Epstein-Barr virus-induced miR-155 attenuates NF-κB signaling and stabilizes latent virus persistence. J. Virol. 82:10436–10443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Merkel O. Identification of differential and functionally active miRNAs in both anaplastic lymphoma kinase ALK+ and ALK− anaplastic large-cell lymphoma. Proc. Natl. Acad. Sci. U. S. A. 107:16228–16233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Minamishima Y. A., et al. 2009. A feedback loop involving the Phd3 prolyl hydroxylase tunes the mammalian hypoxic response in vivo. Mol. Cell. Biol. 29:5729–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Neal C. S., Michael M. Z., Rawlings L. H., Van der Hoek M. B., Gleadle J. M. The VHL-dependent regulation of microRNAs in renal cancer. BMC Med. 8:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. O'Connell R. M., et al. 2008. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J. Exp. Med. 205:585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. O'Connell R. M., Taganov K. D., Boldin M. P., Cheng G., Baltimore D. 2007. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. U. S. A. 104:1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Paszek P. Population robustness arising from cellular heterogeneity. Proc. Natl. Acad. Sci. U. S. A. 107:11644–11649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pulkkinen K., Malm T., Turunen M., Koistinaho J., Yla-Herttuala S. 2008. Hypoxia induces microRNA miR-210 in vitro and in vivo ephrin-A3 and neuronal pentraxin 1 are potentially regulated by miR-210. FEBS Lett. 582:2397–2401 [DOI] [PubMed] [Google Scholar]
- 53. Rai D., Kim S. W., McKeller M. R., Dahia P. L., Aguiar R. C. Targeting of SMAD5 links microRNA-155 to the TGF-beta pathway and lymphomagenesis. Proc. Natl. Acad. Sci. U. S. A. 107:3111–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rane S., et al. 2009. Downregulation of miR-199a derepresses hypoxia-inducible factor-1α and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ. Res. 104:879–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rodriguez A., et al. 2007. Requirement of bic/microRNA-155 for normal immune function. Science 316:608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Romania P., et al. 2008. MicroRNA 155 modulates megakaryopoiesis at progenitor and precursor level by targeting Ets-1 and Meis1 transcription factors. Br. J. Haematol. 143:570–580 [DOI] [PubMed] [Google Scholar]
- 57. Semenza G. L. 2004. Hydroxylation of HIF-1: oxygen sensing at the molecular level. Physiology (Bethesda) 19:176–182 [DOI] [PubMed] [Google Scholar]
- 58. Song X. W. MicroRNAs are dynamically regulated in hypertrophic hearts, and miR-199a is essential for the maintenance of cell size in cardiomyocytes. J. Cell Physiol. 225:437–443 [DOI] [PubMed] [Google Scholar]
- 59. Stiehl D. P., et al. 2006. Increased prolyl 4-hydroxylase domain proteins compensate for decreased oxygen levels: evidence for an autoregulatory oxygen-sensing system. J. Biol. Chem. 281:23482–23491 [DOI] [PubMed] [Google Scholar]
- 60. Sung M. H., et al. 2009. Sustained oscillations of NF-κB produce distinct genome scanning and gene expression profiles. PLoS One 4:e7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Taguchi A., et al. 2008. Identification of hypoxia-inducible factor-1α as a novel target for miR-17-92 microRNA cluster. Cancer Res. 68:5540–5545 [DOI] [PubMed] [Google Scholar]
- 62. Tan M., et al. 2008. SAG/ROC2/RBX2 is a HIF-1 target gene that promotes HIF-1α ubiquitination and degradation. Oncogene 27:1404–1411 [DOI] [PubMed] [Google Scholar]
- 63. Tay S. Single-cell NF-κB dynamics reveal digital activation and analogue information processing. Nature 466:267–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Teng G., et al. 2008. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity 28:621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tili E., Croce C. M., Michaille J. J. 2009. miR-155: on the crosstalk between inflammation and cancer. Int. Rev. Immunol. 28:264–284 [DOI] [PubMed] [Google Scholar]
- 66. Tili E., et al. 2007. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-α stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 179:5082–5089 [DOI] [PubMed] [Google Scholar]
- 67. Valeri N. Modulation of mismatch repair and genomic stability by miR-155. Proc. Natl. Acad. Sci. U. S. A. 107:6982–6987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vigorito E., et al. 2007. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 27:847–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Williams A. E. 2008. Functional aspects of animal microRNAs. Cell. Mol. Life Sci. 65:545–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wu L., Belasco J. G. 2008. Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs. Mol. Cell 29:1–7 [DOI] [PubMed] [Google Scholar]
- 71. Xie X., et al. 2005. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434:338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yamakuchi M. P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc. Natl. Acad. Sci. U. S. A. 107:6334–6339 [DOI] [PMC free article] [PubMed] [Google Scholar]