TEXT
A broken chromosome represents an immediate danger for a living cell; if it is not properly repaired, it may lead to cell death, loss of genetic information, or malignant transformation. Unfortunately, double-strand breaks (DSBs) form constantly, either as a consequence of external insults or as a product of normal cellular processes. Throughout the course of evolution, different repair mechanisms have evolved to repair these extremely dangerous lesions. In this issue, J.-H. Oum and colleagues (11) reveal a central role for the chromatin remodeling complex RSC in the processing and repair of DSBs. Surprisingly, their study links chromatin remodeling with the Rad59 homologous recombination protein and the cohesin complex, which holds chromatids together after DNA replication.
DSB repair mechanisms are usually divided into nonhomologous end joining (NHEJ), which uses little or no homology, and homologous recombination (HR)-based mechanisms, which rely on sequence similarity to achieve repair (reviewed in reference 2). During HR the broken ends are resected, generating single-stranded DNA (ssDNA) that can search the genome and invade similar sequences, copying a short stretch of information, which may result in a gene conversion (GC) event. This can be sometimes associated with a crossing-over, in which the interacting DNA molecules exchange strands. Under some circumstances a replication-like fork that may copy the whole chromosomal arm is created (break-induced replication [BIR]). An alternative pathway consists of the annealing of complementary strands of homology flanking the break to generate a deletion of the intervening sequence, in what is called a single-strand annealing (SSA) reaction (for a recent review of the various mechanisms of HR, see reference 1).
HR reactions are catalyzed by a group of proteins commonly called the RAD52 group, whose most prominent members are Rad51 and Rad52. Rad51 is the eukaryotic ortholog of bacterial RecA and is able to assemble on ssDNA to catalyze a strand exchange (“invasion”) reaction. The role of Rad52 in HR is less understood, although it seems to have two very different roles: on one hand it is necessary for Rad51 loading, whereas on the other hand it promotes annealing between complementary sequences (10). Rad52 plays a central role in HR, and mutations in the RAD52 gene abolish almost all types of recombination events. The Rad52 protein has been shown to form a multisubunit ring. Surprisingly, although all GC, SSA, and BIR events require Rad52 activity, Rad51 is required only for GC and for a subset of BIR events; SSA, on the other hand, is independent of Rad51 and requires Rad52 and in many instances Rad59, a protein that shares sequence similarity with Rad52, as well as a possible role in annealing (4–6).
In the last few years it has become apparent that, in addition to enzymes directly involved in DNA repair itself, additional activities are required in order to deal with the fact that recombination takes place in the context of chromatin. Thus, several chromatin-modifying enzymes have been shown to affect the ability of cells to repair DSBs. One such complex, RSC, is the subject of the paper by J.-H. Oum and colleagues (11). RSC comprises 15 different subunits and can modify the positions of nucleosomes along the chromosome in an ATP-dependent fashion. Surprisingly, this huge complex contains some subunits that are essential for life, whereas a second group of subunits is essential only at high temperatures and still other subunits can be deleted without a significant effect under normal (unstressed) growth conditions. RSC has been shown in the past to play important roles in DSB repair and in the loading of cohesin to the chromosomes (7, 12).
It has been known for some time that mutations in some RSC subunits cause the cells to become sensitive to a variety of DNA-damaging agents. In this paper, the authors show that in the absence of RSC subunits the genome becomes unstable and that cells with fragmented genomes or showing signs of DNA damage response can be seen. The defect in RSC-mutated cells appears to be at the level of DNA repair rather than checkpoint signaling. Indeed, experiments carried out with cells arrested at various stages of the cell cycle showed that rsc (rsc2, rsc7) cells are unable to repair their chromosomes when exposed to methyl methanesulfonate (MMS) in G2, whereas they are less affected at the G1 stage. To better define the mechanism of repair involved, the authors created rsc rad51 and rsc rad59 double mutants. Whereas deletion of RAD51 had an additive effect, the rad59 mutation did not further reduce the repair ability of rsc mutants, suggesting that RSC and Rad59 work together. These results are consistent with the sensitivity of rsc mutants to DNA-damaging agents, which seems to be additive with rad51 but epistatic to rad59.
The authors further strengthen the link between RSC activity and Rad59 by showing that this repair protein physically interacts with the Rsc1 and Rsc2 subunits of the complex. However, mutations in RSC subunits did not have any visible effect in a variety of DNA repair assays, such as assays of DSB-induced SSA, mating type gene conversion, ectopic gene conversion, and heteroallelic recombination.
The strongest repair phenotype for rsc mutants was observed in a genetic screen that selects for unequal sister chromatid recombination. This assay requires a slight misalignment between sister chromatids to allow recombination between nonallelic repeats. Both rad51 and rad59 mutants are proficient for this type of recombination, whereas rad52 mutants are completely defective. rsc mutants were also defective in this assay, and an additional mutation in RAD51 lowered the level of recombination further, whereas deleting RAD59 had no further effect, again suggesting a mechanism in which RSC and Rad59 cooperate. Thus, the authors conclude that the RSC complex plays a central role in facilitating recombination between sister chromatids.
After replication, the two sister chromatids are held together by a large protein circle called cohesin. Since RSC has been previously shown to be required for the loading of cohesin rings onto chromosomal arms (7, 12), the authors investigated whether mutations in RSC impair the loading of cohesin onto the site of a DSB created by an endonuclease. Indeed, they find that rsc mutants are impaired in cohesin loading, suggesting that the defects in DNA repair and the induced DNA damage response observed in the rsc mutants are due to defective recombination between the sister chromatids. They propose a model in which the RSC complex plays a role in chromatin remodeling and/or MRX recruitment that is required for the loading of cohesins, which in turn facilitate sister chromatid recombination.
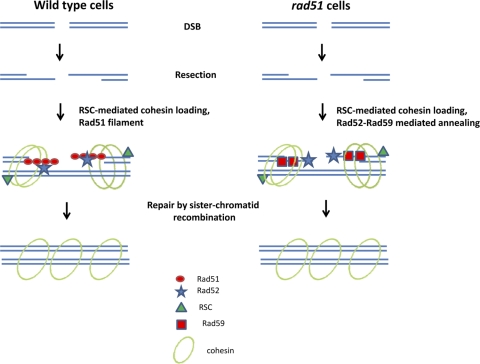
Many questions are raised by this new study. First, what is the relationship between Rad59, cohesin, and RSC and what are their particular roles? The authors demonstrate genetic and physical interactions between RSC subunits and Rad59; such interactions could suggest the possibility that RSC activity is necessary for the loading of Rad59 onto DNA. However, no evidence for such a role could be found: whereas rad59 mutants are defective in both DSB-induced and spontaneous SSA (4, 6, 9, 11), rsc mutants seem to be proficient for this type of repair, negating the possibility that RSC function is a prerequisite for Rad59 activity. The most striking effect of single rsc mutants is on unequal sister chromatid recombination: whereas this process can be carried out efficiently in rad51 or rad59 cells, it absolutely requires RSC. In the absence of RSC, deleting RAD59 has no further effect. This result is probably best explained by a model involving the loading of cohesin by RSC to hold chromatids in position, allowing Rad59-dependent annealing (Fig. 1). In the absence of RSC the chromatids cannot adopt the spatial conformation required for annealing.
Fig. 1.
Possible model for the interactions between RSC, Rad59, and cohesins. In the presence of Rad51 (left) a nucleoprotein is formed and used to repair the break by strand exchange with the sister chromatid (which is held nearby by the cohesin rings, loaded with the help of RSC). In strains lacking Rad51, repair is carried out by an alternative (yet-uncharacterized) mechanism based on Rad52/Rad59-mediated strand annealing. Under these circumstances RSC and cohesin roles in holding sister chromatids together become extremely important.
However, the pattern of sensitivity to MMS and other DNA-damaging agents is puzzling. Some rsc mutants or rad59 mutants are only moderately sensitive or even completely resistant. The importance of these factors can be revealed in the absence of Rad51: rsc rad51 double mutants are as sensitive as rad59 rad51 double mutants or rad52 mutants. This pattern is similar to the one observed previously for Rad59 in GC: its role is visible only in the absence of Rad51 (4, 9). Thus, the Rad59-RSC (and cohesin?) pathway may be important if the Rad51 filament is not created. Despite what seems to be a common pathway, an important distinction between rad59 and rsc mutants has to be made: whereas the former are defective in direct repeat recombination (even in the presence of wild-type levels of Rad51, RSC, and presumably cohesin), rsc mutants have no phenotype if a Rad51 filament is formed.
A second interesting question relates to the cell cycle dependency of the RSC activity required for DNA repair. In a previous publication, the authors showed a role for RSC in NHEJ, a DSB repair process that takes place mainly in G1 cells; the activity of RSC was shown to be due to physical interactions with the MRX and Ku complexes, which bind DSBs (13). In the present study, however, the authors show a role for RSC in sister chromatid recombination, a process that can take place only after DNA replication. Indeed, only G2 rsc cells seem to be defective in repair. A tight cell cycle regulation of the fate of broken chromosomes has been demonstrated (3, 8). This regulation seems to be exerted by allowing or preventing resection, the degradation of one of the strands of the broken arms that leads to the creation of a Rad51 filament. As Rad59 seems to play no role as long as the Rad51 pathway is fully operational (4, 9), it will be interesting to explore the possible role that RSC and cohesin may play in the choice among the various DSB repair avenues (NHEJ, Rad51-driven HR, and Rad59-driven annealing) and their interdependence and how resection and its regulation affect this choice. The study by Oum et al. underscores the importance of chromatin and its modification in cellular decisions and in DNA repair in particular.
Acknowledgments
Related work from our laboratory was supported by grants from the Israeli Ministry of Science and Technology and the Israel Cancer Association.
Footnotes
Published ahead of print on 15 August 2011.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1. Agmon N., Yovel M., Harari Y., Liefshitz B., Kupiec M. 23 May 2011, posting date. The role of Holliday junction resolvases in the repair of spontaneous and induced DNA damage. Nucleic Acids Res. doi:10.1093/nar/gkr277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aylon Y., Kupiec M. 2004. DSB repair: the yeast paradigm. DNA Repair (Amst) 3:797–815 [DOI] [PubMed] [Google Scholar]
- 3. Aylon Y., Liefshitz B., Kupiec M. 2004. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 23:4868–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bai Y., Symington L. S. 1996. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 10:2025–2037 [DOI] [PubMed] [Google Scholar]
- 5. Davis A. P., Symington L. S. 2001. The yeast recombinational repair protein Rad59 interacts with Rad52 and stimulates single-strand annealing. Genetics 159:515–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Feng Q., et al. 2007. Rad52 and Rad59 exhibit both overlapping and distinct functions. DNA Repair (Amst) 6:27–37 [DOI] [PubMed] [Google Scholar]
- 7. Huang J., Hsu J. M., Laurent B. C. 2004. The RSC nucleosome-remodeling complex is required for cohesin's association with chromosome arms. Mol. Cell 13:739–750 [DOI] [PubMed] [Google Scholar]
- 8. Ira G., et al. 2004. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431:1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jablonovich Z., Liefshitz B., Steinlauf R., Kupiec M. 1999. Characterization of the role played by the RAD59 gene of Saccharomyces cerevisiae in ectopic recombination. Curr. Genet. 36:13–20 [DOI] [PubMed] [Google Scholar]
- 10. Mortensen U. H., Bendixen C., Sunjevaric I., Rothstein R. 1996. DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl. Acad. Sci. U. S. A. 93:10729–10734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oum J. H., et al. 2011. RSC facilitates Rad59-dependent homologous recombination between sister chromatids by promoting cohesin loading at DNA double-strand breaks. Mol. Cell. Biol. 31:3924–3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shim E. Y., Hong S. J., Oum J. H., Yanez Y., Zhang Y., Lee S. E. 2007. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol. Cell. Biol. 27:1602–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shim E. Y., Ma J. L., Oum J. H., Yanez Y., Lee S. E. 2005. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol. Cell. Biol. 25:3934–3944 [DOI] [PMC free article] [PubMed] [Google Scholar]