Abstract
The trafficking, membrane localization, and lipid raft association of Ras proteins, which are crucial oncogenic mediators, dictate their isoform-specific biological responses. Accordingly, their spatiotemporal dynamics are tightly regulated. While extensively studied for H- and K-Ras, such information on N-Ras, an etiological oncogenic factor, is limited. Here, we report a novel mechanism regulating the activation-dependent spatiotemporal organization of N-Ras, its modulation by biologically relevant stimuli, and isoform-specific effects on signaling. We combined patching/immobilization of another membrane protein with fluorescence recovery after photobleaching (patch-FRAP) and FRAP beam size analysis to investigate N-Ras membrane interactions. Clustering of raft-associated proteins, either glycosylphosphatidylinositol-anchored influenza virus hemagglutinin (HA-GPI) or fibronectin receptors, selectively enhanced the plasma membrane-cytoplasm exchange of N-Ras–GTP (preferentially associated with raft domains) in a cholesterol-dependent manner. Electron microscopy (EM) analysis showed N-Ras–GTP localization in cholesterol-sensitive clusters, from which it preferentially detached upon HA-GPI cross-linking. HA-GPI clustering enhanced the Golgi compartment (GC) accumulation and signaling of epidermal growth factor (EGF)-stimulated N-Ras–GTP. Notably, the cross-linking-mediated enhancement of N-Ras–GTP exchange and GC accumulation depended strictly on depalmitoylation. We propose that the N-Ras activation pattern (e.g., by EGF) is altered by raft protein clustering, which enhances N-Ras–GTP raft localization and depalmitoylation, entailing its exchange and GC accumulation following repalmitoylation. This mechanism demonstrates a functional signaling role for the activation-dependent differential association of Ras isoforms with raft nanodomains.
INTRODUCTION
Ras proteins regulate cell proliferation, apoptosis, and differentiation (32, 34). Constitutively activating mutations or overactivation of specific Ras isoforms is encountered in different human tumors (7). Ras proteins localize to the plasma membrane (PM) but are also found in endosomes, the endoplasmic reticulum, mitochondria, and the Golgi compartment (GC) (22, 45, 47). The localization to different organelles and membrane domains may determine the effectors encountered by Ras, entail the activation of specific pathways, and regulate cellular responses (9, 33, 46, 48, 50).
The major Ras isoforms—H-Ras, N-Ras, and K-Ras4B (K-Ras)—are highly homologous except for their C-terminal hypervariable region (the last 23 to 24 residues) (19, 22). They share a C-terminal CAAX S-farnesylation motif leading to weak membrane binding but require a second motif for effective membrane association. For N- and H-Ras, the second signal is one (Cys181) or two (Cys181 and Cys184) S-palmitoyl residues, respectively, while K-Ras has a 6-lysine polybasic cluster that interacts with the internal PM leaflet (20, 21, 29). These second signals regulate Ras traffic and distribution between intracellular organelles and the PM and specify Ras localization within PM domains (11, 45, 46). K-Ras does not traffic via the GC, while the labile palmitoylation of N- and H-Ras (4, 31) in the GC (46) enables a de-/repalmitoylation cycle that regulates their PM/GC partitioning and prevents nonselective distribution in endomembranes (2, 46, 48). Palmitoylation also endows N- and H-Ras with affinity to cholesterol-sensitive, dynamic nanoclusters (i.e., rafts) (19, 22, 26, 54). Raft association depends also on the activation state of N- and H-Ras; nonpalmitoylated K-Ras is excluded from rafts (37, 44, 50). The transient association of H-Ras with raft clusters is crucial for signaling via Raf/Erk (37, 43, 44). Using fluorescence recovery after photobleaching (FRAP) combined with patching/immobilization of another membrane protein (patch-FRAP), we have demonstrated (15) that clustering of raft-resident glycosylphosphatidylinositol-anchored influenza virus hemagglutinin (GPI-HA) stabilizes H-Ras–GDP association with raft clusters. The stabilized H-Ras–GDP raft association facilitated GDP/GTP exchange but retarded the exit of H-Ras–GTP from raft to nonraft signaling assemblies, inhibiting the ensuing activation of Raf.
N-Ras palmitoylation also enables its association with rafts. However, unlike H-Ras, activated N-Ras–GTP interacts preferentially with rafts (Fig. 1) (50). Thus, N-Ras localization and activity may be modulated differently from those of H-Ras by clustering raft proteins, providing a mechanism for distinct signaling by different Ras isoforms. This possibility is intriguing in light of the more frequent occurrence of activating N-Ras mutations than of H-Ras mutations in human cancer (39, 49). Here, we show that the targeting of N-Ras–GTP to raft clusters and its PM-GC transport are tightly linked, depend on its depalmitoylation, and are markedly altered by clustering of raft proteins. The analogous effects mediated by the cross-linking of fibronectin receptors by fibronectin support the physiological relevance of this mechanism, which enables multiple costimulators to alter the pattern of N-Ras activation and signaling downstream of the primary stimulus (e.g., epidermal growth factor [EGF]) by clustering raft proteins.
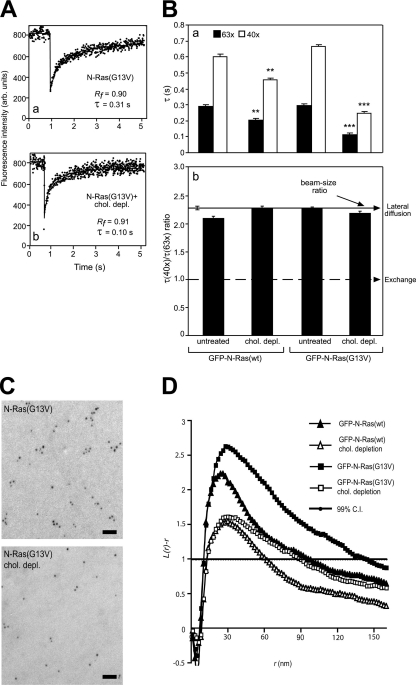
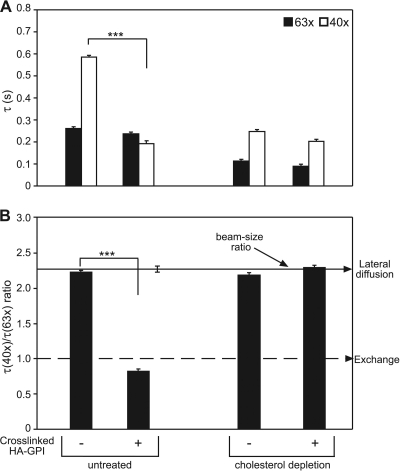
Fig. 1.
FRAP beam size analysis and EM spatial mapping show preferential interactions of constitutively active N-Ras with cholesterol-sensitive assemblies in the PM. (A) Typical FRAP curves (63× objective, 22°C) of GFP–N-Ras(G13V) in untreated (a) or cholesterol-depleted (chol. depl.) (b) COS-7 cells. Solid lines, best fit of a nonlinear regression analysis (Materials and Methods). arb. units, arbitrary units. (B) FRAP beam size analysis. Bars show means ± standard errors of the means (SEM) of 30 to 60 measurements. The studies employed 40× and 63× objectives, yielding a 2.28 ± 0.17 (n = 59) beam size ratio. Thus, this τ(40×)/τ(63×) ratio is expected for FRAP by lateral diffusion (b, upper arrow). A τ ratio of 1 (b, lower arrow) indicates recovery by exchange (24). Comparison between τ values (a) measured with the same beam size showed that cholesterol depletion strongly reduced the τ values of GFP–N-Ras(G13V) (***, P < 10−8; Student's t test), with a mild effect on GFP–N-Ras(wt) (**, P < 10−3). Bootstrap analysis (Materials and Methods) showed that all τ ratios (panel b) are not significantly different from the 2.28 beam size ratio (P > 0.3), suggesting FRAP by lateral diffusion. Calculating D from the τ values before and after cholesterol depletion yielded values of D = 0.52 and 0.74 μm2/s for GFP–N-Ras(wt) and D = 0.51 and 1.35 μm2/s for GFP–N-Ras(G13V). (C) Representative EM images of PM sheets from untreated and cholesterol-depleted HeLa cells expressing GFP–N-Ras (G13V). Bars, 50 nm. (D) Mean univariate K functions of gold–anti-GFP expressed as L(r) − r standardized on the 99% confidence interval (C.I.). Positive deviation from this value indicates clustering for that r. Data were pooled from multiple (n ≥ 21) PM sheets. Cholesterol depletion significantly reduced both GFP–N-Ras(wt) and GFP–N-Ras(G13V) clustering; the effect was stronger for GFP–N-Ras(G13V) (P < 0.001; bootstrap analysis).
MATERIALS AND METHODS
Materials.
COS-7 (American Type Culture Collection) and HeLa cells (CRUK cell bank) were grown as described previously (15, 39). Monovalent rabbit tetramethyl rhodamine isothiocyanate (TRITC)-Fab′ against the HA protein of the X:31 influenza virus strain (anti-X:31 HA) was prepared from IgG donated by J. M. White (University of Virginia, Charlottesville, VA) (56). Rabbit TRITC-Fab′ anti-Japan influenza virus HA was a gift from M. G. Roth, University of Texas Southwestern Medical Center, Dallas, TX (56). IgG of HC3 mouse anti-X:31 HA and Fc125 anti-Japan influenza virus HA were from J. J. Skehel (National Institute for Medical Research, London, United Kingdom) and T. J. Braciale (University of Virginia, Charlottesville, VA), respectively. Rabbit anti-full-length green fluorescent protein (GFP) was affinity purified and conjugated to 5-nm gold beads as described previously (44). Rabbit antifibronectin serum was donated by B. Geiger (Weizmann Institute of Science, Rehovot, Israel). Cy3-goat anti-mouse IgG was from Jackson ImmunoResearch Laboratories. Alexa fluor 546–goat anti-rabbit IgG, Alexa fluor 646–goat anti-mouse IgG, and Alexa fluor 488–goat anti-rabbit IgG were from Invitrogen-Molecular Probes, and rabbit anti-Erk2 (sc-154) from Santa Cruz Biotechnology. Mouse anti-phospho-Erk1/2 (M8159) and rabbit anti-GM130 were from Sigma, mouse pan anti-Ras (Ab-3) from Calbiochem, mouse anti-β-actin from MP Biomedicals, and peroxidase-coupled goat anti-mouse and anti-rabbit IgG from Dianova. EGF was from R&D Systems. Fibronectin and 2-bromopalmitate (2BP) were from Sigma. Palmostatin B was recently described (11). 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid tetra(acetoxymethyl) ester (BAPTA-AM) was from Invitrogen-Molecular Probes, and U73122 from Calbiochem.
Plasmids and cell transfection.
The pEGFP-C3 expression vectors for wild-type GFP–N-Ras (wt) and the constitutively active GFP–N-Ras(G13V) were described previously (40). As shown here and in earlier studies (2, 37), GFP-tagged Ras proteins are biologically active, respond to EGF stimulation, and mediate neurite outgrowth in PC12 cells. Moreover, their localization recapitulates that of endogenous Ras molecules (10). Constructs of mCherry–N-Ras (wt and constitutively active), generated from the GFP-tagged plasmids by replacing the cDNA segment encoding GFP with that of mCherry using restriction enzymes (AgeI and BsrG1), were a gift from L. H. Chamberlain (University of Strathclyde, Glasgow, United Kingdom). Nonpalmitoylable GFP–N-Ras(C181S) (wt and constitutively active) mutants were donated by M. Philips (NYU School of Medicine, New York, NY). The expression vector for HA-GPI (the ectodomain of X:31 HA fused to the GPI anchor addition signal of DAF) in pEE14 (originally designated BHA-PI) (27) was a gift from J. M. White. A pCB6 vector for expression of the Japan influenza virus HA(2A520) mutant, which harbors a GS-to-AA mutation at transmembrane positions 520 to 521 (30, 52), was donated by M. G. Roth. The pGex-2TH bacterial expression vector for the glutathione S-transferase (GST)-fused Raf-1 Ras binding domain (RBD) with an A85K mutation (17) was a gift from A. Burgess (Ludwig Institute for Cancer Research, Melbourne, Australia). Monomeric red fluorescent protein (mRFP)-RBD in pmRFP-C1 (36) was donated by M. Philips. COS-7 or HeLa cells in 35-mm dishes (with glass coverslips for FRAP and confocal studies) were transfected by jetPEI (PolyPlus Transfection) with 0.6 μg of a GFP–N-Ras vector alone or together with an HA vector (0.1 μg) and/or the mRFP-RBD vector (0.3 μg), with the DNA brought to 1.6 μg by empty vector. For electron microscopy (EM) studies, HeLa cells were transfected similarly using GeneJuice (Novagen). The GFP–N-Ras and HA vector cotransfection efficiency was ∼95%.
Cholesterol depletion and palmostatin B treatment.
At 24 h posttransfection, cells were subjected, where indicated below, to metabolic cholesterol depletion by incubation with 50 μM compactin and 50 μM mevalonate in medium supplemented with 10% lipoprotein-deficient fetal calf serum as described previously (30, 55). We have shown that this treatment, which reduces cholesterol by 30 to 33%, has no detectable effect on the cellular phospholipids (measured by the levels of phosphatidylcholine and four different sphingomyelins) or fatty acid composition (55). Notably, this treatment has no significant effects on the general biophysical properties of the PM, as shown by the lack of effect on the FRAP dynamics of nonraft proteins, including K-Ras and the nonraft HA(2A520) mutant (15, 55).
Palmostatin B treatment was as described previously (11), with cells incubated in growth medium (15 min for 37°C) with 10 μM palmostatin B (diluted from dimethyl sulfoxide [DMSO] stock solution to 0.5% [vol/vol] DMSO). The drug concentration was kept during the ensuing measurements.
Cross-linking of cell-surface HA proteins and fibronectin receptors.
Cells were transfected with combinations of GFP–N-Ras and HA expression vectors. After 24 h, they were subjected (or not) to cholesterol depletion. Cell-surface HA proteins were cross-linked at 4°C by IgGs (30 μg/ml HC3 anti-X:31 HA or Fc123 anti-Japan influenza virus HA followed by 30 μg/ml Cy3-goat anti-mouse IgG). All incubations (30 min each) and washes were in Hanks' balanced salt solution (HBSS) supplemented with 20 mM HEPES, 2% bovine serum albumin (BSA), pH 7.2 (HBSS-HEPES-BSA). In control experiments, the IgGs were replaced by rabbit TRITC-Fab′ of anti-X:31 HA or anti-Japan influenza virus HA (50 μg/ml). Where indicated below, cells were serum starved for 12 h, and IgG cross-linking was replaced by incubation with fibronectin (20 μg/ml for 30 min at 4°C) in the HBSS buffer, followed where indicated below by further cross-linking with antifibronectin (1:300 for 20 min at 4°C).
FRAP beam size analysis and patch-FRAP.
FRAP studies were conducted in HBSS-HEPES-BSA on COS-7 or HeLa cells transfected with GFP–N-Ras and HA expression vectors. In some experiments, the cells were first subjected to one or more of the following treatments: cholesterol depletion, cross-linking with anti-HA IgGs or fibronectin (alone or followed by antifibronectin), palmostatin B, or serum starvation followed by incubation with EGF (see “EGF stimulation” and figure legends). FRAP studies were at 22°C or 37°C, with samples being replaced within 10 min (22°C) or 5 min (37°C) to minimize internalization. Since these studies are performed on single cells selected under the microscope according to the GFP–N-Ras expression levels, we selected cells with low (10,000 to 20,000 molecules/cell) to medium (∼50,000/cell) expression levels. Cells on the low-expression end (which is below the level of endogenous Ras) yielded results similar to those for the higher-expression cells, suggesting that the FRAP results are not due to overexpression. For the FRAP studies, an argon ion laser beam (Innova 70C; Coherent) was focused through a fluorescence microscope (AxioImager.D1; Carl Zeiss MicroImaging) to a Gaussian spot of 0.77 ± 0.03 μm (mean ± standard deviation) (Plan apochromat 63×/1.4 numeric aperture [NA] oil immersion objective) or 1.17 ± 0.05 μm (C apochromat 40×/1.2 NA water immersion objective), and experiments were conducted with each (beam size analysis) (24). The ratio between the bleach areas was 2.28 ± 0.17 (n = 59). After a brief measurement at the monitoring intensity (488 nm and 1 μW), a 5-mW pulse (5 to 10 ms) bleached 60 to 75% of the fluorescence in the spot, and recovery was followed by the monitoring beam. The characteristic fluorescence recovery time (τ) and mobile fraction (Rf) were extracted from the FRAP curves by nonlinear regression analysis with fitting to a lateral diffusion process (24). Due to the very weak (logarithmic) dependence of the lateral diffusion rate of membrane-anchored proteins on the mass of the membrane-embedded segment (51), the addition of GFP to Ras has a negligible effect on Ras lateral diffusion. Patch-FRAP studies were performed similarly, except that cross-linking of an HA protein or fibronectin receptors preceded the measurement (15, 23). The Rf values for GFP–N-Ras proteins were above 0.90 in all cases.
Statistical analysis of FRAP data.
The significance of differences between τ values measured with the same beam size was evaluated by Student's t test. To compare ratio measurements [τ(40×)/τ(63×) and ω2(40×)/ω2(63×) (see Results)], we employed bootstrap analysis, which is preferable for comparison between ratios (13). The bootstrap analysis was performed exactly as described by us earlier (18), using 1,000 bootstrap samples.
EM and statistical analysis.
PM sheets of transfected HeLa cells were prepared from untreated or cholesterol-depleted cells as described previously (44). In some cases, cell-surface HA-GPI was IgG cross-linked as described above prior to the preparation of PM sheets. The membrane sheets were fixed with 4% paraformaldehyde, 0.1% glutaraldehyde and labeled with anti-GFP–5-nm gold beads as described previously (44). Following imaging at 100 kV in an FEI Tecnai G2 transmission electron microscope, digital images were analyzed for spatial mapping by Ripley's univariate K function to determine the extent of clustering within the immunogold patterns (44). Bootstrap tests to examine differences between replicated point patterns were constructed exactly as described earlier (12); statistical variance was evaluated against 1,000 bootstrap samples.
Data acquisition and processing for live-cell imaging.
Images were acquired with a spinning-disk (confocal head Yokogawa CSU-22) microscope (Axiovert 200 M [Carl Zeiss MicroImaging] with Photometrics HQ2 charge-coupled device) under the control of SlideBook (Intelligent Imaging Innovations), using a Plan apochromat 63×/1.4 NA oil immersion objective. Three-dimensional (3-D) image stacks were generated by sequential recordings along the z axis by the position of a piezo-controlled stage. A step size of 0.4 μm was used for single-time-point acquisitions. GFP excitation was with a 40-mW solid-state 473-nm laser, and mRFP was excited with a 10-mW solid-state 561-nm laser. Typical exposure times were 0.5 to 1 s. Alexa fluor 646 was visualized with a 175-W Xenon light source with the appropriate filters.
The mRFP-RBD GC fluorescence was quantified as described previously (14). 3-D images were restored by Nearest Neighbor deconvolution (1) using SlideBook. The GC was identified as a concentrated GFP–N-Ras signal at the perinuclear region, as validated by colocalization with the GM130 GC marker (see Fig. 5A). The total fluorescence intensity of mRFP-RBD per cell and the proportion of GC-localized mRFP fluorescence were measured in the deconvoluted images after projection onto 2 dimensions, using the sum of pixel intensities in each plane.
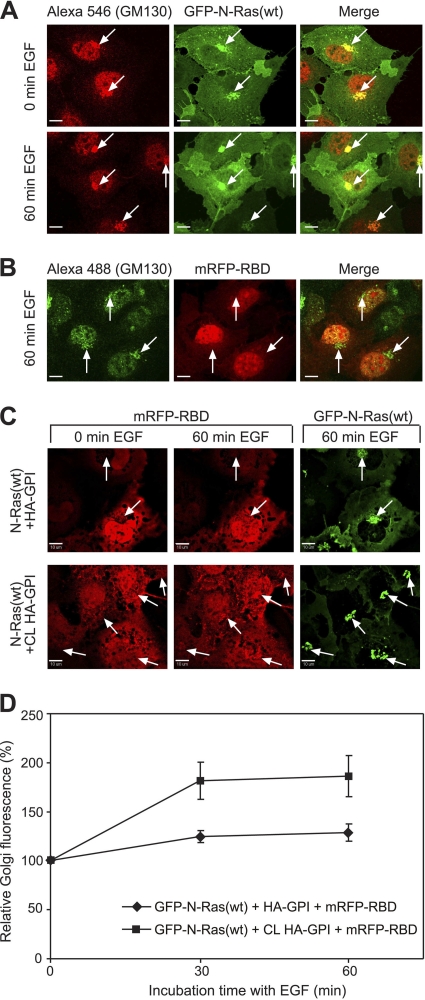
Fig. 5.
HA-GPI clustering enhances the accumulation of activated N-Ras in the Golgi compartment (GC) following EGF stimulation. COS-7 cells were transfected with GFP–N-Ras(wt) alone (A), mRFP-RBD alone (B), or both together with HA-GPI (C, D). They were serum starved, subjected (or not) to HA-GPI cross-linking at 4°C, and either left untreated (0 min EGF) or incubated at 37°C with 100 ng/ml EGF. Images were taken with a spinning-disk confocal microscope as described in Materials and Methods. Bars, 10 μm. (A) GFP–N-Ras and GM130 colocalization. After fixation (4% paraformaldehyde), cells were permeabilized (0.2% Triton X-100), labeled with rabbit anti-GM130 (0.2 μg/ml) and Alexa fluor 546–goat anti-rabbit IgG (3 μg/ml), and imaged at a plane with distinct GM130 GC labeling. Arrows point at GC staining (GM130 labeling) in GFP–N-Ras-expressing cells. (B) Endogenous Ras levels are insufficient to detect mRFP-RBD GC accumulation. The experiment was performed as described for panel A on cells transfected with mRFP-RBD. After stimulation with EGF (60 min), cells were fixed, permeabilized, and stained for GM130 using Alexa fluor 488–goat anti-rabbit IgG. Arrows indicate GM130 GC labeling in cells expressing mRFP-RBD. (C) Live- cell 3-D imaging of EGF-stimulated triple-transfected cells. Typical midplane images of cells at 0 and 60 min of EGF stimulation, without (top) or with (bottom row) HA-GPI cross-linking (CL). The GC was identified by the dense GFP–N-Ras fluorescence (arrows). (D) Quantification (means ± SEM, 9 to 10 cells) of the relative GC mRFP-RBD fluorescence in 3-D image stacks of live cells. The intensity in the GC at time zero was taken as 100%.
EGF stimulation.
Transfected cells were grown for 24 h, followed by a 12-h serum starvation. The HA proteins were cross-linked at 4°C by IgGs or labeled by TRITC-Fab′ (non-cross-linked controls). In FRAP studies of EGF-stimulated GFP–N-Ras(wt), the serum-starved cells were stimulated with EGF (100 ng/ml for 4 min at 37°C), retaining EGF during the measurement (up to 10 min after the addition of EGF). For confocal studies on mRFP-RBD GC accumulation, Alexa fluor 646–goat anti-mouse IgG was used as the secondary antibody for HA-GPI cross-linking. EGF (100 ng/ml) was then added at 37°C, and the cells were subjected to live-cell imaging for up to 60 min.
Immunoblotting.
At 24 h posttransfection, cells were serum starved (12 h), stimulated with EGF (100 ng/ml at 37°C) for 5 or 60 min, lysed, and subjected to SDS-PAGE and immunoblotting as described previously (28), with 20 μg protein loaded per lane. Blots were probed with anti-phospho-Erk (1:10,000 for 12 h at 22°C) followed by peroxidase-coupled goat-anti-mouse IgG (1:5,000 for 1 h at 22°C), acid stripped (28), and reprobed for total Erk with rabbit anti-Erk (1:1,500) and peroxidase-coupled anti-rabbit IgG. The bands were visualized by enhanced chemiluminescence (ECL; Amersham) and quantified by densitometry (EZQuant-Gel 2.2; EZQuant Ltd.). A second cycle of reprobing was used to visualize GFP-Ras by pan anti-Ras (1:2,500) and peroxidase-coupled secondary IgG (1:5,000).
Determination of the levels of GFP-Ras-GTP and GFP-Ras.
Cells were transfected, serum starved, cross-linked with anti-HA IgG, and/or stimulated by EGF as described above under “EGF stimulation.” In studies employing BAPTA-AM or U73122 to inhibit direct Ras activation in the GC, the inhibitors were added after serum starvation (10 μM for 30 min at 37°C) and kept at the same concentration during the ensuing steps. Aliquots (20 μg of protein) of the lysates were resolved by immunoblotting to determine the total level of the GFP-Ras proteins using pan anti-Ras antibodies as described above or anti-β-actin (1:10,000) as a loading control. To determine the levels of GFP-Ras-GTP, cell lysates (200 μg) were precipitated using glutathione-Sepharose beads coupled to GST-RBD (28). The GST-RBD precipitates were dissolved, resolved by SDS-PAGE, and immunoblotted with pan-anti-Ras (1:2,500) to visualize GFP-Ras-GTP. ECL and densitometry were performed as described above.
RESULTS
Constitutively active N-Ras interacts preferentially with cholesterol-dependent assemblies in live cells.
To investigate the dynamics of N-Ras interactions with rafts in the PM of live cells and their dependence on its activation state, we expressed GFP–N-Ras(wt) or constitutively active GFP–N-Ras(G13V) in COS-7 cells and conducted FRAP-beam size analysis using two beam sizes (Fig. 1A and B), employing a 63× (smaller Gaussian radius, ω) or a 40× (larger ω) objective (24). The ratio between the illuminated areas, ω2(40×)/ω2(63×), was 2.28. If FRAP occurs by lateral diffusion, τ (half-life [t1/2] for recovery) is the characteristic diffusion time τD, proportional to the bleached area (τD = ω2/4D, where D is the lateral diffusion coefficient). Therefore, for FRAP by lateral diffusion, the expected τ(40×)/τ(63×) ratio equals the beam size ratio (2.28). A τ ratio of 1 is indicative of FRAP by exchange between membrane-associated and cytoplasmic pools; here, τ is the characteristic exchange time, τex, and is independent of the beam size because it is a chemical relaxation time. Intermediate τ ratios suggest mixed recovery (24).
At 22°C, GFP–N-Ras(wt) and GFP–N-Ras(G13V) displayed similar FRAP kinetics in untreated cells (typical FRAP curves are shown in Fig. 1A, and averaged data in Fig. 1B), yielding similar τ values with high (≥0.90) mobile fractions (Rf). The τ(40×)/τ(63×) ratios for both N-Ras(wt) and N-Ras(G13V) in untreated cells were similar to the 2.28 ratio expected for FRAP by lateral diffusion (Fig. 1Bb), suggesting that their exchange is slow relative to their lateral diffusion. The diffusion-dependent FRAP enables the direct derivation of D from τ (for D values, see the Fig. 1 legend). The low capacity of GFP for dimerization has no effect on the FRAP mode, since essentially similar results were obtained with the equivalent mCherry–N-Ras constructs (data not shown).
To explore whether the N-Ras activation state affects its raft association, we disrupted rafts by cholesterol depletion using metabolic inhibition of its synthesis (Materials and Methods) (30) and measured the effects on the FRAP kinetics of GFP–N-Ras (wt and G13V) (Fig. 1A and B). This treatment significantly reduced the τ values of both GFP–N-Ras proteins, with a stronger effect on the GTP-loaded N-Ras(G13V) (Fig. 1B). However, both proteins retained relatively stable PM association, as their τ(40×)/(63×) ratios remained similar to the 2.28 value indicative of FRAP by lateral diffusion (Fig. 1Bb). Notably, analogous FRAP results were obtained at 37°C (see Fig. 4A and B). We conclude that cholesterol depletion releases N-Ras(wt) and N-Ras(G13V) from mobility-restricting interactions with cholesterol-dependent assemblies. An increase in the lateral diffusion following similar cholesterol depletion treatment was also observed for other raft interactors [HA-GPI or HA(wt)] but not for nonraft molecules [K-Ras or HA(2A520)] (15, 55), demonstrating the specificity of the observed effects. The stronger effect on N-Ras(G13V) suggests that it has a higher affinity to rafts. However, the similar τ values of N-Ras(G13V) and N-Ras(wt) prior to cholesterol depletion indicate that the lateral diffusion of the latter is restricted by transient interactions with nonraft domains/protein clusters, as shown for H- and K-Ras (37, 44).
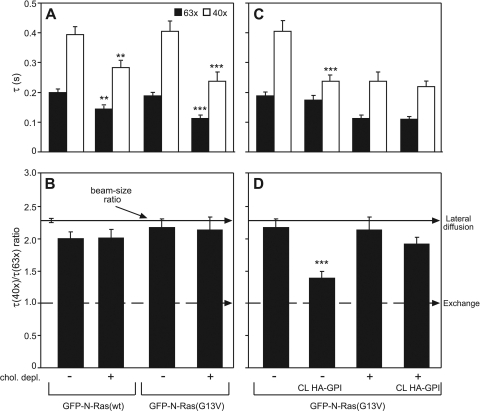
Fig. 4.
The preferential interactions of activated N-Ras with rafts and its shift to exchange in response to HA-GPI cross-linking are retained at 37°C. COS-7 cells expressing GFP–N-Ras (wt or G13V) alone or with HA-GPI were subjected (or not) to cholesterol depletion, followed by HA-GPI cross-linking (or Fab′ labeling) and FRAP beam size analysis at 37°C as described in the Fig. 2 legend. Bars show means ± SEM (n = 30 to 60). (A and B) Effects of cholesterol depletion. Comparison between τ values (A) measured with the same beam size shows that cholesterol depletion significantly reduces the τ of GFP–N-Ras(G13V) (***, P < 10−8; Student's t test), with a milder effect on GFP–N-Ras(wt) (**, P < 10−5). (C and D) HA-GPI cross-linking induces a cholesterol-dependent shift of N-Ras(G13V) to exchange. The significant effects of HA-GPI cross-linking on the τ(40×) of N-Ras(G13V) (***, P < 10−8; Student's t test) (C) and on its τ ratio (***, P < 10−12; bootstrap analysis) (B) were abolished in cholesterol-depleted cells.
To obtain an independent measure for the interactions of GFP–N-Ras proteins with PM rafts, we employed EM spatial analysis to measure the clustering of immunogold-labeled GFP–N-Ras proteins (44). These studies employed HeLa cells due to the difficulty in preparing PM sheets from COS-7 cells; therefore, we validated that the effects of cholesterol depletion on the lateral diffusion of N-Ras (G13V and wt) in HeLa cells resemble those in COS-7 cells (data available upon request). For the EM studies, HeLa cells expressing GFP–N-Ras(wt) or GFP–N-Ras(G13V) (untreated or cholesterol depleted) were subjected to preparation of PM sheets and immunogold labeling and analyzed for clustered versus random distribution using Ripley's K function (Materials and Methods) (44). Figure 1C and D show that in untreated cells, both GFP–N-Ras(wt) and GFP–N-Ras(G13V) have clustered distribution. The curves from the equation L(r) − r (a linear transformation of the K function [42]) display positive deviations over the 99% confidence interval for complete spatial randomness; the peak radius (r) value minus the gold probe r of ∼10 nm (42) represents the most common cluster size (∼20 nm). The clustering of GFP–N-Ras(G13V) was greater and showed a longer tail, which may reflect some aggregation of the minimal cluster unit. It was more sensitive than GFP–N-Ras(wt) to cholesterol depletion, which significantly reduced the clustering of both proteins. The stronger effect on N-Ras(G13V) is qualitatively similar to the cholesterol sensitivity of constitutively active N-Ras nanoclusters measured by EM in BHK cells (50). However, while GFP–N-Ras(wt) clustering was not affected by cholesterol depletion in the latter study, it showed partial cholesterol sensitivity in our experiments, potentially due to cell type differences and/or some baseline stimulation of N-Ras(wt) by serum in the current studies. A negative control is supplied by the cholesterol independence of K-Ras clusters demonstrated by the same technique (44). These findings corroborate the FRAP results and suggest that clusters of GTP-loaded GFP–N-Ras(G13V) display higher sensitivity to cholesterol depletion than those of GFP–N-Ras(wt).
Cross-linking raft-interacting HA-GPI increases the exchange of activated N-Ras between the PM and the cytoplasm.
We have shown that IgG cross-linking of raft-associated HA-GPI (27, 56) stabilizes H-Ras association with the raft clusters, modulating its activation and signaling (15). In view of the preferential targeting of activated N-Ras to rafts (Fig. 1), we combined FRAP beam size analysis with patch-FRAP (15, 23) to investigate the modulation of N-Ras membrane interactions by clustering raft-associated proteins and its dependence on N-Ras activation. We clustered HA-GPI with IgGs, leading to its lateral immobilization (Fig. 2A and B), and measured the effects on the FRAP kinetics of GFP–N-Ras (wt or G13V) at the PM (Fig. 2C to F). Coexpression with HA-GPI did not affect the FRAP parameters of GFP–N-Ras(G13V), but IgG cross-linking of HA-GPI dramatically reduced the τ(40×) of N-Ras(G13V), shifting its FRAP mechanism from pure lateral diffusion [τ(40×)/τ(63×) = 2.28] to exchange (τ ratio of ∼1) (Fig. 2C and E). This implies that interactions with immobilized raft clusters retard the lateral diffusion of N-Ras(G13V), resulting in a minimal contribution to the FRAP, which becomes dominated by the exchange kinetics (see reference 24). Analogous results were obtained when the FRAP measurements were conducted at 37°C (see Fig. 4C and D). Importantly, cross-linking of the nonraft HA-2A520 mutant (15, 30) had no significant effect on the GFP–N-Ras(G13V) FRAP parameters (Fig. 2C and E), providing a direct demonstration independent of cholesterol-reducing treatments that the effects of HA-GPI clustering require raft association of the cross-linked protein. In contrast, cross-linking HA-GPI failed to alter the FRAP parameters of unactivated GFP–N-Ras(wt) (Fig. 2D and F). Activation of GFP–N-Ras(wt) at the PM by a short (4 to 5 min) stimulation with EGF had no effect on its FRAP parameters, demonstrating that EGF activation alone is not sufficient to enhance N-Ras–GTP exchange. However, EGF stimulation sensitized the membrane interactions of GFP–N-Ras(wt) to HA-GPI clustering, which together with EGF stimulation shifted GFP–N-Ras(wt) to exchange (Fig. 2D and F). HA-GPI cross-linking shifted GFP–N-Ras(G13V) to exchange in HeLa cells also, as measured by FRAP beam size analysis at either 22°C or 37°C (data available upon request). Taken together, these findings suggest that cross-linking raft-associated HA-GPI specifically modulates the PM interactions of activated N-Ras, enhancing its PM-cytoplasm exchange. This notion is supported by EM clustering analysis of the distribution of GFP–N-Ras in PM sheets derived from HeLa cells (Fig. 2G): cross-linking HA-GPI had no effect on the clustering of GFP–N-Ras(wt) but markedly reduced GFP–N-Ras(G13V) clustering without altering the cluster radius. Together with the preferential localization of N-Ras(G13V) in cholesterol-sensitive clusters (Fig. 1), this suggests that GFP–N-Ras(G13V) is lost mainly from these clusters following HA-GPI cross-linking, in line with the enhancement of its exchange rate. To confirm that the effect of HA-GPI cross-linking on the membrane interactions of activated N-Ras depends on interactions with cholesterol-sensitive assemblies, we measured the effects of cholesterol depletion on the modulation of the FRAP kinetics of GFP–N-Ras(G13V) by HA-GPI cross-linking. As shown in Fig. 3, cholesterol depletion abolished the modulation of GFP–N-Ras(G13V) membrane interactions in response to cross-linking of HA-GPI; a similar effect was obtained at 37°C (Fig. 4C and D).
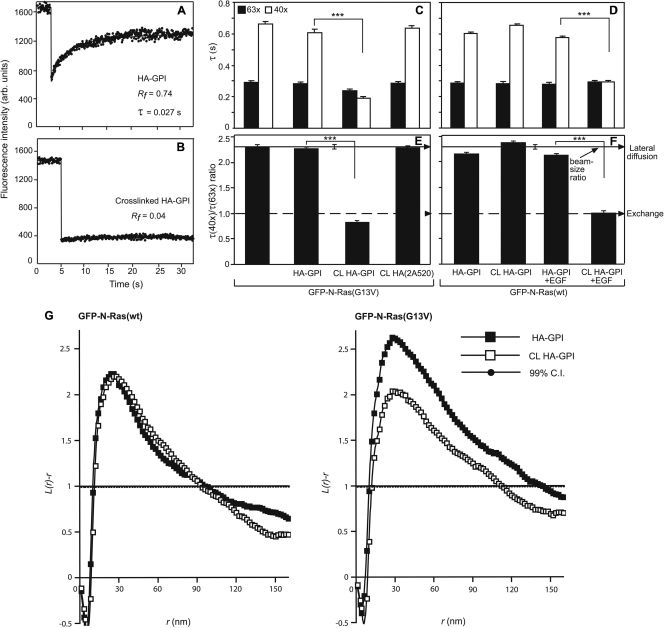
Fig. 2.
The results of patch-FRAP and EM demonstrate that cross-linking raft-resident HA-GPI reduces the PM association of activated N-Ras. COS-7 cells expressing GFP–N-Ras (wt or G13V) with or without HA-GPI (raft) or HA(2A520) (nonraft) were subjected to HA cross-linking (CL) at 4°C by IgGs or to TRITC-Fab′ labeling (control). For EGF stimulation (D and F), cells were serum starved, HA cross-linked, incubated (or not) with EGF (100 ng/ml, 4 min, 37°C), and subjected to FRAP studies at 22°C in EGF-containing buffer within 10 min. FRAP and EM studies were performed as described in the Fig. 1 legend. (A and B) Typical FRAP curves (63× objective) of HA-GPI without (A) or with (B) IgG cross-linking. (C to F) FRAP beam size analysis of GFP–N-Ras proteins. Bars show means ± SEM of 30 to 60 measurements. IgG cross-linking (CL) of HA-GPI but not of HA(2A520) dramatically reduced the τ(40×) (C and D) of activated (but not unactivated) N-Ras (***, P < 10−13; Student's t test). Bootstrap analysis of the τ ratios (E, F) showed that they are all similar (P > 0.3) to the 2.28 beam size ratio indicative of lateral diffusion, except for constitutively active or EGF-activated N-Ras in cells with cross-linked HA-GPI. In the latter cases, the τ ratios were ∼1, as predicted for FRAP by exchange (P > 0.06), highly different from their τ ratios without cross-linking (***, P < 10−20; bootstrap analysis). (G) EM spatial distribution analysis. HeLa cells coexpressing HA-GPI and GFP–N-Ras (wt or G13V) were pretreated as described above for HA-GPI cross-linking before generation of PM sheets and K function analysis (n ≥ 21). Only GFP–N-Ras (G13V) clustering was significantly reduced by HA-GPI cross-linking (P < 0.05; bootstrap analysis).
Fig. 3.
Cholesterol depletion abolishes the shift of GFP–N-Ras(G13V) to exchange by cross-linking HA-GPI. COS-7 cells coexpressing GFP–N-Ras(G13V) and HA-GPI were subjected (or not) to cholesterol depletion, followed by HA-GPI cross-linking (or Fab′ labeling; control). FRAP beam size analysis (22°C) was as described in the Fig. 2 legend. Bars depict means ± SEM (n = 30 to 60). The highly significant effects of HA-GPI cross-linking on the τ(40×) of N-Ras(G13V) (***, P < 10−13; Student's t test) (A) and on its τ ratio (***, P < 10−20; bootstrap analysis) (B) were abolished by cholesterol depletion.
Accumulation of EGF-stimulated GFP–N-Ras(wt) at the GC and its prolonged downstream signaling are enhanced by HA-GPI cross-linking.
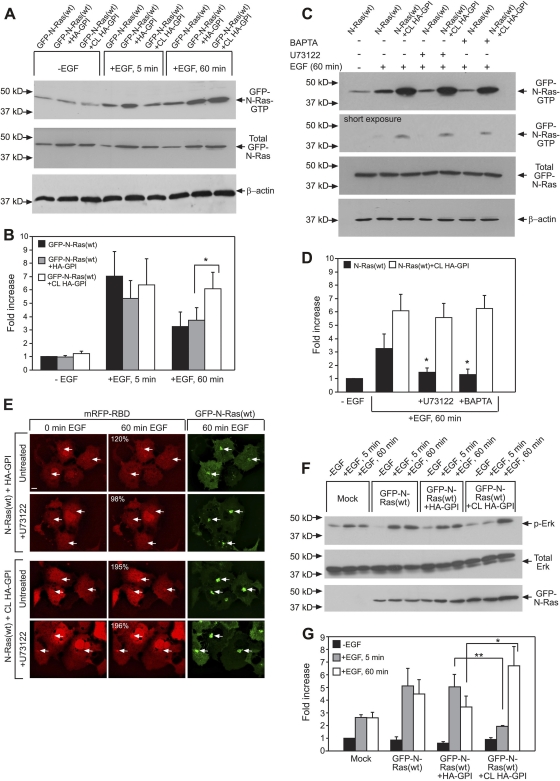
The enhanced PM-cytoplasm exchange of activated N-Ras following HA-GPI clustering may increase the accumulation of activated N-Ras in the GC. To explore this possibility, we first employed quantitative live-cell confocal microscopy (Fig. 5). COS-7 cells coexpressing GFP–N-Ras(wt), mRFP-RBD, and HA-GPI were serum starved and subjected (or not) to IgG cross-linking of HA-GPI. EGF was added, and the same cells were imaged at 37°C. Figure 5C shows representative images of mRFP-RBD, which binds specifically to GTP-loaded Ras, at a focal plane with distinct GC fluorescence; the GC is identified by the high concentration of GFP–N-Ras(wt), validated by colocalization with the GC marker GM130 (Fig. 5A). Since GFP–N-Ras is the major Ras protein in the transfected cells and the endogenous Ras level is insufficient to detect GC accumulation of mRFP-RBD (Fig. 5B), the increase in GC-localized mRFP-RBD reflects GFP–N-Ras–GTP localization. To quantify this increase and enable statistical comparisons, we averaged multiple independent measurements (n = 9 or 10 in each case), calculating the relative increase in GC-localized mRFP-RBD fluorescence from the z-stacks of the 3-D images (Fig. 5D). These studies show that HA-GPI clustering markedly enhanced the GC accumulation of EGF-activated (30 and 60 min) GFP–N-Ras. This is in line with the delayed and sustained activation of N- and H-Ras in the GC by EGF versus their rapid and transient activation (2 to 10 min) at the PM of fibroblasts and epithelial cells (6, 9, 48).
We took advantage of these different activation time scales to obtain a biochemical corollary to the microscopy studies, probing for effects of HA-GPI clustering on EGF-induced GFP–N-Ras activation. Ras-GTP was pulled down by beads coupled to GST-RBD from unstimulated or EGF-stimulated (5 or 60 min) cells coexpressing GFP–N-Ras(wt) and HA-GPI, with or without HA clustering. As shown in Fig. 6A and B, after a short EGF activation (5 min; signaling mainly from the PM), HA-GPI cross-linking had no significant effect on the GTP loading of GFP–N-Ras(wt). In contrast, after prolonged EGF stimulation (60 min), when N-Ras signals mainly from the GC, HA-GPI clustering significantly enhanced GFP–N-Ras–GTP, concomitant with the accumulation of activated N-Ras in the GC demonstrated by mRFP-RBD (Fig. 5C and D). Since EGF may also activate N- and H-Ras directly in the GC via Src-dependent activation of phospholipase Cγ1 (PLCγ1) by a mechanism involving Ca2+ and the exchange factor RasGRP1 (6, 9, 45), the accumulation of N-Ras–GTP at the GC could also be due to direct activation of N-Ras in the GC. We therefore conducted analogous studies on cells treated with inhibitors of the above pathway, the Ca2+ chelator BAPTA-AM and the PLC inhibitor U73122 (6). Both inhibitors partially reduced GFP–N-Ras activation at the GC following prolonged incubation with EGF but failed to inhibit GFP–N-Ras–GTP GC accumulation in response to HA-GPI cross-linking (Fig. 6C and D). These findings were validated by the results of live-cell confocal microscopy (Fig. 6E). This indicates that the major portion of the enhanced GFP–N-Ras–GTP accumulation in the GC following HA-GPI cross-linking is not due to direct activation in the GC.
Fig. 6.
Cross-linking of HA-GPI differentially modulates GFP–N-Ras(wt) activation and signaling after short and long EGF stimulation. For biochemical assays (A to D, F, and G), COS-7 cells were transfected with GFP–N-Ras(wt), GFP–N-Ras(wt) plus HA-GPI, or empty vector (Mock). They were serum starved, subjected (or not) to HA-GPI cross-linking (CL), and/or stimulated with EGF, followed by GST-RBD pulldown and immunoblotting (see Materials and Methods). (A and B) Representative blots (A) and quantification (means ± SEM, n = 3) of the fold increase [relative to the results for unstimulated cells expressing GFP–N-Ras(wt)] in the ratio of GTP-bound to total GFP–N-Ras (transfection control) (B). β-Actin served as loading control. An asterisk indicates a significant difference between the results for similarly treated HA-GPI-expressing cells with or without cross-linking (P < 0.02; Student's t test). (B and C) Representative blots (C) and quantification (means ± SEM, n = 4) of the effects of BAPTA-AM (BAPTA) and U73122 on GFP–N-Ras–GTP pulldown (D). The inhibitors (or 0.1% DMSO; control) were added (30 min, 37°C, 10 μM) before HA-GPI cross-linking and retained. The inhibitors significantly reduced GFP–N-Ras–GTP in non-cross-linked cells stimulated with EGF (60 min) (black bars) (*, P < 0.02; Student's t test) but had no effect in cells subjected to HA-GPI cross-linking (white bars). The lack of effect of the inhibitors was validated by the results of short exposure of the blots (C, second panel). (E) Live-cell confocal analysis demonstrates no effect of U73122 on mRFP-RBD GC accumulation. Transfection and experimental details were as described in the Fig. 5C legend. Quantification of the relative percentages of GC accumulation of mRFP-RBD (upper left corner of the middle panels) was as described in the Fig. 5D legend, extracting the values from the cells indicated by the arrows pointing at the GC. The images show typical fields. Bar, 10 μm. Inhibition with BAPTA gave similar results (not shown). (F and G) Representative blots (F) and quantification of EGF-stimulated phospho-Erk (p-Erk) formation (means ± SEM, n = 4) normalized to total Erk (G). Results depict fold increase in calibrated p-Erk level relative to that in unstimulated control. Asterisks indicate significant differences between the results for HA-GPI-expressing cells with or without cross-linking after EGF stimulation, comparing samples stimulated with EGF for the same duration (*, P < 0.02; **, P < 0.004).
To investigate the effects of cross-linking HA-GPI on the downstream signaling of EGF-stimulated N-Ras via Raf/Erk, we employed a similar experimental setup, probing the cell lysates for Erk phosphorylation. Figure 6F depicts the results of a representative experiment; the quantification of several independent experiments is shown in Fig. 6G. In mock-transfected cells, EGF mildly stimulated phospho-Erk (p-Erk) formation, reflecting endogenous Ras activity. Under these conditions, EGF stimulation for 5 or 60 min yielded similar p-Erk levels. The expression of GFP–N-Ras(wt) markedly increased p-Erk formation following 5 or 60 min of EGF stimulation, augmenting p-Erk to similar extents. Coexpression with HA-GPI did not affect Erk activation at either 5 or 60 min of EGF stimulation. However, cross-linking of HA-GPI significantly reduced p-Erk formation at 5 min of EGF stimulation and strongly augmented p-Erk after 60 min of stimulation (Fig. 6G). Thus, HA-GPI cross-linking reduces EGF-induced N-Ras PM signaling while enhancing its prolonged signaling from the GC, in accord with the enhanced PM-cytoplasm exchange of activated N-Ras (Fig. 2) and its accumulation in the GC (Fig. 5 and 6) in response to HA-GPI clustering.
Inhibition of depalmitoylation abrogates the shift of activated N-Ras to exchange and its accumulation in the GC in response to cross-linking HA-GPI.
The GC-PM shuttling of N-Ras and H-Ras is regulated by palmitoylation in the GC and depalmitoylation at the PM and other membranes (38, 46, 48, 59). To examine whether the effect of HA-GPI cross-linking on N-Ras–GTP association with the PM depends on depalmitoylation, we employed palmostatin B, a recently developed inhibitor of APT1 (acyl protein thioesterase 1), which inhibits N-Ras depalmitoylation (11). Cells coexpressing GFP–N-Ras (G13V or wt) and HA-GPI were treated with 10 μM palmostatin B, a concentration that inhibits depalmitoylation in a short time (15 min at 37°C) such that N-Ras still retains its overall localization in the PM and GC (11). This was followed by HA-GPI cross-linking and FRAP beam size analysis. Palmostatin B fully abolished the shift of GFP–N-Ras(G13V) and EGF-stimulated (4 min) GFP–N-Ras(wt) to exchange in response to HA-GPI cross-linking (Fig. 7A and B). The vehicle (0.5% DMSO) did not affect the membrane interactions of the GFP–N-Ras isoforms, while treatment with palmostatin B without HA-GPI cross-linking induced only a mild shift toward exchange, potentially due to some alterations in the PM composition or biophysical properties following inhibition of depalmitoylation. Importantly, palmostatin B also abrogated the ability of HA-GPI cross-linking to enhance the GC accumulation of GTP-loaded GFP–N-Ras(wt) following EGF stimulation (Fig. 7C). The notion that the shift of activated N-Ras to exchange following HA-GPI clustering is due to enhanced depalmitoylation is supported by the finding that nonpalmitoylated N-Ras shows FRAP by exchange. Thus, nonpalmitoylable GFP–N-Ras(C181S) (wt or constitutively active) or GFP–N-Ras (wt or G13V) in cells incubated for 24 h with 50 μM palmitoylation inhibitor 2BP (treatment was as described in reference 2) exhibited largely decreased (but measurable) PM localization. In all these cases, the τ(40×)/τ(63×) ratio (n = 30 for each protein) was not significantly different from 1 (P > 0.1, bootstrap analysis).
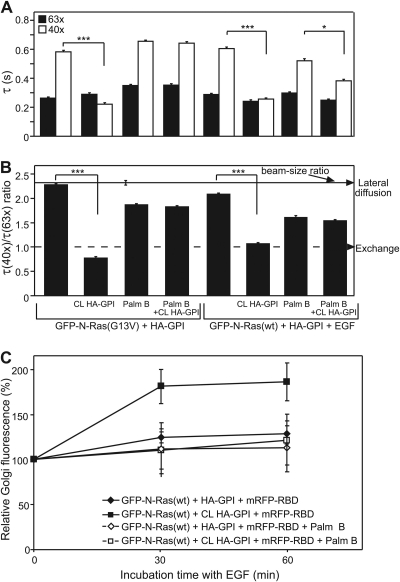
Fig. 7.
Effects of HA-GPI cross-linking on the PM interactions and GC accumulation of N-Ras–GTP are blocked by palmostatin B (Palm B). (A and B) Patch-FRAP studies. COS-7 cells were transfected with GFP–N-Ras (wt or G13V) plus HA-GPI, serum starved, and EGF stimulated (4 min) as described in the Fig. 2 legend. Palmostatin B (10 μM, 15 min, 37°C) was added prior to HA-GPI cross-linking (CL). DMSO (0.5%, control samples) had no effect. Palmostatin B at 50 μM yielded identical results. Bars, means ± SEM (n = 30 to 60). (A) τ values. Pairs of similarly treated cells expressing the same GFP–N-Ras protein with or without HA-GPI cross-linking were compared. The reduction in the τ(40×) of activated GFP–N-Ras induced by HA-GPI cross-linking (***, P < 10−12; Student's t test) was abolished by palmostatin B for N-Ras(G13V) (P > 0.6) and strongly compromised for EGF-stimulated N-Ras(wt) (*, P < 0.005). (B) τ(40×)/τ(63×) ratios. Bootstrap analysis showed that the significant effect of HA-GPI cross-linking on the τ ratio of activated N-Ras (***, P < 10−20) was abolished by palmostatin B (P > 0.12). (C) GC accumulation of GFP–N-Ras–GTP (means ± SEM, 9 to 10 cells per time point/condition). COS-7 cells triple transfected with GFP–N-Ras(wt), HA-GPI, and mRFP-RBD (see the Fig. 5 legend) were treated with palmostatin B as described above. EGF stimulation, 3-D live-cell imaging, and quantification of GC mRFP-RBD fluorescence were as described in the Fig. 5 legend.
Cross-linking by fibronectin induces a cholesterol- and depalmitoylation-dependent shift of N-Ras–GTP to exchange.
To explore whether the modulation of N-Ras-PM interactions by HA-GPI cross-linking extends to clustering of endogenous cell-surface proteins by natural ligands, we studied the effects of cross-linking by fibronectin. Fibronectin binds to α5β1, αVβ1, and α8β1 integrins, and numerous reports have tied β1 integrin to rafts/caveolae (25, 35, 57, 61). Fibronectin binding markedly reduced the τ(40×) and the τ(40×)/τ(63×) ratio of N-Ras(G13V) in COS-7 cells (Fig. 8A and C), indicating a significant increase in its exchange rate. The clustering-based nature of the effect is exemplified by the augmented shift to exchange after further cross-linking of fibronectin by antifibronectin antibodies (Fig. 8A and C).
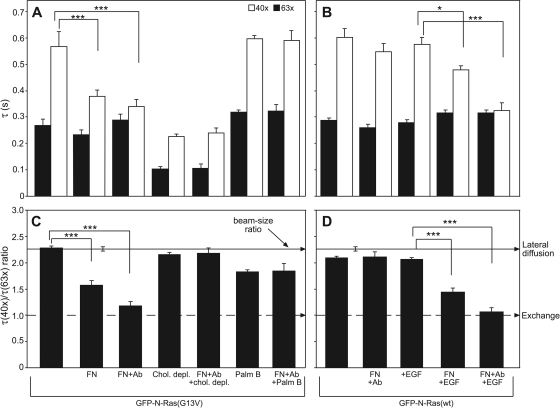
Fig. 8.
Fibronectin (FN) binding to its surface receptors recapitulates the effects of HA-GPI clustering on GFP–N-Ras-PM interactions. FRAP beam size analysis (means ± SEM, n = 30 to 60) was performed on COS-7 cells expressing GFP–N-Ras (G13V or wt) as described in the Fig. 2 legend. (A and C) Fibronectin effects on GFP–N-Ras(G13V). Where indicated, cells were cholesterol depleted (as described in the Fig. 3 legend) or treated with palmostatin B (Palm B) (see the Fig. 7 legend). They were then incubated with fibronectin alone or followed by antifibronectin (+Ab [antibody]). (A) Fibronectin significantly reduced the τ(40×) of N-Ras(G13V), an effect augmented by antifibronectin (***, P < 10−12; Student's t test). These effects were abrogated by cholesterol depletion or palmostatin B (P > 0.1). (C) Concomitantly, fibronectin (with or without Ab) significantly reduced the τ ratios of GFP–N-Ras(G13V) (***, P < 10−12; bootstrap analysis). These effects were abolished by cholesterol depletion or palmostatin B (P > 0.4). (B and D) Fibronectin effects on GFP–N-Ras(wt). Cells were serum starved, incubated at 4°C with fibronectin followed by antifibronectin (+Ab), and stimulated by EGF (100 ng/ml, 4 min, 37°C) where indicated. Fibronectin or EGF stimulation alone did not affect the τ value or the τ ratio of N-Ras(wt); however, combining EGF with fibronectin cross-linking significantly reduced the τ(40×) (***, P < 10−12; *, P < 0.01 [Student's t test]) and the τ ratio (***, P < 10−12; bootstrap analysis).
Analogous studies on cells expressing GFP–N-Ras(wt) without and with EGF activation (Fig. 8B and D) recapitulated the requirement for GTP loading of N-Ras, as in the case of HA-GPI cross-linking. Cross-linking with fibronectin and antifibronectin, as well as EGF stimulation without fibronectin, failed to enhance the exchange of GFP–N-Ras(wt). In contrast, EGF stimulation (which induced a 7-fold increase in GFP–N-Ras–GTP) (Fig. 6B) on top of fibronectin strongly enhanced the exchange of GFP–N-Ras(wt) (Fig. 8B and D). Importantly, cholesterol depletion or treatment with palmostatin B abolished the effects of fibronectin and antifibronectin on the interactions of GFP–N-Ras–GTP with the PM (Fig. 8A and C), in complete analogy to modulation by HA-GPI cross-linking.
DISCUSSION
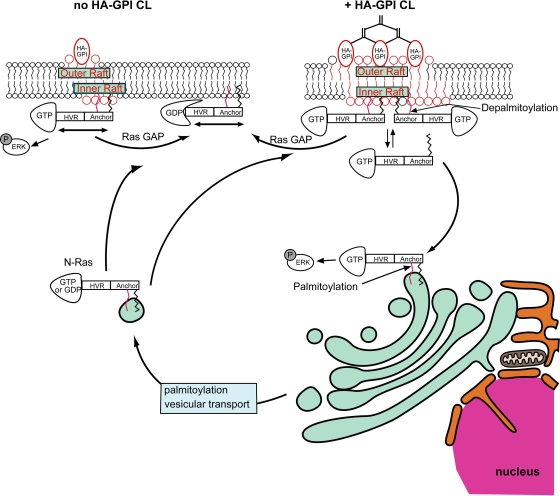
The current studies demonstrate interdependence between the raft association of activated N-Ras, its depalmitoylation, and its signaling from distinct cellular locations (PM versus GC). We show that cross-linking of raft proteins stimulates N-Ras–GTP depalmitoylation, enhancing its dissociation from PM raft clusters, culminating in its accumulation in and signaling from the GC. We propose a novel mechanism (Fig. 9) whereby costimulation by clustering of raft proteins by multivalent ligands or antibodies can alter the pattern of N-Ras activation by EGF or other stimuli.
Fig. 9.
A model for the modulation of N-Ras membrane interactions and PM/GC signaling by clustering raft proteins. The model depicts cells expressing N-Ras and HA-GPI as a representative raft-associated protein. Similar effects can be induced by clustering fibronectin receptors or other raft proteins. N-Ras interacts with the membrane mainly by the farnesyl (black) and single palmitoyl (red) residues at the C-terminal anchor; additional interactions (not shown) are provided by the hypervariable (HVR) region. N-Ras–GTP has preferential dynamic interactions with rafts (Fig. 1) in the inner leaflet (upper left; no HA-GPI cross-linking), from which most of its PM signaling (i.e., p-Erk formation) emanates. Cross-linking of (CL) raft proteins by IgG (HA-GPI) or ligands (e.g., fibronectin) clusters and stabilizes raft domains. This enhances the recruitment of N-Ras–GTP to raft clusters and increases its susceptibility to depalmitoylation, either by association with raft protein(s) that confer a topological orientation/conformation that is highly sensitive to depalmitoylation or by increased proximity to depalmitoylating enzymes. The depalmitoylation weakens the interactions of N-Ras–GTP with the PM, enhancing its PM-cytoplasm exchange (Fig. 2 to 4, 7, and 8). The depalmitoylated N-Ras–GTP diffuses through the cytoplasm and interacts with all cellular membranes. With time, repalmitoylation in the GC results in N-Ras–GTP accumulation in and signaling from the GC (Fig. 5 to 7). This mechanism links costimulation by ligands that cross-link raft proteins with the reshaping of the response of N-Ras to a primary stimulus (e.g., EGF).
Preferential association of N-Ras–GTP with raft assemblies is essential for the model. Figure 1B demonstrates that the τ(40×)/τ(63×) ratios for GFP–N-Ras(G13V) and GFP–N-Ras(wt) are similar to the beam size ratio (2.28), suggesting FRAP by lateral diffusion. This implies that the characteristic PM-cytoplasm exchange times (τex) are at least 10-fold higher than the τD (i.e., τex ≥ 3 s). Cholesterol depletion did not affect the FRAP mechanism, but it enhanced the lateral diffusion rates of GFP–N-Ras(G13V) (2.7-fold) and GDP-loaded GFP–N-Ras(wt) (1.4-fold). Thus, disruption of rafts releases GFP–N-Ras from mobility-restricting interactions, as demonstrated by us earlier for the raft-interacting H-Ras but not for the nonraft K-Ras (37, 55). These observations also hold at 37°C (Fig. 4) and for HeLa cells. The stronger effect on GTP-loaded N-Ras(G13V) (Fig. 1B) suggests that the affinity of N-Ras–GTP to rafts is higher than that of N-Ras–GDP. Accordingly, EM spatial mapping (Fig. 1C and D) shows higher nanoclustering for GFP–N-Ras(G13V), which is more sensitive to cholesterol depletion.
Cross-linking GPI-anchored raft proteins was shown to stabilize raft clusters, enhancing transient, cholesterol-dependent recruitment of signaling proteins in the internal PM leaflet to the clusters (8, 15, 58). Analogous enhanced recruitment of activated N-Ras to raft clusters is an inherent feature of the mechanism depicted in Fig. 9. However, for N-Ras–GTP, this recruitment dramatically increases the PM-cytoplasm exchange (Fig. 2), due to enhanced depalmitoylation (Fig. 7). Thus, HA-GPI cross-linking shifts GFP–N-Ras–GTP but not GFP–N-Ras–GDP to FRAP by exchange (Fig. 2C to F). The measured τ (0.2 to 0.3 s) under these conditions is τex, which is at least 10-fold shorter than the estimated τex (≥3 s) prior to HA-GPI cross-linking, suggesting a faster dissociation rate (62) and implying weaker PM association. This view is supported by the results of EM studies showing reduced levels of GFP–N-Ras(G13V) in clusters (which are mainly cholesterol sensitive) following HA-GPI cross-linking, in accord with preferential loss of N-Ras–GTP from cholesterol-dependent clusters (Fig. 2G). Importantly, the cross-linking-mediated enhanced exchange of N-Ras–GTP requires raft association, as evidenced by (i) the failure of cross-linking the nonraft HA(2A520) mutant to enhance the exchange (Fig. 2C and D) and (ii) the disruption of the shift of GFP–N-Ras(G13V) to exchange by cholesterol depletion (Fig. 3 and 4). Notably, the ability to enhance the exchange of N-Ras–GTP is not limited to cross-linking HA-GPI; as shown in Fig. 8, a cholesterol-dependent shift to exchange of activated GFP–N-Ras can be mediated by cross-linking with the natural integrin ligand fibronectin. For cross-linking of either HA-GPI or fibronectin receptors, all three parameters (formation of N-Ras–GTP, existence of raft domains, and raft protein cross-linking) have to be met in order to shift N-Ras to exchange. Thus, the cross-linking is a costimulatory signal, which operates only when combined with a bona fide N-Ras-activating stimulus.
Stimulation by EGF without cross-linking HA-GPI is not sufficient to enhance GFP–N-Ras exchange (Fig. 2D and E), in line with the FRAP by lateral diffusion of the GTP-loaded GFP–N-Ras(G13V) in non-cross-linked cells (Fig. 1 and 2). Notably, HA-GPI (Fig. 2C and E) or fibronectin receptor cross-linking (Fig. 8A and C) shift GFP–N-Ras(G13V) to exchange without EGF stimulation. This suggests that the exchange-enhancing mechanism (Fig. 9) is distinct from pathways that require EGF activation of phospholipase D2 (PLD2) to produce phosphatidic acid. Such stimulation by EGF in nonlymphoid cells was shown to enhance H-Ras activation in the PM via Sos recruitment (63) and to increase the number and size of cholesterol-sensitive EGF receptor nanoclusters (3).
The enhanced exchange of N-Ras–GTP after HA-GPI cross-linking modulates both its PM/GC distribution and its signaling in response to EGF (Fig. 5 and 6). The weaker PM association leads to N-Ras–GTP accumulation at the GC in response to prolonged EGF stimulation (Fig. 5 and 6A and B). Notably, inhibition of the Src/PLCγ1/RasGRP1 pathway, which directly activates N- and H-Ras in the GC (6, 41), by BAPTA-AM or U73122 did not reduce the enhanced GC accumulation of N-Ras–GTP in response to HA-GPI cross-linking together with long EGF stimulation (Fig. 6C to E). Thus, the latter accumulation is mainly due to PM-to-GC transfer of N-Ras–GTP rather than in situ activation of N-Ras at the GC. Moreover, HA-GPI cross-linking reduced p-Erk formation following short (5-min) EGF stimulation (i.e., transient N-Ras activation in the PM) but increased p-Erk after prolonged EGF stimulation (delayed and sustained activation in the GC) (6, 48). Thus, HA-GPI cross-linking augments N-Ras/Mek/Erk signaling from the GC at the expense of N-Ras PM signaling, parallel to the enhanced GC accumulation of N-Ras–GTP. The prolonged Ras signaling from the GC was proposed to arise due to the susceptibility of Ras in the PM but not the GC to the Ras GTPase-activating protein CAPRI (6). Specific biological functions have been attributed to GC-localized activated Ras, as positive thymocyte selection was shown to require endogenous GC Ras activation (36). Alternatively, sequestration of Ras effectors in the GC has been proposed as a potential mechanism for reducing Ras output (16, 60).
The last requirement for shifting N-Ras–GTP to exchange by raft clustering is enhancement of its depalmitoylation (Fig. 9). This is demonstrated by the ability of palmostatin B to block the shift of N-Ras–GTP to exchange following cross-linking of HA-GPI or fibronectin receptors (Fig. 7 and 8). Concomitantly, palmostatin B abrogates the enhanced accumulation of EGF-stimulated N-Ras(wt) in the GC (Fig. 7C), in line with the regulation of the PM/GC distribution of N- and H-Ras by de-/repalmitoylation (46, 48). The notion that depalmitoylated N-Ras shifts to exchange is supported by the exchange-based FRAP mechanism of unpalmitoylated N-Ras (C181S mutants or GFP–N-Ras in cells treated with 2BP). The undetectable shift of GFP–N-Ras to exchange in the absence of raft protein cross-linking may seem at odds with the de-/repalmitoylation cycle that continually regulates N-Ras PM/GC distribution (46). However, FRAP by lateral diffusion does not imply that there is no exchange; rather, it indicates that the exchange is significantly slower (24). In addition, the FRAP studies measure the transient interaction kinetics of GFP–N-Ras proteins with the PM, which occur on a much shorter timescale than N-Ras accumulation at the GC (46, 48). The lower rate of the latter process may reflect the random redistribution of depalmitoylated N-Ras proteins over all membranes, increasing the time required for their kinetic trapping (46, 53) in the GC upon repalmitoylation (Fig. 9). Finally, enhanced susceptibility of raft-localized N-Ras–GTP to depalmitoylation may provide the link between the preferential localization of N-Ras–GTP in raft clusters and its selective shift to exchange following raft-protein clustering (Fig. 9). Although this localization may increase its proximity to raft-associated depalmitoylating enzymes, the finding that depalmitoylation occurs everywhere in the cell (46) argues against it. We therefore propose that binding of N-Ras–GTP to proteins or scaffolds associated with raft clusters can alter the membrane topology of N-Ras–GTP and/or induce a conformational change that renders the thioester group highly accessible to depalmitoylating enzymes, in line with reports on enhanced palmitate turnover for activated Ras proteins (4, 31).
The differential association of Ras proteins with raft clusters may underlie the specificity of the raft-clustering effects. Only N-Ras displays preferential raft targeting of the GTP-loaded form; in H-Ras, the preference is for H-Ras–GDP, while K-Ras does not interact with rafts (43, 44, 50) and is therefore insensitive to HA-GPI cross-linking (15). For N-Ras, both GTP-loading and association with raft clusters are required to enhance its depalmitoylation and exchange. On the other hand, H-Ras membrane interactions and signaling are modulated differently by raft clustering; cross-linking HA-GPI slows the dynamic H-Ras-raft cluster interaction (without a shift to exchange), inhibiting the transport of H-Ras to its nonraft signaling sites (15). This difference may be due to the lower affinity of H-Ras–GTP to rafts, assisted by its association with nonraft nanoclusters and scaffold proteins, e.g., galectin-1 (5). Moreover, the lower probability for the concomitant removal of two palmitates (H-Ras) may result in slower dissociation from the PM, in line with its slower transport to the GC (48).
In summary, the current studies reveal a novel mechanism that links together raft clustering, selective association with GTP-loaded palmitoylated Ras isoforms, and depalmitoylation (Fig. 9). This mechanism allows costimulation by cross-linked raft proteins to alter the kinetic and spatial pattern of N-Ras activation and signaling in response to primary stimuli (e.g., by EGF), reducing short-term signaling from the PM and increasing long-term signaling from the GC.
ACKNOWLEDGMENTS
We thank T. J. Braciale, A. Burgess, L. H. Chamberlain, B. Geiger, M. Philips, M. G. Roth, J. J. Skehel, and J. M. White for reagents and P. I. Bastiaens and A. Mor for advice.
This work was supported by a grant from the Ministry of Science & Technology, Israel, and the Ministry of Research, France (to Y.I.H.). S.E. was supported by a Nehemia Levtzion Ph.D. Fellowship (Council of Higher Education, Israel). Y.I.H. is an incumbent of the Zalman Weinberg Chair in Cell Biology.
Footnotes
Published ahead of print on 1 August 2011.
REFERENCES
- 1. Agard D. A., Hiraoka Y., Shaw P., Sedat J. W. 1989. Fluorescence microscopy in three dimensions. Methods Cell Biol. 30:353–377 [DOI] [PubMed] [Google Scholar]
- 2. Ahearn I. M., et al. 2011. FKBP12 binds to acylated H-ras and promotes depalmitoylation. Mol. Cell 41:173–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ariotti N., et al. 2010. Epidermal growth factor receptor activation remodels the plasma membrane lipid environment to induce nanocluster formation. Mol. Cell. Biol. 30:3795–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baker T. L., Zheng H., Walker J., Coloff J. L., Buss J. E. 2003. Distinct rates of palmitate turnover on membrane-bound cellular and oncogenic H-ras. J. Biol. Chem. 278:19292–19300 [DOI] [PubMed] [Google Scholar]
- 5. Belanis L., Plowman S. J., Rotblat B., Hancock J. F., Kloog Y. 2008. Galectin-1 is a novel structural component and a major regulator of H-Ras nanoclusters. Mol. Biol. Cell 19:1404–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bivona T. G., et al. 2003. Phospholipase Cγ activates Ras on the Golgi apparatus by means of RasGRP1. Nature 424:694–698 [DOI] [PubMed] [Google Scholar]
- 7. Bos J. L. 1989. Ras oncogenes in human cancer: a review. Cancer Res. 49:4682–4689 [PubMed] [Google Scholar]
- 8. Chen Y., Veracini L., Benistant C., Jacobson K. 2009. The transmembrane protein CBP plays a role in transiently anchoring small clusters of Thy-1, a GPI-anchored protein, to the cytoskeleton. J. Cell Sci. 122:3966–3972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chiu V. K., et al. 2002. Ras signalling on the endoplasmic reticulum and the Golgi. Nat. Cell Biol. 4:343–350 [DOI] [PubMed] [Google Scholar]
- 10. Choy E., et al. 1999. Endomembrane trafficking of Ras: the CAAX motif targets proteins to the ER and Golgi. Cell 98:69–80 [DOI] [PubMed] [Google Scholar]
- 11. Dekker F. J., et al. 2010. Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat. Chem. Biol. 6:449–456 [DOI] [PubMed] [Google Scholar]
- 12. Diggle P. J., Morris S. E., Wakefield J. C. 2000. Point-source modelling using matched case-control data. Biostatistics 1:89–105 [DOI] [PubMed] [Google Scholar]
- 13. Efron B., Tibshirani R. 1993. Estimates of bias, p. 124–130In Cox D. R., Hinkley D. V., Reid N., Rubin D. B., Silverman B. W.(ed.), An introduction to the bootstrap. Chapman & Hall, Boca Raton, FL [Google Scholar]
- 14. Eisenberg S., Giehl K., Henis Y. I., Ehrlich M. 2008. Differential interference of chlorpromazine with the membrane interactions of oncogenic K-Ras and its effects on cell growth. J. Biol. Chem. 283:27279–27288 [DOI] [PubMed] [Google Scholar]
- 15. Eisenberg S., Shvartsman D. E., Ehrlich M., Henis Y. I. 2006. Clustering of raft-associated proteins in the external membrane leaflet modulates internal leaflet H-Ras diffusion and signaling. Mol. Cell. Biol. 26:7190–7200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feng L., et al. 2007. Spatial regulation of Raf kinase signaling by RKTG. Proc. Natl. Acad. Sci. U. S. A. 104:14348–14353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fridman M., et al. 2000. Point mutants of c-raf-1 RBD with elevated binding to v-Ha-Ras. J. Biol. Chem. 275:30363–30371 [DOI] [PubMed] [Google Scholar]
- 18. Gutman O., Walliser C., Piechulek T., Gierschik P., Henis Y. I. 2010. Differential regulation of phospholipase C-β2 activity and membrane interaction by Gαq, Gβ1γ2, and Rac2. J. Biol. Chem. 285:3905–3915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hancock J. F. 2006. Lipid rafts: contentious only from simplistic standpoints. Nat. Rev. Mol. Cell Biol. 7:456–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hancock J. F., Cadwallader K., Paterson H., Marshall C. J. 1991. A CAAX or a CAAL motif and a second signal are sufficient for plasma membrane targeting of ras proteins. EMBO J. 10:4033–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hancock J. F., Paterson H., Marshall C. J. 1990. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell 63:133–139 [DOI] [PubMed] [Google Scholar]
- 22. Henis Y. I., Hancock J. F., Prior I. A. 2009. Ras acylation, compartmentalization and signaling nanoclusters. Mol. Membr. Biol. 26:80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Henis Y. I., Katzir Z., Shia M. A., Lodish H. F. 1990. Oligomeric structure of the human asialoglycoprotein receptor: nature and stoichiometry of mutual complexes containing H1 and H2 polypeptides assessed by fluorescence photobleaching recovery. J. Cell Biol. 111:1409–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Henis Y. I., Rotblat B., Kloog Y. 2006. FRAP beam-size analysis to measure palmitoylation-dependent membrane association dynamics and microdomain partitioning of Ras proteins. Methods 40:183–190 [DOI] [PubMed] [Google Scholar]
- 25. Ichikawa N., et al. 2009. Binding of laminin-1 to monosialoganglioside GM1 in lipid rafts is crucial for neurite outgrowth. J. Cell Sci. 122:289–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jacobson K., Mouritsen O. G., Anderson R. G. 2007. Lipid rafts: at a crossroad between cell biology and physics. Nat. Cell Biol. 9:7–14 [DOI] [PubMed] [Google Scholar]
- 27. Kemble G. W., Danieli T., White J. M. 1994. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell 76:383–391 [DOI] [PubMed] [Google Scholar]
- 28. Kfir S., et al. 2005. Pathway- and expression level-dependent effects of oncogenic N-Ras: p27Kip1 mislocalization by the Ral-GEF pathway and Erk-mediated interference with Smad signaling. Mol. Cell. Biol. 25:8239–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Laude A. J., Prior I. A. 2008. Palmitoylation and localisation of RAS isoforms are modulated by the hypervariable linker domain. J. Cell Sci. 121:421–427 [DOI] [PubMed] [Google Scholar]
- 30. Lin S., Naim H. Y., Rodriguez A. C., Roth M. G. 1998. Mutations in the middle of the transmembrane domain reverse the polarity of transport of the influenza virus hemagglutinin in MDCK epithelial cells. J. Cell Biol. 142:51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Magee A. I., Gutierrez L., McKay I. A., Marshall C. J., Hall A. 1987. Dynamic fatty acylation of p21N-ras. EMBO J. 6:3353–3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Malumbres M., Barbacid M. 2003. RAS oncogenes: the first 30 years. Nat. Rev. Cancer. 3:459–465 [DOI] [PubMed] [Google Scholar]
- 33. Matallanas D., et al. 2006. Distinct utilization of effectors and biological outcomes resulting from site-specific Ras activation: Ras functions in lipid rafts and Golgi complex are dispensable for proliferation and transformation. Mol. Cell. Biol. 26:100–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mitin N., Rossman K. L., Der C. J. 2005. Signaling interplay in Ras superfamily function. Curr. Biol. 15:R563–R574 [DOI] [PubMed] [Google Scholar]
- 35. Mocanu M. M., et al. 2005. Associations of ErbB2, beta1-integrin and lipid rafts on Herceptin (trastuzumab) resistant and sensitive tumor cell lines. Cancer Lett. 227:201–212 [DOI] [PubMed] [Google Scholar]
- 36. Mor A., et al. 2007. The lymphocyte function-associated antigen-1 receptor costimulates plasma membrane Ras via phospholipase D2. Nat. Cell Biol. 9:713–719 [DOI] [PubMed] [Google Scholar]
- 37. Niv H., Gutman O., Kloog Y., Henis Y. I. 2002. Activated K-Ras and H-Ras display different interactions with saturable nonraft sites at the surface of live cells. J. Cell Biol. 157:865–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ohno Y., Kihara A., Sano T., Igarashi Y. 2006. Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim. Biophys. Acta 1761:474–483 [DOI] [PubMed] [Google Scholar]
- 39. Omerovic J., Hammond D. E., Clague M. J., Prior I. A. 2008. Ras isoform abundance and signalling in human cancer cell lines. Oncogene 27:2754–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Paz A., Haklai R., Elad-Sfadia G., Ballan E., Kloog Y. 2001. Galectin-1 binds oncogenic H-Ras to mediate Ras membrane anchorage and cell transformation. Oncogene 20:7486–7493 [DOI] [PubMed] [Google Scholar]
- 41. Perez de Castro I., Bivona T. G., Philips M. R., Pellicer A. 2004. Ras activation in Jurkat T cells following low-grade stimulation of the T-cell receptor is specific to N-Ras and occurs only on the Golgi apparatus. Mol. Cell. Biol. 24:3485–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Plowman S. J., Muncke C., Parton R. G., Hancock J. F. 2005. H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc. Natl. Acad. Sci. U. S. A. 102:15500–15505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Prior I. A., et al. 2001. GTP-dependent segregation of H-ras from lipid rafts is required for biological activity. Nat. Cell Biol. 3:368–375 [DOI] [PubMed] [Google Scholar]
- 44. Prior I. A., Muncke C., Parton R. G., Hancock J. F. 2003. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 160:165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Quatela S. E., Philips M. R. 2006. Ras signaling on the Golgi. Curr. Opin. Cell Biol. 18:162–167 [DOI] [PubMed] [Google Scholar]
- 46. Rocks O., et al. 2010. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 141:458–471 [DOI] [PubMed] [Google Scholar]
- 47. Rocks O., Peyker A., Bastiaens P. I. 2006. Spatio-temporal segregation of Ras signals: one ship, three anchors, many harbors. Curr. Opin. Cell Biol. 18:351–357 [DOI] [PubMed] [Google Scholar]
- 48. Rocks O., et al. 2005. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 307:1746–1752 [DOI] [PubMed] [Google Scholar]
- 49. Rodenhuis S. 1992. ras and human tumors. Semin. Cancer Biol. 3:241–247 [PubMed] [Google Scholar]
- 50. Roy S., et al. 2005. Individual palmitoyl residues serve distinct roles in H-ras trafficking, microlocalization, and signaling. Mol. Cell. Biol. 25:6722–6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Saffman P. G., Delbruck M. 1975. Brownian motion in biological membranes. Proc. Natl. Acad. Sci. U. S. A. 72:3111–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Scheiffele P., Roth M. G., Simons K. 1997. Interaction of influenza virus haemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domain. EMBO J. 16:5501–5508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shahinian S., Silvius J. R. 1995. Doubly-lipid-modified protein sequence motifs exhibit long-lived anchorage to lipid bilayer membranes. Biochemistry 34:3813–3822 [DOI] [PubMed] [Google Scholar]
- 54. Sharma P., et al. 2004. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell 116:577–590 [DOI] [PubMed] [Google Scholar]
- 55. Shvartsman D. E., Gutman O., Tietz A., Henis Y. I. 2006. Cyclodextrins but not compactin inhibit the lateral diffusion of membrane proteins independent of cholesterol. Traffic 7:917–926 [DOI] [PubMed] [Google Scholar]
- 56. Shvartsman D. E., Kotler M., Tall R. D., Roth M. G., Henis Y. I. 2003. Differently-anchored influenza hemagglutinin mutants display distinct interaction dynamics with mutual rafts. J. Cell Biol. 163:879–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Singh R. D., et al. 2010. Gangliosides and β1-integrin are required for caveolae and membrane domains. Traffic 11:348–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Suzuki K. G., et al. 2007. GPI-anchored receptor clusters transiently recruit Lyn and Gα for temporary cluster immobilization and Lyn activation: single-molecule tracking study 1. J. Cell Biol. 177:717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Swarthout J. T., et al. 2005. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J. Biol. Chem. 280:31141–31148 [DOI] [PubMed] [Google Scholar]
- 60. Torii S., Kusakabe M., Yamamoto T., Maekawa M., Nishida E. 2004. Sef is a spatial regulator for Ras/MAP kinase signaling. Dev. Cell 7:33–44 [DOI] [PubMed] [Google Scholar]
- 61. Wang C., et al. 2010. Regulation of integrin β1 recycling to lipid rafts by Rab1a to promote cell migration. J. Biol. Chem. 285:29398–29405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wolfenson H., et al. 2009. A role for the juxtamembrane cytoplasm in the molecular dynamics of focal adhesions. PLoS One 4:e4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhao C., Du G., Skowronek K., Frohman M. A., Bar-Sagi D. 2007. Phospholipase D2-generated phosphatidic acid couples EGFR stimulation to Ras activation by Sos. Nat. Cell Biol. 9:706–712 [DOI] [PubMed] [Google Scholar]