Abstract
The mammalian HIRA/UBN1/ASF1a complex is a histone chaperone complex that is conserved from yeast (Saccharomyces cerevisiae) to humans. This complex preferentially deposits the histone variant H3.3 into chromatin in a DNA replication-independent manner and is implicated in diverse chromatin regulatory events from gene activation to heterochromatinization. In yeast, the orthologous complex consists of three Hir proteins (Hir1p, Hir2p, and Hir3p), Hpc2p, and Asf1p. Yeast Hir3p has weak homology to CABIN1, a fourth member of the human complex, suggesting that Hir3p and CABIN1 may be orthologs. Here we show that HIRA and CABIN1 interact at ectopic and endogenous levels of expression in cells, and we isolate the quaternary HIRA/UBN1/CABIN1/ASF1a (HUCA) complex, assembled from recombinant proteins. Mutational analyses support the view that HIRA acts as a scaffold to bring together UBN1, ASF1a, and CABIN1 into a quaternary complex. We show that, like HIRA, UBN1, and ASF1a, CABIN1 is involved in heterochromatinization of the genome of senescent human cells. Moreover, in proliferating cells, HIRA and CABIN1 regulate overlapping sets of genes, and these genes are enriched in the histone variant H3.3. In sum, these data demonstrate that CABIN1 is a functional member of the human HUCA complex and so is the likely ortholog of yeast Hir3p.
INTRODUCTION
The human HIRA/UBN1/ASF1a complex is an evolutionarily conserved histone chaperone complex. This complex preferentially deposits the histone variant H3.3 into chromatin in a DNA replication-independent manner (28, 33). For example, in Drosophila melanogaster, HIRA activity is required for DNA replication-independent H3.3 deposition and nucleosome assembly during sperm nucleus decondensation after fertilization of the oocyte (24). Depending on species, there are 4 or 5 subunits of the core complex. In Saccharomyces cerevisiae, these are Hir1p, Hir2p, Hir3p, Hpc2p, and Asf1p (30, 32, 39). Hir1p and Hir2p are both orthologous to metazoan HIRA, although both Hir1p and Hir2p are components of the same multisubunit chaperone complex (13, 25, 27). Hpc2p is orthologous to two related proteins in mammals, UBN1 and UBN2 (4, 5). Asf1p also has two counterparts in mammals, ASF1a and ASF1b (31), of which only ASF1a is included in the HIRA-containing complex (33, 47). Yeast Hir3p has some homology to a fourth member of the human complex, CABIN1 (4, 33), suggesting that Hir3p and CABIN1 are orthologs. Structural and mutagenesis studies indicate that HIRA serves as a scaffold protein for recruitment of ASF1a and UBN1 into the mammalian complex (5, 34).
Incorporation of the HIRA/UBN1/ASF1a substrate, histone H3.3, into nucleosomes contributes to nucleosome destabilization (19). Accordingly, H3.3 is enriched at nucleosomes at transcription start sites (TSSs) of genes and gene bodies of actively transcribed genes, where it is thought to facilitate nucleosome dynamics associated with transcription activation and ongoing transcription (12, 20). The HIRA protein is required for deposition of histone H3.3 at these regions (12). Consistent with this, HIRA is required for gene activation in viral genomes after virus infection and during angiogenesis and myogenesis (7, 36, 41).
However, histone H3.3 is also linked to transcription silencing and is incorporated at regions of the genome typically thought to be relatively transcriptionally inactive (12, 35, 37, 38, 45). Likewise, the HIRA/UBN1/ASF1a complex is also implicated in heterochromatin formation. In S. cerevisiae, the genes encoding Hir1p, Hir2p, Hir3p, and Hpc2p were first identified as repressors of histone gene expression (30, 39) that are recruited to histone gene regulatory regions (9), likely via interaction with other proteins. In addition, Hir1p, Hir2p, and Asf1p contribute to heterochromatin-mediated silencing of telomeres and mating loci (21, 23, 29). In Schizosaccharomyces pombe, the HIRA orthologs Hip1 and Slm9 are involved in silencing some genes, long terminal repeat (LTR) retrotransposons, and transcription from cryptic promoters associated with transcribed regions (2, 3, 40). Indeed, Hip1/Asf1 contributes to heterochromatinization by promoting histone deacetylation and promotion of HP1 binding (40). In plants, HIRA facilitates transcriptional repression of the Knox genes and therefore maintenance of a pluripotent undifferentiated stem cell phenotype (14, 26). In mammals, histone variant H3.3 is incorporated into transcriptionally silent chromatin during meiotic sex chromosome inactivation, and this is thought to depend on HIRA (35). In human cells, HIRA and ASF1a have also been reported to contribute to silencing of a latent modified HIV virus after its integration into the genome (11). Also in human cells, HIRA, UBN1, and ASF1a are implicated in heterochromatinization of the genome in proliferation-arrested senescent cells. Specifically, this chaperone complex is thought to play a role in assembly of domains of heterochromatin in senescent cells, senescence-associated heterochromatin foci (SAHF) (36, 47).
In light of the diverse roles of this chaperone complex in various biological contexts, it is important to define the functioning members of the complex and, ultimately, their respective molecular roles. Although a calcineurin-binding protein (CABIN1) has been reported to be a fourth component of the HIRA/UBN1/ASF1a complex based on affinity and chromatographic purification (33), its mode of interaction with the rest of the complex is unknown, and to date, there has been no molecular or cellular analysis to test whether CABIN1 is a functional member of the complex. Therefore, in this work, we set out to define the molecular basis of the interaction between CABIN1 and the remainder of the complex and to ask whether CABIN1 shares function with other members of the complex.
MATERIALS AND METHODS
Cell culture.
IMR90 fibroblasts and U2OS and HeLa cells were grown as described by ATCC, Manassas, VA.
Immunofluorescence, SAHF, and SA β-Gal staining.
Two-color indirect immunofluorescence and SAHF assays were performed as described previously (42, 47). Senescence-associated β-galactosidase (SA β-Gal) staining was performed as described previously (6).
Plasmids, siRNAs, and antibodies.
pSG5-myc-CABIN1 was a gift from Jun O. Liu, Johns Hopkins University School of Medicine. CABIN1 was cloned into pLXSN and pBABE retroviruses by standard molecular biology procedures; details are available on request. HIRA plasmids were defined previously (15). pBABE-RasV12 was a gift from William Hahn (Dana-Farber Cancer Institute). Small interfering RNAs (siRNAs) to HIRA and CABIN1 and a nontargeting (NTG) siRNA were purchased from Dharmacon (catalog no. L-013610-00-0005, L-012454-01-0005, and D-001810-10-05, respectively). The following reagents have been described previously: anti-HIRA (15) and rabbit polyclonal anti-UBN1 (5). The following were obtained from the indicated suppliers: anti-CABIN1 (ab3349; Abcam), antihemagglutinin (anti-HA) (sc805; Santa Cruz), anti-glutathione S-transferase (anti-GST) (B-14 sc138; Santa Cruz), anti-Flag (F3165; Sigma), anti-myc (9E10 sc40; Santa Cruz), anti-HIRA (H-300; Santa Cruz 48774), anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) (ab9484; Abcam), anti-promyelocytic leukemia body (anti-PML) (sc-966 and sc5621; Santa Cruz), anti-β-actin (A1978; Sigma), anti-green fluorescent protein (anti-GFP) (G1546; Sigma), anti-RB (9309; Cell Signaling), anti-phosphoserine 807/811 (9308; Cell Signaling), and p16INK4a (51-1325GR; BD).
Purification of the recombinant HUCA complex.
The four subunits of the recombinant HIRA/UBN1/CABIN1/ASF1a quaternary complex were expressed via baculovirus infection of insect Sf9 cells. Sequence-confirmed pFastBac baculovirus transfer vectors for human HIRA, UBN1, ASF1a, and CABIN1 were transformed into DH10Bac cells. Bacmid DNA was screened by PCR with M13 sequencing primers for proper transposition of the transfer vector sequences into the baculovirus genome, and positive bacmid DNAs were transfected into Sf9 cells. Passage 1 (P1) virus stocks were recovered 96 h posttransfection. A high-titer P2 virus stock was generated by infecting Sf9 at a multiplicity of infection (MOI) of ∼0.1, followed by incubation for 120 h. For production, in Sf900-III medium (Invitrogen), 1 × 106 Sf9 cells/ml were coinfected with viruses for each complex subunit at an MOI of 1. Infected cells were harvested 48 h postinfection. To purify protein complexes, cell pellets were lysed by Dounce homogenization in 20 mM HEPES (pH 8.0) plus 500 mM NaCl, supplemented with 5 mM 2-mercaptothanol and protease inhibitors (phenylmethylsulfonyl fluoride [PMSF], aprotinin, leupeptin, and pepstatin). Clarified supernatants were incubated with anti-Flag (M2) agarose (Sigma) for 1 h. Bound proteins were washed with 60 column volumes of lysis buffer prior to elution with 800 μg/ml of Flag (M2) peptide (Sigma) for 2 h at 4°C. Eluted proteins were analyzed on a 4 to 12% NuPAGE gel (Invitrogen) run in 1× MOPS (morpholinepropanesulfonic acid) running buffer and stained with R250 Coomassie blue stain. Subunit expression and complex composition were confirmed by Western blot analysis.
Retrovirus infections.
Retrovirus-mediated gene transfer was performed as described previously (47), using Phoenix cells to make the infectious viruses (Gary Nolan, Stanford University). Cells infected with viruses encoding resistance to puromycin were selected in 1 μg/ml of the selection agent.
Microarray and qRT-PCR.
Total RNA was prepared using the RNeasy kit (Qiagen catalog no. 74104), according to the manufacturer's instructions. Microarray expression analysis was performed on the Affymetrix platform human U133 plus2. For statistical analysis, the CEL files (Affymetrix raw data files) for all GeneChips were imported into the R program and analyzed using the Bioconductor package (http://www.bioconductor.org). Microarrays were quality controlled using the Affymetrix package, and expression values were calculated using the GCRMA background correcting and quantile normalization approach. Differentially expressed genes were detected using a t test followed by fold change (FC) threshold (1.2-fold) and P value threshold (0.05). Quantitative reverse transcription-PCR (qRT-PCR) was performed using the Dynamo SYBR green kit according to the manufacturer's instructions. GAPDH, β-actin, and 18S rRNA were used as housekeeping controls. Primer sequences are available on request.
Computational histone H3.3 analysis.
The distribution of H3.3 through the genome was determined from the previously published data set of Jin and Felsenfeld (20). We used SICER, an algorithm that identifies chromatin immunoprecipitation (ChIP)-enriched regions by looking for clusters of reads unlikely to occur by chance. Briefly, the genome is partitioned into nonoverlapping summary windows of 200 bp. Within each region, the number of reads is counted, with the location of each Watson (Crick) read shifted by +75bp (−75 bp) from its 5′ start to represent the center of the DNA fragment associated with the read. Windows showing enrichment (P value of 0.2 based on a Poisson background model) were identified, and islands were defined as clusters of enriched windows, allowing gaps of at most two unenriched windows (400 bp). H3.3 enriched regions were identified as islands whose read counts were above a threshold, determined by a very stringent E value requirement of 0.1, representing the expected number of islands whose counts are above the threshold (under a background model of random reads). The Ensembl gene set was obtained by downloading the transcript information from Ensembl version 54 (NCBI36) using the GeneMart service. Affymetrix U133P2 probes were mapped to Ensembl probe identification numbers (IDs) using the NetAffx annotation version 29 from Affymetrix. A total of 18,603 unique gene IDs were identified in this way.
ChIP assays.
HIRA ChIP assays were performed by a standard protocol (46) using an additional ethylene glycol disuccinate bis(sulfo-N-succinimidyl) ester (EGS) cross-linking step (44).
RESULTS
CABIN1 interacts with HIRA and UBN1.
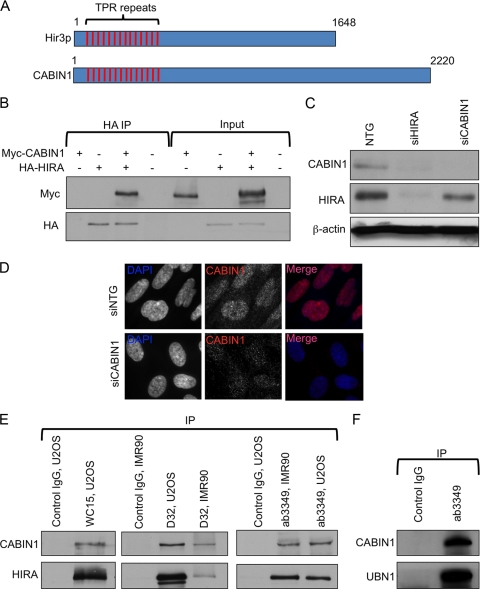
CABIN1 has been previously reported to copurify with HIRA, UBN1, and ASF1a from human cell extracts (33). In addition, CABIN1 has been suggested to be the human ortholog of yeast Hir3p (4). However, outside about 30 tetratricopeptide repeats (TPRs) within the N-terminal region of both Hir3p and CABIN1 (Fig. 1A), the homology between Hir3p and CABIN1 is quite low. The TPR is a structural motif present in a wide range of proteins mediating protein-protein interactions and assembly of multiprotein complexes. To test whether HIRA and CABIN1 physically interact, we ectopically expressed HA-tagged HIRA and myc-tagged CABIN1 in U2OS cells and performed immunoprecipitation-Western blot analysis. An interaction was readily detected under these conditions (Fig. 1B). To test whether HIRA and CABIN1 interact at endogenous levels of expression, we first confirmed the specificity of a commercially available antibody to CABIN1. In Western blot analysis, a polyclonal antibody recognized a 248-kDa polypeptide that was abolished by siRNA to CABIN1 (Fig. 1C). Furthermore, siRNA to CABIN1 abolished the nuclear signal in an immunofluorescence assay (Fig. 1D). We used this antibody and two different antibodies to HIRA (a mouse monoclonal antibody, WC15, and a rabbit polyclonal antibody, D32, raised by us and described previously [15]) to show that CABIN1 coimmunoprecipitates with human HIRA in primary (IMR90) and transformed (U2OS) human cell types, in the absence of any ectopic overexpression (Fig. 1E). Similarly, CABIN1 coimmunoprecipitated with another member of the complex, UBN1 (Fig. 1F). Consistent with CABIN1 being in complex with HIRA and UBN1, we observed that siRNA-mediated knockdown of HIRA decreased the steady-state abundance of CABIN1 (Fig. 1C). To a lesser extent, knockdown of CABIN1 also downregulated HIRA. Similarly, we previously showed that the stabilities of the HIRA and UBN1 proteins are mutually dependent on each other (5). This is consistent with a close physical interaction between HIRA, UBN1, and CABIN1, such that removal of one protein destabilizes the other. Together, these data confirm that HIRA, CABIN1, and UBN1 are members of the same multiprotein complex, like yeast Hir1p, Hir2p, Hir3, and Hpc2p (13, 27). Hereafter, the quaternary complex is referred to as HIRA/UBN1/CABIN1/ASF1a, abbreviated as HUCA.
Fig. 1.
Physical interaction of HIRA, UBN1, and CABIN1. (A) Schematic alignment of CABIN1 and S. cerevisiae Hir3p showing conserved N-terminal TPRs. (B) Ectopically expressed HA-HIRA and myc-CABIN1 interaction, detected by coimmunoprecipitation (IP) with anti-HA antibodies. (C) siRNA-mediated knockdown of endogenous HIRA and CABIN1 confirms specificity of HIRA and CABIN1 antibodies. (D) siRNA-mediated knockdown of endogenous CABIN1 confirms specificity of the CABIN1 antibody by immunofluorescence. (E) Interaction of endogenous HIRA and CABIN1 in osteosarcoma cells (U2OS) and primary fibroblasts (IMR90), detected by coimmunoprecipitation with antibodies to HIRA (WC15 and D32) and CABIN1 (ab3349). Anti-mouse IgG (anti-myc 9E10; Santa Cruz) and anti-rabbit IgG (M7023; Sigma) were used as control IgGs for mouse and rabbit antibodies, respectively. (F) Interaction of endogenous UBN1 and CABIN1, detected by coimmunoprecipitation with antibodies to CABIN1 (ab3349).
The HIRA C domain interacts with the N terminus of CABIN1.
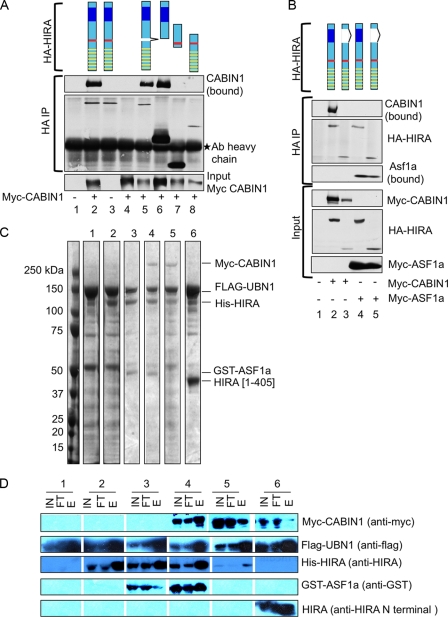
We have previously demonstrated that the N-terminal WD40 repeat region of HIRA interacts with the N-terminal Hpc2-related domain (HRD) of UBN1 (5) and that another conserved region of HIRA (the B domain, residues 425 to 475) interacts with ASF1a (22, 34, 47). This leaves the function of the conserved C-terminal domain of HIRA (the C domain, residues 763 to 793) (22) undefined. We hypothesized that this C domain might bind to CABIN1. To test this, we proceeded to define the interaction domains between HIRA and CABIN1 by immunoprecipitation and immunoblot analyses of cell lysates containing ectopically expressed wild-type and mutant proteins. First, deletion analysis of HIRA showed that binding of HIRA to CABIN1 does not require the ASF1a-binding B domain nor the N-terminal UBN1-binding HRD, but does require the C-terminal region of the protein containing the C domain (Fig. 2A). More specifically, a HIRA protein lacking only the C domain failed to bind to CABIN1 but still bound to ASF1a (Fig. 2B), showing that the inability of this fragment to bind to CABIN1 was not due to gross misfolding of the protein. Confirming the role of the C domain, a fragment of HIRA encompassing not much more than this domain was sufficient for binding to CABIN1 (Fig. 2A). These results demonstrate the importance of the HIRA C domain in recruiting CABIN1 into the HIRA/UBN1/ASF1a complex in crude mammalian cell lysates. To begin to define the HIRA/CABIN1 interaction domains and architecture of the quaternary complex using purified recombinant proteins, we ectopically coexpressed all four members of the complex in Sf9 insect cells as full-length, epitope-tagged proteins (specifically, myc-CABIN1, Flag-UBN1, His-HIRA, and GST-ASF1a). Remarkably, by purification over an anti-Flag immunoaffinity agarose, we were able to isolate the complete quaternary complex, confirmed by SDS-PAGE followed by Coomassie blue staining and immunoblot analysis (Fig. 2C and D). Consistent with previous results, expression and purification of partial complexes showed that His-HIRA interacted with Flag-UBN1 alone (5). Similarly, we have previously shown that ASF1a binds directly to the B domain of HIRA (34). The ability of HIRA to directly interact with both ASF1a and UBN1 supports a central role for HIRA in assembly of the quaternary complex. Consistent with this idea, myc-CABIN1 did not depend on the presence of GST-ASF1a for its incorporation into the complex (Fig. 2C and D). However, myc-CABIN1 failed to enter a complex containing only the N-terminal 405 residues of HIRA [His-HIRA(1-405)]. This supports a role for the C-terminal half of HIRA, presumably through the C domain, in directly recruiting CABIN1 into the quaternary complex.
Fig. 2.
Mapping of CABIN1 interaction domain on HIRA. (A) U2OS cells were transiently transfected with the indicated plasmids and immunoprecipitated (IP) with anti-HA. Lane 1, mock; lane 2, wild-type HA-HIRA and wild-type myc-CABIN1; lane 3, wild-type HA-HIRA only; lane 4, wild-type myc-CABIN1 only; lanes 5 to 8, wild-type myc-CABIN1 and indicated HA-HIRA mutants [lane 5, HA-HIRA(del 439-475); lane 6, HA-HIRA(del520-1017); lane 7, HA-HIRA(421-729); lane 8, HA-HIRA(1-600)]. Schematics are color coded as follows: yellow bars, WD40 repeats; red bars, B domain; blue boxes, C domain. Ab, antibody. (B) U2OS cells were transiently transfected with the indicated plasmids and immunoprecipitated with anti-HA. Lane 1, mock; lane 2, wild-type myc-CABIN1 and wild-type HA-HIRA; lane 3, wild-type myc-CABIN1 and HA-HIRA(del737-963); lane 4, wild-type myc-ASF1a and wild-type HA-HIRA; lane 5, wild-type myc-ASF1a and HA-HIRA(del737-963). Schematics are color coded as in panel A. (C) Insect Sf9 cells were infected with baculoviruses encoding the indicated proteins, and complexes were purified by anti-Flag affinity chromatography. Shown is a Coomassie blue (R250) stain of recombinant proteins. Lanes correspond to following proteins: 1, flag-UBN1; 2, Flag-UBN1 and His–HIRA; 3, Flag-UBN1, His–HIRA, and GST-ASF1a; 4, Flag-UBN1, His–HIRA, GST-ASF1a, and myc-CABIN1; 5, Flag-UBN1, His–HIRA, and myc-CABIN1; 6, Flag-UBN1, HIRA(1-405), and myc-CABIN1. (D) Western blot analysis with indicated antibodies of input lysate (IN), column flowthrough (FT), and elution (E) from panel C.
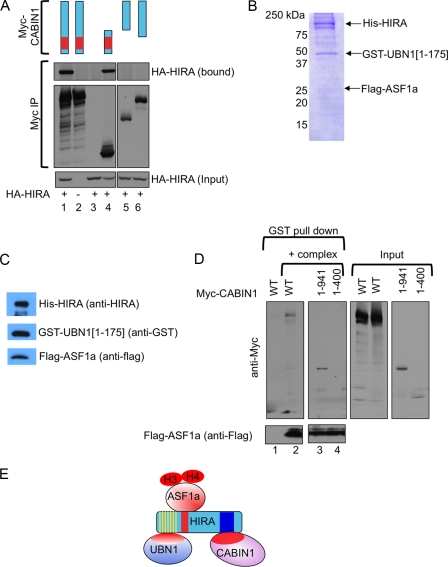
Next, we set out to define the region of CABIN1 that mediates binding to HIRA (Fig. 3A), initially by immunoprecipitation and immunoblot analyses in cell lysates containing ectopically expressed wild-type and mutant proteins. Deletion of the C terminus of CABIN1 did not affect binding to HIRA. Conversely, deletion of the N-terminal region of CABIN1 containing the TPRs abolished binding to HIRA (Fig. 3A), and the same region of CABIN1 (residues 1 to 941) was sufficient for binding to HIRA (Fig. 3A). To confirm this in an alternative assay, we expressed full-length His-HIRA [His-HIRA(1-1017)], Flag-ASF1a, and a GST-tagged fragment of UBN1 [GST-UBN1(1-175)] containing the HIRA-binding HRD in insect cells using baculovirus infection. The trimeric complex was recovered by nickel affinity chromatography and assessed by SDS-PAGE followed by Coomassie blue staining and immunoblotting (Fig. 3B and C). This recombinant complex was then immobilized on glutathione beads and incubated with cell lysates containing ectopically expressed full-length myc-CABIN1 protein, myc-CABIN1(1-941), and myc-CABIN1(1-400), and bound proteins were analyzed in a pulldown assay. Bound myc-CABIN1 proteins were detected by SDS-PAGE and immunoblotting to detect the myc epitope tag. Consistent with results obtained in Fig. 3A, full-length myc-CABIN1 bound to the HIRA/UBN1/ASF1a complex, as did myc-CABIN1(1-941) containing all of the TPRs (Fig. 3D). The shorter N-terminal CABIN1 fragment lacking some of the TPRs [myc-CABIN1(1-400)] did not bind to the complex. Taken together, these results indicate that CABIN1 interacts with the HIRA/UBN1/ASF1a complex through its N-terminal domain harboring the TPRs. Specifically, CABIN1 interacts with the C domain of HIRA (Fig. 3E). Together, these results support the view of HIRA as a scaffold for the HUCA complex.
Fig. 3.
Mapping of HIRA interaction domain on CABIN1. (A) U2OS cells were transiently transfected with the indicated plasmids and immunoprecipitated with anti-myc. Lane 1, wild-type myc-CABIN1 and wild-type HA-HIRA; lane 2, wild-type myc-CABIN1; lane 3, wild-type HA-HIRA; lane 4, myc-CABIN1(1-941) and wild-type HA-HIRA; lane 5, myc-CABIN1(942-2220) and wild-type HA-HIRA; lane 6, myc-CABIN1(401-2220) and wild-type HA-HIRA. (B) Insect Sf9 cells were infected with baculoviruses encoding His-HIRA, GST-UBN1(1-175), and Flag-ASF1a, and recombinant complex was purified by nickel affinity chromatography. Shown in the figure is a Coomassie blue (R250) stain of recombinant proteins. (C) Western blot of complex from panel B with indicated antibodies. (D) The trimeric protein complex from panel B was incubated with lysates from U2OS cells transfected with wild-type myc-CABIN1, myc-CABIN1(1-941), or myc-CABIN1(1-400), as indicated. Bound proteins were Western blotted as indicated. (E) Schematic of the HUCA complex based upon binding studies reported here and previously published literature.
CABIN1 is involved in cell senescence.
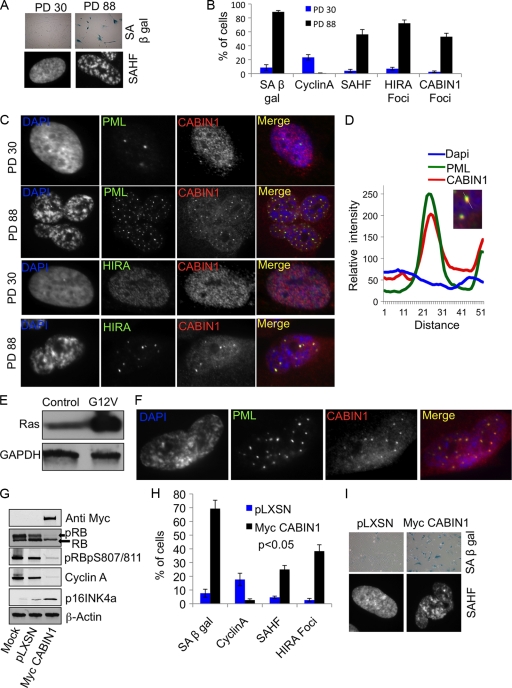
Having confirmed that CABIN1 is the fourth member of the HIRA/UBN1/ASF1a/CABIN1 complex, we set out to compare HIRA and CABIN1 in cell biological and functional assays. First, we asked whether CABIN1 is involved in formation of SAHF in senescent human cells. Since HIRA's localization to PML bodies is a prerequisite for formation of SAHF (42), we asked whether CABIN1 is also recruited to PML bodies in senescent cells. Primary human IMR90 fibroblasts were passaged in culture to the end of their proliferative life span, and replicative senescence was confirmed by detection of known markers of senescence (1), specifically SAHF, SA β-Gal, localization of HIRA to PML bodies, and absence of the S-phase marker cyclin A (Fig. 4A and B). We found that, like HIRA, CABIN1 is localized throughout the nucleoplasm of proliferating cells in a fine speckled pattern (Fig. 4C). However, CABIN1 colocalized with both HIRA and PML in PML bodies in senescent cells (Fig. 4C). A line trace drawn through one of these CABIN1/HIRA foci confirmed good overlap of CABIN1 and HIRA immunofluorescence signals, supporting colocalization of these proteins (Fig. 4D). Similarly, CABIN1 was recruited to PML bodies in cells made senescent through expression of an activated Ras oncogene (oncogene-induced senescence) (Fig. 4E and F). To more directly test a role for CABIN1 in formation of SAHF, we asked whether ectopic expression of CABIN1 is able to accelerate cell senescence and induce SAHF formation. Primary human IMR90 fibroblasts at population doubling 30 (PD 30) were infected with a retrovirus encoding wild-type CABIN1 or a control virus and assayed for cellular and molecular hallmarks of senescence, including formation of SAHF. Ectopic expression of CABIN1 markedly reduced proliferation of primary human fibroblasts over a 10-day period (data not shown), and this was accompanied by biochemical changes consistent with proliferation arrest and senescence, notably decreased pRB phosphorylation (detected by increased mobility in SDS-PAGE and using a phospho-specific antibody to phosphoserine 807/811), upregulation of p16INK4a, and downregulation of cyclin A (Fig. 4G and H). Proliferation arrest was accompanied by a dramatic increase in the number of cells displaying SA β-Gal activity (Fig. 4I). In addition, cells ectopically expressing CABIN1 showed enhanced localization of HIRA to PML bodies and formation of SAHF (Fig. 4H and I). We conclude that ectopic expression of CABIN1 activates the HIRA-driven SAHF assembly pathway and accelerates cell senescence. In sum, these results show that, like its binding partner HIRA, CABIN1 is recruited to PML bodies in both replicative and oncogene-induced senescent cells. HIRA localized to PML bodies is implicated in formation of SAHF, and here we show that CABIN1 similarly impinges on the SAHF assembly process.
Fig. 4.
CABIN1 regulates senescence. (A) PD 30 and PD 88 IMR90 fibroblasts stained for markers of senescence, SA β-Gal and SAHF. (B) Cells from panel A were scored for the percentage of cells expressing SA β-Gal, cyclin A, SAHF, HIRA foci, and CABIN1 foci. Values are means ± standard errors of the means (SEM) of three independent experiments. (C) Cells from panel A stained with antibodies to HIRA, PML bodies, and CABIN1 and with DAPI to visualize SAHF. (D) Relative intensity of DAPI, PML body, and CABIN1 fluorescence along a straight line through the CABIN1/PML focus. (E) Western blot showing expression of oncogenic H-RasG12V in IMR90 cells. (F) H-RasG12V-expressing cells stained with antibodies to PML bodies, CABIN1, and with DAPI (4′,6-diamidino-2-phenylindole). (G) IMR90 cells were infected with pLXSN or pLXSN-myc-CABIN1, and lysates were Western blotted with the indicated antibodies. (H) Cells from panel G were scored for SA β-Gal, cyclin A, HIRA foci, and SAHF. Values are means ± SEM of three independent experiments. (I) Cells from panel G stained to detect SA β-Gal and with DAPI to detect SAHF.
CABIN1 and HIRA regulate many of the same genes.
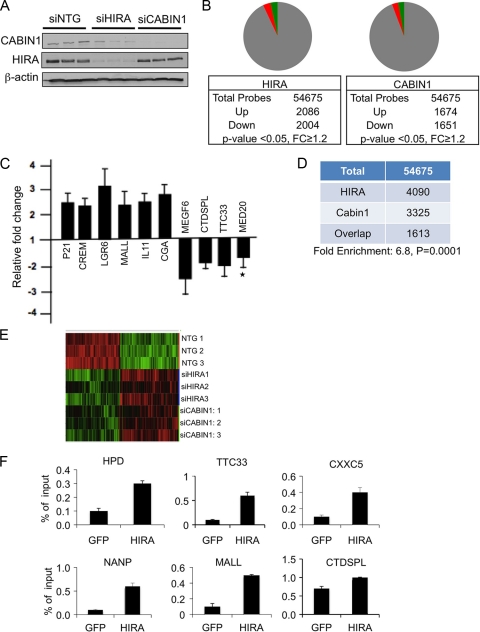
As members of the same histone chaperone complex, HIRA and CABIN1 ought to regulate many of the same genes. To test this, we used siRNA to separately knockdown HIRA or CABIN1 in HeLa cells (Fig. 5A) and then performed gene microarray expression analysis on the Affymetrix platform. Compared to control cells, cells lacking HIRA showed upregulation of 2,086 probes and downregulation of 2,004 probes (P < 0.05, fold change [FC] ≥ 1.2) (Fig. 5B). Cells lacking CABIN1 showed upregulation of 1,674 probes and downregulation of 1,651 probes, by the same criteria (Fig. 5B). The microarray results were validated using qRT-PCR; 9 out of 10 genes tested by qRT-PCR were confirmed to change significantly after HIRA knockdown (Fig. 5C). Consistent with many of the genes regulated by HIRA and CABIN1 being specific targets of the HUCA complex (as opposed to off-target effects of the siRNA), the overlapping set of genes regulated by both HIRA and CABIN1 (1,613 probes) was 6.8-fold larger than expected from chance overlap (P < 0.001) (Fig. 5D). Underscoring the significance of this overlap, 1,597 of the 1,613 probes changed in the same direction after HIRA or CABIN1 knockdown (770 increased and 827 decreased). The overlap of HIRA- and CABIN1-regulated genes was also apparent from hierarchical clustering (Fig. 5E). The set of genes regulated by both HIRA and CABIN1 included up- and downregulated genes in approximately equal proportions (data not shown). Consistent with many of these genes being direct targets of the HUCA complex, endogenous HIRA in HeLa cells was detected at the TSS of 6 out of 6 of these genes analyzed by ChIP assay (Fig. 5F, P < 0.05). In this analysis, we specifically chose to analyze TSS because HIRA has previously been shown to be required for recruitment of histone H3.3 at these regions in mouse embryonic stem (ES) cells (although in this study, inactivation of HIRA did not affect gene expression) (12). In sum, regulation of a common set of genes by HIRA and CABIN1 supports our hypothesis that both proteins are functional members of the same gene regulatory complex.
Fig. 5.
HIRA and CABIN1 regulate overlapping sets of genes. (A) HeLa cells were nucleofected with siRNAs to HIRA, CABIN1, or the nontargeting (NTG) control as indicated. Three independent nucleofections were preformed with Smartpool siRNAs to HIRA or CABIN1. Lysates were Western blotted to detect HIRA, CABIN1, and β-actin as indicated. (B) Pie charts showing significantly upregulated and downregulated probes after HIRA or CABIN1 knockdown. Green indicates upregulated probes, and red indicates downregulated probes. (C) qRT-PCR analysis to confirm expression changes detected by microarray after siHIRA knockdown. Fold change is calculated by dividing normalized (to housekeeping gene) expression in siHIRA cells by normalized (to housekeeping gene) expression in siNTG cells. *, P > 0.05. P < 0.05 for the other 9 genes. Values are means ± SEM of four independent experiments. (D) Table showing overlap of changing probes after HIRA and CABIN1 knockdown. (E) Heat maps showing hierarchical clustering of genes whose expression changes after three independent siHIRA or siCABIN1 knockdowns, compared to three independent siNTG nucleofections. Green indicates upregulated probes, and red indicates downregulated probes. (F) Sonicated chromatin from HeLa cells was immunoprecipitated with antibodies to GFP or HIRA, and the indicated target genes were detected by qPCR. Expression of HPD and MALL increased on HIRA knockdown, while expression of all others decreased. Genes were selected at random from a list of genes with a fold change (FC) of >1.2-fold and P < 0.05 from siHIRA knockdown cells. Values are means ± SEM of three independent ChIP experiments. P < 0.05 for all comparisons.
HIRA- and CABIN1-regulated genes are both enriched in histone H3.3.
If HIRA and CABIN1 both regulate gene expression through deposition of histone H3.3, then the genes that they each regulate should be enriched in histone H3.3. To test this, we used the publically available ChIP-chip data set from Jin et al. (20), describing the genomic distribution of histone H3.3 in HeLa cells.
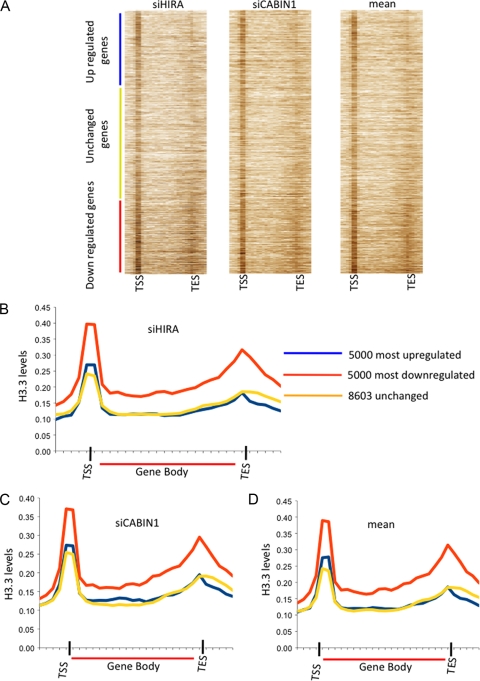
First we rank ordered all probes regulated by HUCA complex according to fold change and then plotted the genomic H3.3 distribution on each probe. The genes that are upregulated after HIRA or CABIN1 knockdown are at the top of the heat map, and the genes that are downregulated after HIRA or CABIN1 knockdown are at the bottom (Fig. 6A). From the heat maps, it is apparent that genes that are upregulated after HIRA or CABIN1 knockdown are slightly enriched in H3.3 at the TSS, compared to genes that don't change. Most strikingly, genes that are downregulated after HIRA or CABIN1 knockdown are markedly enriched in histone H3.3 at the TSS, in the gene body, and especially at the 3′ end of the gene body (Fig. 6A). A similar distribution was observed when genes were rank ordered according to the mean effect of HIRA and CABIN1 knockdown.
Fig. 6.
Genes activated by HIRA are enriched in histone H3.3. (A) Heat map showing H3.3 enrichment on genes ordered by fold change after HIRA or CABIN1 knockdown versus siNTG and mean fold change of the two independent knockdowns versus siNTG. On the y axis, genes are rank ordered by fold change; on the x axis, positions along the gene are shown in 1-kb windows from 5 kb upstream of the gene to the transcription start site (TSS) and 1-kb windows from the transcription end site (TES) to 5 kb downstream of the gene. Within the gene bodies, windows of 5% of the length of the gene were used. (B) Average histone H3.3 enrichment over three classes of genes, defined according to their response to HIRA knockdown. x axis, position along the gene in 1-kb windows from 5 kb upstream of the gene to the transcription start site (TSS) and 1-kb windows from the TES to 5 kb downstream of the gene. Within the gene bodies, windows of 5% of the length of the gene were used. (C) Average histone H3.3 enrichment over three classes of genes, defined according to their response to CABIN1 knockdown. Results are color coded as in panel B. (D) Average histone H3.3 enrichment over three classes of genes, defined according to their averaged response to HIRA and CABIN1 knockdown. Results are color coded as in panel B.
To perform a more quantitative analysis, composite histone H3.3 profiles were plotted for the three gene sets identified by microarray analysis (upregulated, downregulated, or unchanged) (Fig. 6B to D). For each gene set, H3.3 island read counts were summed in 1-kb windows, from 5 kb upstream of the TSS to the TSS and from the transcription end site (TES) to 5 kb downstream of the TES. Within the gene body, the island read counts were summed in windows equal to 5% of the gene length. Island read counts were normalized by the total number of bases in the windows. Consistent with the analysis of Jin et al. (20), we found that histone H3.3 is enriched around the TSS and the 3′ end of the gene body (Fig. 6B to D) of all gene sets. Genes whose expression increased or did not change after HIRA or CABIN1 knockdown showed comparable H3.3 distributions. However, genes whose expression decreased after HIRA or CABIN1 knockdown were even more enriched in histone H3.3 around the TSS and in the gene body and toward the 3′ end of the gene (Fig. 6B and C). Again, a similar effect was observed when H3.3 distribution was analyzed in the three gene sets defined by the mean response to HIRA and CABIN1 knockdown (Fig. 6D). Taken together, these results indicate that HIRA and CABIN1 both contribute to gene activation in a manner that is linked to H3.3 deposition, consistent with both of these proteins functioning in a histone H3.3 deposition complex.
DISCUSSION
In this study, we have shown that CABIN1 is a functional member of the HUCA histone chaperone complex. Our data confirm CABIN1 as the human ortholog of the yeast Hir3p protein.
To start, we have confirmed that CABIN1 physically interacts with other members of the HUCA complex in cells. The interaction with the complex is most likely direct, because we have successfully reconstituted a quaternary complex comprised of HIRA, UBN1, CABIN1, and ASF1a. While there are likely other proteins that interact with this complex in vivo, based on previous complex purification studies (10, 27, 33) and our own unpublished efforts in this regard, this quaternary complex appears to be a particularly stable core entity.
We have defined the region of CABIN1 that interacts with HIRA and vice versa. Specifically, HIRA binds to the conserved N-terminal TPRs of CABIN1, and CABIN1 binds to the conserved C domain of HIRA. The latter result is particularly significant in light of a previous analysis that highlighted three conserved domains in HIRA: the N-terminal WD40 repeats, a central B domain, and a C-terminal C domain (22). We now know that the N-terminal WD40 repeats bind to UBN1 (5), the B domain binds to ASF1a (34, 47), and the C domain binds to CABIN1. By this view, HIRA forms a scaffold for the HUCA complex, acting as a binding platform to recruit UBN1, ASF1a, and CABIN1. This model is supported by our previous demonstration of heterodimeric complexes formed between purified recombinant HIRA and UBN1 (5) and also HIRA and ASF1a (34). While we have not yet achieved this for HIRA and CABIN1, we have shown that the C-terminal C domain of HIRA is required to recruit CABIN1 to the quaternary complex. While HIRA is the scaffold for the complex, UBN1, CABIN1, and ASF1a presumably have their own specialized functions. Indeed, ASF1a appears to be the primary histone binding subunit of this histone chaperone complex (8). In sum, HUCA is a four-member quaternary complex comprised of HIRA, UBN1, CABIN1, and ASF1a, in which HIRA serves as a platform to bring the other members together.
Presumably, the other members of the complex, UBN1 and CABIN1, bring specific functionalities to the complex. In this regard, CABIN1 has been previously shown to facilitate transcriptional repression by transcription factors, including MEF2 and p53, by recruiting histone-modifying enzymes, mSin3, histone deacetylases (HDACs), and SUV39H1 (16–18, 43). These enzymes modify chromatin to achieve a more transcriptionally repressed state. Interestingly, CABIN1 is regulated by intracellular calcium, to link calcium signaling to control of gene expression (16). Whether or not CABIN1 serves to recruit similar histone-modifying enzymes to the HUCA complex and whether HUCA is also regulated by calcium signaling remain to be established.
In addition to showing that CABIN1 is a member of some members of the HUCA complex, we have demonstrated that CABIN1 shares functional outputs with other members of the complex. Like other members of the complex, CABIN1 is involved in chromatin heterochromatinization in senescent human cells (5, 47). Two lines of evidence show this. First, CABIN1 is recruited to PML nuclear bodies, together with HIRA and UBN1, in senescent cells. Second, ectopic expression of CABIN1 induces a senescent-like state, as indicated by proliferation arrest, activation of the pRB tumor suppressor pathway, expression of SA β-Gal, and formation of SAHF. Previous studies from our lab have shown that recruitment of some members of the HUCA complex to PML bodies is linked to formation of SAHF (5, 47). While PML bodies and SAHF do not colocalize, it is thought that PML bodies serve as sites to modify or assemble the HUCA complex into higher-order complexes prior to its role in SAHF formation.
HUCA's role in formation of SAHF is likely to involve histone deposition and nucleosome assembly. Specifically, the HUCA complex is thought to preferentially deposit the histone variant H3.3 into chromatin in a DNA replication-independent manner (33). Histone H3.3 is best viewed as a replacement variant histone, whose incorporation into chromatin outside DNA replication is associated with both transcription activation and repression (see the introduction). Here, we have underscored the link between HIRA and histone H3.3 deposition and similarly established a link between CABIN1 and histone H3.3 deposition. siRNA-mediated knockdown of HIRA and CABIN1 in HeLa cells modestly affects cellular gene expression (in terms of magnitude of the changes in RNA abundance) programs in both cases. The relatively subtle effect of HIRA and CABIN1 knockdown is in line with a previous study that failed to identify a marked effect on global gene expression after HIRA inactivation in mouse embryonic stem (ES) cells (12). Presumably, the function of the HUCA complex at genes is redundant with other chromatin regulators. More important than the modest effect on gene expression, two results point to the shared function of HIRA and CABIN1. First, there is significant overlap of the genes regulated by HIRA and CABIN1. Second, genes that are downregulated by inactivation of HIRA and CABIN1 are both especially enriched in histone variant H3.3 at the TSS, gene body, and 3′ end of gene body, compared to genes that are upregulated or do not change. This supports the role of histone H3.3 and HUCA in gene activation. The presence of histone H3.3 at the gene's TSS is thought to promote gene expression, because nucleosomes containing histone H3.3 and variant H2AZ are more labile than canonical nucleosomes (20). These labile nucleosomes are thought to facilitate access of transcription factors and chromatin remodeling events associated with transcription. Since HUCA is largely responsible for deposition of histone H3.3 at the TSS (12), knockdown of HIRA or CABIN1 is likely to repress transcription at these genes by generating a more static, less transcriptionally permissive, and perhaps histone H3.1-containing, nucleosome structure at the TSS. Regardless of the precise mechanism, the similar link between both HIRA and CABIN1 and histone H3.3 supports the notion that CABIN1 is a functional member of the HUCA complex.
ACKNOWLEDGMENTS
The labs of R.M., D.S., and P.D.A. are funded by NIA program project P01 AG031862.
We thank members of the Adams, Schultz, and Marmorstein labs and the rest of the PO1 group for critical discussion.
Footnotes
Published ahead of print on 1 August 2011.
REFERENCES
- 1. Adams P. D. 2009. Healing and hurting: molecular mechanisms, functions, and pathologies of cellular senescence. Mol. Cell 36:2–14 [DOI] [PubMed] [Google Scholar]
- 2. Anderson H. E., et al. 2010. Silencing mediated by the Schizosaccharomyces pombe HIRA complex is dependent upon the Hpc2-like protein, Hip4. PLoS One 5:e13488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson H. E., et al. 2009. The fission yeast HIRA histone chaperone is required for promoter silencing and the suppression of cryptic antisense transcripts. Mol. Cell. Biol. 29:5158–5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Balaji S., Iyer L. M., Aravind L. 2009. HPC2 and ubinuclein define a novel family of histone chaperones conserved throughout eukaryotes. Mol. Biosyst. 5:269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Banumathy G., et al. 2009. Human UBN1 is an ortholog of yeast Hpc2p and has an essential role in the HIRA/ASF1a chromatin-remodeling pathway in senescent cells. Mol. Cell. Biol. 29:758–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dimri G. P., et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. U. S. A. 92:9363–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dutta D., et al. 2010. Regulation of angiogenesis by histone chaperone HIRA-mediated Incorporation of lysine 56-acetylated histone H3.3 at chromatin domains of endothelial genes. J. Biol. Chem. 285:41567–41577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. English C. M., Adkins M. W., Carson J. J., Churchill M. E., Tyler J. K. 2006. Structural basis for the histone chaperone activity of Asf1. Cell 127:495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fillingham J., et al. 2009. Two-color cell array screen reveals interdependent roles for histone chaperones and a chromatin boundary regulator in histone gene repression. Mol. Cell 35:340–351 [DOI] [PubMed] [Google Scholar]
- 10. Franco A. A., Lam W. M., Burgers P. M., Kaufman P. D. 2005. Histone deposition protein Asf1 maintains DNA replisome integrity and interacts with replication factor C. Genes Dev. 19:1365–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gallastegui E., Millan-Zambrano G., Terme J. M., Chavez S., Jordan A. 2011. Chromatin reassembly factors are involved in transcriptional interference promoting HIV latency. J. Virol. 85:3187–3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goldberg A. D., et al. 2010. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140:678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Green E. M., et al. 2005. Replication-independent histone deposition by the HIR complex and Asf1. Curr. Biol. 15:2044–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guo M., Thomas J., Collins G., Timmermans M. C. 2008. Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20:48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hall C., et al. 2001. HIRA, the human homologue of yeast Hir1p and Hir2p, is a novel cyclin-cdk2 substrate whose expression blocks S-phase progression. Mol. Cell. Biol. 21:1854–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Han A., et al. 2003. Sequence-specific recruitment of transcriptional co-repressor Cabin1 by myocyte enhancer factor-2. Nature 422:730–734 [DOI] [PubMed] [Google Scholar]
- 17. Jang H., Choi D. E., Kim H., Cho E. J., Youn H. D. 2007. Cabin1 represses MEF2 transcriptional activity by association with a methyltransferase, SUV39H1. J. Biol. Chem. 282:11172–11179 [DOI] [PubMed] [Google Scholar]
- 18. Jang H., Choi S. Y., Cho E. J., Youn H. D. 2009. Cabin1 restrains p53 activity on chromatin. Nat. Struct. Mol. Biol. 16:910–915 [DOI] [PubMed] [Google Scholar]
- 19. Jin C., Felsenfeld G. 2007. Nucleosome stability mediated by histone variants H3.3 and H2A. Z. Genes Dev. 21:1519–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jin C., et al. 2009. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat. Genet. 41:941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaufman P. D., Cohen J. L., Osley M. A. 1998. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol. Cell. Biol. 18:4793–4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kirov N., Shtilbans A., Rushlow C. 1998. Isolation and characterization of a new gene encoding a member of the HIRA family of proteins from Drosophila melanogaster. Gene 212:323–332 [DOI] [PubMed] [Google Scholar]
- 23. Krawitz D. C., Kama T., Kaufman P. D. 2002. Chromatin assembly factor I mutants defective for PCNA binding require Asf1/Hir proteins for silencing. Mol. Cell. Biol. 22:614–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Loppin B., et al. 2005. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature 437:1386–1390 [DOI] [PubMed] [Google Scholar]
- 25. Lorain S., et al. 1996. Structural organization of the WD repeat protein-encoding gene HIRA in the DiGeorge syndrome critical region of human chromosome 22. Genome Res. 6:43–50 [DOI] [PubMed] [Google Scholar]
- 26. Phelps-Durr T. L., Thomas J., Vahab P., Timmermans M. C. 2005. Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell 17:2886–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Prochasson P., Florens L., Swanson S. K., Washburn M. P., Workman J. L. 2005. The HIR corepressor complex binds to nucleosomes generating a distinct protein/DNA complex resistant to remodeling by SWI/SNF. Genes Dev. 19:2534–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ray-Gallet D., et al. 2002. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9:1091–1100 [DOI] [PubMed] [Google Scholar]
- 29. Sharp J. A., Fouts E. T., Krawitz D. C., Kaufman P. D. 2001. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr. Biol. 11:463–473 [DOI] [PubMed] [Google Scholar]
- 30. Sherwood P. W., Tsang S. V., Osley M. A. 1993. Characterization of HIR1 and HIR2, two genes required for regulation of histone gene transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sillje H. H., Nigg E. A. 2001. Identification of human Asf1 chromatin assembly factors as substrates of Tousled-like kinases. Curr. Biol. 11:1068–1073 [DOI] [PubMed] [Google Scholar]
- 32. Sutton A., Bucaria J., Osley M. A., Sternglanz R. 2001. Yeast ASF1 protein is required for cell cycle regulation of histone gene transcription. Genetics 158:587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116:51–61 [DOI] [PubMed] [Google Scholar]
- 34. Tang Y., et al. 2006. Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly. Nat. Struct. Mol. Biol. 13:921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van der Heijden G. W., et al. 2007. Chromosome-wide nucleosome replacement and H3.3 incorporation during mammalian meiotic sex chromosome inactivation. Nat. Genet. 39:251–258 [DOI] [PubMed] [Google Scholar]
- 36. Wajapeyee N., Serra R. W., Zhu X., Mahalingam M., Green M. R. 2008. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell 132:363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wong L. H., et al. 2010. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 20:351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wong L. H., et al. 2009. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res. 19:404–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xu H., Kim U. J., Schuster T., Grunstein M. 1992. Identification of a new set of cell cycle-regulatory genes that regulate S-phase transcription of histone genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:5249–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yamane K., et al. 2011. Asf1/HIRA facilitate global histone deacetylation and associate with HP1 to promote nucleosome occupancy at heterochromatic loci. Mol. Cell 41:56–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang J. H., et al. 2011. Myogenic transcriptional activation of MyoD mediated by replication-independent histone deposition. Proc. Natl. Acad. Sci. U. S. A. 108:85–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ye X., et al. 2007. Definition of pRB- and p53-dependent and -independent steps in HIRA/ASF1a-mediated formation of senescence-associated heterochromatin foci. Mol. Cell. Biol. 27:2452–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Youn H. D., Sun L., Prywes R., Liu J. O. 1999. Apoptosis of T cells mediated by Ca2+-induced release of the transcription factor MEF2. Science 286:790–793 [DOI] [PubMed] [Google Scholar]
- 44. Zeng P. Y., Vakoc C. R., Chen Z. C., Blobel G. A., Berger S. L. 2006. In vivo dual cross-linking for identification of indirect DNA-associated proteins by chromatin immunoprecipitation. Biotechniques 41:694, 696, 698. [DOI] [PubMed] [Google Scholar]
- 45. Zhang R., Chen W., Adams P. D. 2007. Molecular dissection of formation of senescent associated heterochromatin foci. Mol. Cell. Biol. 27:2343–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang R., et al. 2007. HP1 proteins are essential for a dynamic nuclear response that rescues the function of perturbed heterochromatin in primary human cells. Mol. Cell. Biol. 27:949–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang R., et al. 2005. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 8:19–30 [DOI] [PubMed] [Google Scholar]