Abstract
The production of water-insoluble glucan (WIG) enables Streptococcus mutans to survive and persist in the oral niche. WIG is produced from sucrose by glucosyltransferase encoded tandemly by the highly homologous gtfB and gtfC genes. Conversely, a single hybrid gene from the endogenous recombination of gtfB and gtfC is easily generated using RecA, resulting in S. mutans UA159 WIG− (rate of ∼1.0 × 10−3). The pneumococcus recA gene is regulated as a late competence gene. comX gene mutations did not lead to the appearance of WIG− cells. The biofilm collected from the flow cell had more WIG− cells than among the planktonic cells. Among the planktonic cells, WIG− cells appeared after 16 h and increased ∼10-fold after 32 h of cultivation, suggesting an increase in planktonic WIG− cells after longer culture. The strain may be derived from the biofilm environment. In coculture with donor WIG+ and recipient WIG− cells, the recipient cells reverted to WIG+ and acquired an intact gtfBC region from the environment, indicating that the uptake of extracellular DNA resulted in the phenotypic change. Here we demonstrate that endogenous DNA rearrangement and uptake of extracellular DNA generate WIG− cells and that both are induced by the same signal transducer, the com system. Our findings may help in understanding how S. mutans can adapt to the oral environment and may explain the evolution of S. mutans.
INTRODUCTION
Biofilms are defined as matrix-enclosed bacterial populations that are adherent to each other and/or to surfaces or interfaces (8). Biofilms are studied due to their significance in medical, industrial, and environmental settings. Biofilm formation consists of several phases: (i) initial adherence of planktonic cells to a surface, (ii) irreversible attachment to the surface, (iii) unstructured biofilm growth through cell division and exopolysaccharide synthesis, and (iv) maturation into a complex three-dimensional architecture of microcolonies and void spaces (13, 37). As biofilm accumulation progresses, the cells within biofilms can disperse in response to environmental changes. Biofilms are resistant to environmental stresses (e.g., nutritional, oxidative, and antibiotic stress) and to host-mediated responses (e.g., complement proteins and phagocytes) (11, 16, 33). Therefore, understanding the molecular mechanisms underlying the life cycle of biofilm-forming bacteria may assist in the development of prevention and treatment strategies.
Streptococcus mutans is an oral bacterium that depends on biofilms for survival and persistence in its natural ecosystem. Under favorable environmental conditions, S. mutans can rapidly produce acid from fermentable dietary carbohydrates and initiate demineralization of the tooth surface. Therefore, S. mutans is an important etiological agent for dental caries. S. mutans is capable of forming biofilms using various mechanisms, e.g., surface adhesion- and cell density-dependent gene expression (9, 18). Clinically relevant in caries development is the ability of S. mutans to metabolize sucrose. Sucrose is the substrate for glucosyltransferase-mediated sucrose-dependent glucan production that promotes adhesion of S. mutans to the tooth surface. Water-insoluble glucan (WIG) is synthesized using GtfB- and GtfC-glucosyltransferases and promotes adhesion and biofilm maturation. Conversely, reports describe the spontaneous occurrence of a naturally derived WIG− strain (1, 40). This deletion is a recombination of gtfB and gtfC (which are proximate and have a high homology) (40), resulting in the generation of the single hybrid gtfBC gene. This mutation decreases the synthesis of WIG and reduces its biofilm-forming ability (40). Conversely, the growth rate of the WIG− strain in media supplemented with sucrose is greater than that of the wild type (31). Additionally, the appearance of an S. mutans WIG− strain exerts pleiotropic effects, e.g., reduced WIG in a gnotobiotic rat model using the gtfBC recombinant strain showed lower cariogenicity, inactivating the glucan-binding protein and thus changing the plaque structure (15). Further, Nomura et al. (32) reported the detection of S. mutans in human heart valve tissues with a reduced biofilm due to gtfB-gtfC recombination and with a lower susceptibility to antibiotics. With this adaptive ability, the WIG− variant may play a role in survival and development of the microorganism in its environmental niche; however, its appearance and ecological significance are not understood.
Previous studies have shown that recombination of gtfB and gtfC causes the formation of the WIG− strain and is RecA dependent (3). Previous studies with Streptococcus pneumoniae found the that recA gene was induced at competence (23, 24), and competence-specific induction of recA was also demonstrated in Bacillus subtilis (22). The genetics and physiology of the competence cascade in S. mutans have been reported (9, 24). In brief, the development of competence requires transcriptional activation of the com regulon, which is induced when the competence-stimulating peptide (CSP) (encoded by comC) stimulates its receptor, the membrane-bound histidine kinase ComD. ComD then autophosphorylates its cognate response regulator ComE and then activates the expression of early com genes, including comCDE and comX. The latter encodes an alternative sigma factor (19). The ComDE signal transduction system is among the primary sensory-regulatory mechanisms that mediate bacterial adaptation processes in response to environmental perturbation (24). The environmental conditions encountered by S. mutans in dental biofilms are highly variable, including frequent shifts in pH from above 7.0 to as low as 3.0 during the ingestion of dietary carbohydrates. In addition, S. mutans is subject to various environmental stresses, such as temperature fluctuation, nutritional limitation, antibiotic agents, and variation in oxygen tension (6). Therefore, the appearance of the WIG− strain may result from environmental perturbation. In this study, we investigated the mechanisms underlying expansion of the WIG− strain in a clonal population and the ecological significance of this variant.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are described in Table 1. Brain heart infusion (BHI) (Becton Dickinson) was used, or pH-buffered BHI (prepared with phosphate-buffered saline [pH 7.2]) was used for counting WIG− cells and studying natural competency (see below). The clinical isolate S. mutans FSM-11 (29) was cultured at 37°C with 5% CO2. Escherichia coli DH5α was used for cloning and plasmid amplification and was grown in Luria-Bertani broth or 1.5% agar at 37°C. When required, sucrose (0.25% [wt/vol]) or horse serum (10% [vol/vol]) was added to the medium. Bactericidal agents tested for the appearance of WIG− cells and natural competency were used under different conditions at a 10% to 20% reduction in optical density (OD) after 20 h of culture.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant properties | Source or reference |
|---|---|---|
| E. coli DH5α | Cloning host | Takara |
| S. mutans | ||
| FSM-11 WIG+ | Clinical isolate, wild type; Amps Erms Spcs Kms, serotype e | 29 |
| WIG+recA mutant | FSM-11 WIG+::pSAR; recA Ermr | This study |
| WIG+comC mutant | FSM-11 WIG+::pSAC; comC Ermr | This study |
| WIG+comD mutant | FSM-11 WIG+::pSAD; comD Ermr | This study |
| WIG+comE mutant | FSM-11 WIG+::pSAE; comE Ermr | This study |
| WIG+comX mutant | FSM-11 WIG+::pSAX; comX Ermr | This study |
| WIG+recA donor strain | FSM-11 WIG+::pSAR, pSABC; recA gtfBC Ermr Spcs Kanr | This study |
| FSM-11 WIG− | Naturally derived variant of FSM-11 WIG+; Amps Erms Spcs Kans; serotype e | This study |
| WIG− recipient strain | FSM-11 WIG−::pSAL; ldh Erms Kans Spcr | This study |
| Plasmids | ||
| pGEM-T | PCR cloning vector; Ampr | Promega |
| pUC19 | PCR cloning vector; Ampr | 43 |
| pDL276 | E. coli-Streptococcus shuttle vector; Kanr | 10 |
| pResEmMCS10 | Streptococcal integration plasmid; Ermr | 38 |
| pFW5 | Streptococcal integration plasmid; Spcr | 35 |
| pSAR | pUC19 containing recA fragment; Ampr Ermr | This study |
| pSAC | pUC19 containing comC fragment; Ampr Ermr | This study |
| pSAD | pUC19 containing comC fragment; Ampr Ermr | This study |
| pSAE | pUC19 containing comE fragment; Ampr Ermr | This study |
| pSAX | pUC19 containing comX fragment; Ampr Ermr | This study |
| pSABC | pUC19 containing gtfBC fragment; Ampr Kanr | This study |
| pSAL | pUC19 containing ldh fragment; Ampr Spcr | This study |
Frequency of WIG− cells.
S. mutans was precultivated to the stationary phase. To eliminate extracellular DNA in the bacterial preculture, the culture was washed twice in distilled water (D/W) using centrifugation. An aliquot of the precultured solution with a reduced cell number (∼100 to 500 cells) was inoculated in pH-buffered BHI medium with or without bactericidal agents. These cultures were incubated with 5% CO2 at 37°C statically for 20 h. The cultures were suspended in D/W, serially diluted, and spread-plated on BHI agar to 300 colonies/plate. The morphology of the samples were determined after 48 h on plates with 0.25% sucrose, and the ratio of smooth colonies to total viable cells was determined. Over 1,000 colonies were selected from at least three independent experiments.
Biofilm formation. (i) Biofilm formation in flow cells.
To show the appearance of the WIG− variant, S. mutans was grown in continuous-culture flow cells as described previously with modifications (30). Flow cells were inoculated with reduced numbers of precultured S. mutans (∼100 to 500). After 1 h of incubation at 37°C without flow, pH-buffered BHI with 0.25% sucrose was added at a rate of 3.0 ml/h using a peristaltic pump. The detection cell at outflow was a silicon tube 10 cm in length and 1 mm in inner diameter. After 16 h and 32 h of cultivation, the viable cells in the biofilm and the effluent were collected and plated on BHI agar with 0.25% sucrose. Over 1,000 colonies were selected from at least three independent experiments.
(ii) Microtiter plate biofilm formation.
Quantification of biofilm formation was performed using cells grown in the wells of polystyrene microtiter plates (Sumitomo Bakelite, Tokyo, Japan) (29). Strains were grown to an OD at 600 nm (OD600) of 0.4, diluted 1:100 in BHI with 0.25% sucrose, and added to the wells of the plates. The plates were incubated at 37°C with 5% CO2 for 20 h. After incubation, the plates were washed two times with D/W to remove planktonic and loosely bound bacteria, and adherent cells were stained with safranin for 10 min. Biofilm formation was then quantified by measuring the OD of the solubilized stain at 492 nm.
DNA extraction.
DNA from bacterial cells was extracted using the benzyl chloride method (47). The isolated DNA was used as the template for the PCR experiments described below.
Construction of mutants.
Insertion-inactivation of the gene in S. mutans FSM-11 was performed with insertion of erythromycin (Erm), kanamycin (Kan), and/or spectinomycin (Spc) resistance genes using homologous recombination. Sequence information was obtained from the S. mutans UA159 genome (http://www.ncbi.nlm.nih.gov/genomes). The method for homologous recombination was described previously (29). The recA and comE gene fragments from chromosomal DNA of S. mutans FSM-11 were ligated into the pGEM-T Easy vector (Promega, Madison, WI). The constructed pSAR and pSAE, harboring the recA and comE genes, respectively, were disrupted using digestion with BamHI and ligation with the Erm fragment from pResEmMCS10 (38). To construct comC, comD, and comX mutant strains and to insert an antibiotic resistance marker in the ldh and gtf genes, a PCR fragment from the upstream region of each gene from the chromosomal DNA of FSM-11 was amplified using PCR. The amplified fragment was inserted into the respective sites of cloning vector pUC19 (43). A PCR fragment from the downstream region was ligated to pUC19 from the upstream region. The resultant plasmid was digested with BamHI, and either an Erm fragment from pResEmMCS10 (38), a Kan fragment from pDL276 (10), or a Spc fragment from pFW5 (35) was inserted. Each strain was transformed with the resultant plasmids pSAC, pSAD, and pSAX for disruption and with comC, comD, and comX for insertion of the antibiotic resistance marker in the ldh gene. Further, each strain was transformed with the resultant plasmids pSEL and pSEBC to insert the antibiotic resistance marker in the gtf gene. Erm-resistant strains, Kan-resistant strains, and Spc-resistant strains were selected on agar containing 10 μg/ml Erm, 800 μg/ml Kan, and 800 μg/ml Spc, respectively. The mutations were confirmed using PCR.
Long-range PCR.
Amplification of the gtfB and gtfC region was achieved with the primers GtfB186F and GtfC9269R (Table 2) with LA polymerase (Takara). Amplification conditions were 30 cycles of 30 s at 95°C, 1 min at 56°C, and 9 min at 70°C. PCR products were visualized after gel electrophoresis in 0.8% agarose using ethidium bromide staining.
Table 2.
Amplicons and oligonucleotide primers used in this study
| Primer | Sequence (5′→3′)a | Amplicon |
|---|---|---|
| Long-range PCR | ||
| GtfB186F | TTGGAGGTTCCTAATGGAC | gtfBC |
| GtfC9269R | GAAATTTACAGCTCAGACTTC | |
| Mutants | ||
| RecA2F | GAGTTGCAGGTCCGGATAGC | recA |
| RecA2R | GCAATGGAGAGCCTTAGCATAGCGG | |
| ComE1F | CCTGAAAAGGGCAATCACCAG | comE |
| ComE1R | GCGATGGCACTGAAAAAGTCTC | |
| ComCuF | cccccgaattcAAATCTGAACAAGCAGGGG | comC1 |
| ComCuR | cccccggtaccGATAGTGTTTTTTTCATTTTATATCTCC | |
| ComCdF | ccccctctagaGCCTATCAACATTTTTCCGGC | comC2 |
| ComCdR | cccccaagcttCCACTAAAGGCTCCAATCGC | |
| ComDuF | cccccgaattcCCATTCATCTGAAACTCAGT | comD1 |
| ComDuR | cccccggtaccAACAGGCAGCAGACCATAA | |
| ComDdF | ccccctctagaGGCGGGCAATCATATTCTT | comD2 |
| ComDdR | cccccaagcttTCCTGCAATTGATGTCCTG | |
| ComX2uF | cccccgaattcATTGGGCGTGTTCGACATAC | comX1 |
| ComX2uR | cccccggtaccTGACTTTCTTGCTGACGC | |
| ComX2dF | ccccctctagaTTGGTAGCAGGAGAGCAC | comX2 |
| ComX2dR | cccccaagcttGAGATATGGTATCTCCTC | |
| LdhuF | cccccgaattcATCTGGAAGAGCCCGAGCAAC | ldh1 |
| LdhuR | cccccggtaccGGTGAAGTAAAAGCAAGTGC | |
| LdhdF | ccccctctagaGGTATCTTCCTCGTTGCTGC | ldh2 |
| LdhdR | cccccaagcttGGAATATTTACTGGGCGAAC | |
| GtfuF | cccccgaattcTGAGTGGTGTATGGCGTCAC | gtfB |
| GtfuR | cccccggtaccGACCGTTAATGGTTCTGGC | |
| GtfdF | ccccctctagaAAACTCTGACTGCTACTGATAC | gtfC |
| GtfdR | cccccaagcttGAGCAAAGCTGTTAGTGTTATCA |
Sequences homologous to the S. mutans genome sequences are in uppercase; synthetic restriction sites added for cloning purposes are in lowercase.
Transformation assay.
After being washed in D/W, S. mutans cultures were diluted 1:20 in BHI with horse serum. To determine the transformation frequency under stress conditions of a 10% to 20% OD reduction, S. mutans was grown to an OD600 of 0.5 without horse serum. Aliquots (0.2 ml) of culture were incubated at 37°C in equal volumes of fresh medium with 100 μg purified ldh (∼2 kb) containing the Spcr gene. After 2 h of incubation, transformants and total cells were plated on BHI agar with and without spectinomycin, respectively. The transformation efficiency was determined after 48 h of incubation and was expressed as the percentage of transformants compared to the total number of viable recipient cells.
Coculture experiments.
WIG+ (Kanr and Ermr) and WIG− (Spcr) strains were grown separately overnight in BHI. The cultures were centrifuged and washed two times to eliminate released extracellular DNA. The cells were resuspended in fresh BHI to an OD600 of 0.1. Both cultures were allowed to grow to an OD600 of 0.3 and mixed at a 1:4 ratio of donor to recipient strains. An aliquot (500 μl) of the mixed sample was centrifuged and the supernatant discarded. The sample was resuspended and incubated at 37°C for 8 h in 1 ml of fresh BHI medium with 10% horse serum. The culture was plated on BHI with 0.25% sucrose containing various antibiotics. Transformation efficiency was determined after 48 h of incubation and was expressed as the percentage of Spc- and Kan-resistant cells.
Bacteriocin production assay.
To detect bacteriocin produced by S. mutans FSM-11 WIG+ and the WIG− variant, 5 μl of an overnight culture of each strain adjusted to an OD600 of 0.4 was inoculated on BHI plates. After overnight culture, 5 μl each of the indicator Streptococcus species, S. gordonii, S. mitis, and S. sanguinis, at the same concentration was inoculated near the early colonies and further incubated anaerobically at 37°C. Bacteriocin production was determined by measuring the diameter of the zone inhibited compared to the indicator strains.
RESULTS
WIG− strains require a complete competence cascade.
Colonies with the parental WIG+ phenotype show a hard and rough colony morphology on sucrose agar medium, whereas the naturally derived WIG− colonies show a smooth morphology. We used colony morphology as a criterion for the WIG− phenotype. Using reduced inocula (∼100 to 500 cells) decreased the number of naturally derived variants in the bacterial preculture. After 20 h of culture in pH buffer (pH 7.2), the cultures were plated on BHI agar plates containing sucrose. Laboratory strains (MT8148, UA159, and GS5) and 16 clinical isolates (29) all showed WIG− cells at a rate of ∼1.0 × 10−3 to 1.0 × 10−2. Interestingly, several clinical isolates (FSC-1, FSM-2, and FSM-11) produced 10-fold more WIG− cells than the laboratory strains (31). Thus, we used the FSM-11 isolate to determine the mechanisms underlying the appearance of the WIG− variant.
Previous studies demonstrated that the recombination of gtfB and gtfC causes the WIG− phenotype and is RecA dependent (3). We found the S. mutans FSM-11 recA mutant did not produce WIG− cells (Table 3). The recA gene in pneumococci regulates a late competence gene (24). The comX gene encodes an alternative sigma factor that recognizes a com box (TACGAATA) that is conserved in the putative promoter regions of the late com genes (19). The recA gene in S. mutans is located immediately upstream of the cinA gene, the location of the putative promoter com box. Additionally, using microarray analysis, Perry et al. (34) showed that recA gene expression was regulated by ComX. We therefore determined the appearance of the WIG− variant using various com gene mutations. comX, comC, and comD mutations completely inhibited WIG− strain formation, and the inactivation of comE decreased the appearance of WIG− cells compared to that with the wild type (Table 3). This indicates that the generation of WIG− cells is regulated by the com system.
Table 3.
Appearance of WIG− cells among various com gene mutant cells
| Genotype | No. of colonies | WIG− cells |
|
|---|---|---|---|
| No. of colonies | Frequency (×10−3)a | ||
| Wild type | 3,468 | 23 | 6.63 |
| recA | 4,530 | NDb | ND |
| comC | 2,953 | ND | ND |
| comD | 3,087 | ND | ND |
| comE | 2,839 | 1 | 0.34 |
| comX | 3,513 | ND | ND |
The WIG− frequency was expressed as the ratio of cells with smooth colony morphology to total viable cells on a sucrose-containing BHI plate. More than 1,000 colonies were selected from at least five independent experiments.
ND, not detected.
Environmental stress induces the WIG− phenotype.
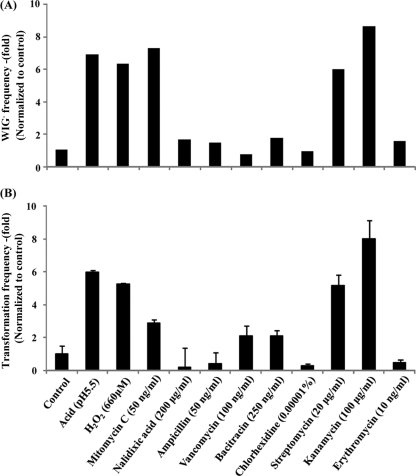
Environmental stress induces the competence cascade (36); we therefore determined the effect of various environmental stresses on the appearance of the WIG− variant. Acid stress is an important factor in the oral environment. A pH of 5.5 induced a 10% to 20% OD reduction where the final pH values were pH 5.8 and pH 4.6, respectively. WIG− cells were present under acidic conditions at ∼7-fold-higher levels than at neutral pH (Fig. 1A). Oxidative stress and bactericidal agents were tested because a number of antibiotics are linked to the development of natural competence (34, 36). The presence of 660 μM H2O2 in the pH-buffered medium induced the appearance of WIG− cells at an increased rate. Cell wall synthesis inhibitors (ampicillin, vancomycin, bacitracin, and chlorhexidine) had no effect. The protein synthesis inhibitors kanamycin and streptomycin (not erythromycin) induced the appearance of WIG− cells. The DNA synthesis inhibitor mitomycin C induced the appearance of WIG− cells, whereas nalidixic acid did not. Acidic conditions and sublethal concentrations of H2O2, mitomycin C, streptomycin, and kanamycin induced the transformation frequency >7- to 9-fold compared to that in the culture at neutral pH (Fig. 1B), and these compounds induced the WIG− strain, indicating that the activation of natural competence is related to the appearance of the WIG− phenotype.
Fig. 1.
Effect of environmental stresses on the appearance of WIG− cells and the transformation frequency. (A) Appearance of WIG− cells. More than 1,000 colonies were selected from at least three independent experiments. (B) Transformation frequency. Transformation efficiency was determined after 48 h of incubation and is expressed as the percentage of transformants among the total viable recipient cells. Data are averages from three independent experiments. Bactericidal agents tested for the appearance of WIG− cells and transformation frequency were used under different conditions at a 10% to 20% OD reduction after 20 h of cultivation. Values are expressed as percentages of the control values.
The biofilm environment induces the WIG− phenotype.
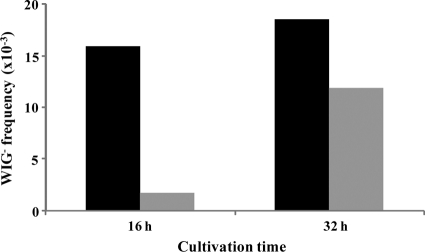
The biofilm has a higher natural competence than planktonic cells (21). To show the appearance of WIG− biofilm cells, we introduced a diluted inoculum into the flow cell (see Materials and Methods). After 16 h and 32 h of cultivation, the biofilm was directly collected from the flow cell and plated on sucrose agar medium. We also plated the outflow at each time point (planktonic cells). The biofilm biomasses after 16 h and 32 h were 3.78 × 108 CFU/ml and 7.76 × 108 CFU/ml, respectively. The biofilm cells at 16 h and 32 h included greater numbers of WIG− cells than in the planktonic cells (Fig. 2). Among planktonic cells, the presence of WIG− cells was slightly induced at of 16 h culture and increased after 32 h, suggesting that the increase in the numbers of WIG− cells among planktonic cells after the longer cultivation period was derived from the biofilm.
Fig. 2.
Presence of WIG− cells in the biofilm and among planktonic cells. Frequency is expressed as the ratio of the WIG− colony number to the total colony number. The presence of WIG− cells in the biofilm (black bars) and among planktonic cells (gray bars) is expressed as a percentage of biofilm cells at 16 h. More than 1,000 colonies were selected from at least three independent experiments.
DNA uptake affects the appearance of WIG− cells.
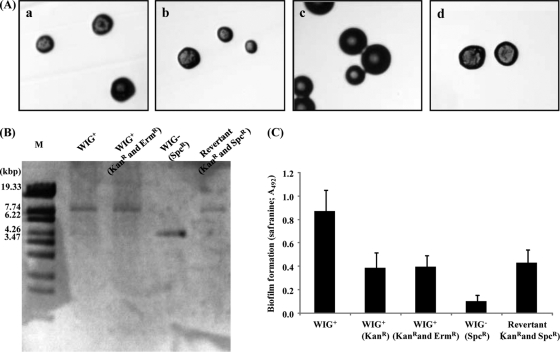
Activation of the com cascade allows endogenous chromosomal DNA to be transported and allows incorporation of exogenous DNA (9). We therefore considered that incorporation of extracellular DNA may induce the presence of WIG. This was difficult to evaluate quantitatively because both exogenous DNA and endogenous DNA affect the appearance of WIG− recombination. WIG− cells were capable of transformation (data not shown). Therefore, the appearance of the revertant WIG− to WIG+ with an intact gtfBC may indicate exogenous transformation. To test this, we used a change of phenotype in a coculture experiment. To monitor DNA transfer, a WIG+ strain with a Kanr marker in the intergenic region between gtfB and gtfC gene was constructed as the donor cell. The WIG+ strain with Kanr also was disrupted in the recA gene to eliminate exogenous DNA integration into its chromosomal DNA. The constructed WIG+ strain (Kanr and Ermr) lost the integration of exogenous DNA into the genome (Table 4). Additionally, the WIG+ cells (Kanr and Ermr) showed a rough morphology on plates including sucrose (Fig. 3A, panel b); however they showed a decrease in safranin-stained biofilm biomass compared to that for the parental WIG+ strain in the microtiter plate biofilm assay with sucrose (Fig. 3C). We also inserted a spectinomycin resistance gene into the ldh gene of the WIG− strain. The constructed WIG− (Spcr) strain maintained a natural competency as did the wild type (Table 4), with a stable smooth colony morphology (Fig. 3A, panel c); no revertants were detected after several passages (data not shown). Both the WIG+ (Kanr and Ermr) and WIG− (Spcr) strains showed strong inhibitory activity against S. gordonii (data not shown). We prepared a mixed 1:4 inoculum containing the donor WIG+ and the recipient WIG− strains and inoculated them into BHI with horse serum. After 8 h of coculture, we plated on Kan-Spc to select revertants from the recipient WIG− cells and on Spc to detect the nontransformed recipient cells. After 2 days of culture, Kan-Spc-resistant cells with a rough morphology were found at a level of 8.45 × 10−6 using medium with sucrose (Table 4). The gtfBC region of the revertants was determined using long-range PCR. PCR products of ∼9.0 kb were amplified from the chromosomal DNAs of the parent WIG+ cells (Fig. 3B, lane 2) and the donor WIG+ cells (Fig. 3B, lane 3), whereas a 5.0-kb product was amplified from the chromosomal DNA of the recipient WIG− cells (Fig. 3B, lane 4). A randomly selected rough-morphology colony on the Kan-Spc-sucrose plate was used for replacement with a 9.0-kb gene fragment (Fig. 3B, lane 5). In connection with its gene replacement, biofilm formation of the revertant was restored to the level of the donor WIG+ strain (Fig. 3C).
Table 4.
Transformation efficiencies of WIG+ and WIG− strains in single- and two-species cultures
| DNA recipient strain | Donor DNAa | Target transformant gene | Transformant frequencyb |
|---|---|---|---|
| WIG+ | ldh (Spc) gene | Spcr | 3.87 × 10−4 ± 1.45 × 10−4 |
| WIG+ (gtfBC and recA) | ldh (Spc) gene | Spcr | NDc |
| WIG+ (gtfBC and recA) | WIG− (Spcr) strain | Spcr | ND |
| WIG− | ldh (Spc) gene | Spcr | 3.34 × 104 ± 2.48 × 104 |
| WIG− (ldh) | WIG+ (Kanr and Ermr) strain | Kanr | 8.45 × 10−6 ± 3.81 × 10−6 |
The ldh gene with Spcr in the donor DNA was amplified from the S. mutans FSM-11 ldh mutant. An aliquot (100 ng) of purified PCR product was added to the BHI medium with horse serum. The WIG− and WIG+ donor DNA strains were cocultured with the recipient strain in BHI with horse serum.
Transformation efficiency in single cultures was determined and was expressed as the percentage of Spc-resistant cells compared to total viable cells after 2 days of culture. With coculture, the number of recipient WIG+ cells was expressed as the percentage of Spc-Erm-resistant cells compared to Erm-resistant cells, and the number of recipient WIG− cells was expressed as the percentage of Spc-Kan-resistant cells compared to Spc-resistant cells. The data are the averages and standard deviations from three independent experiments.
ND, not detected.
Fig. 3.
Characterization of parental WIG+ cells, donor WIG+ cells, recipient WIG− cells, and WIG+ revertant cells. (A) Colony morphologies of WIG+ (a), WIG+ donor (Kanr and Ermr) (b), WIG− recipient (Spcr) (c), and revertant (Kanr Spcr) (d) cells on BHI agar plates with 0.25% sucrose. Colonies were observed using a stereomicroscope (Nikon). (B) Amplification of the gtfB and gtfC regions. Lane M, Sty1-digested lambda DNA; lane 2, WIG+ cells; lane 3, donor WIG+ (Kanr and Ermr) cells; lane 4, recipient WIG− (Spcr) cells; lane 5, revertant (Kanr Spcr) cells. (C) Biofilm formation assay on a microtiter plate. Cells were grown in BHI medium with 0.25% sucrose at 37°C. Quantification of the biofilm after 20 h of cultivation was performed using the safranin staining method (see Materials and Methods). The data are expressed as means ± standard deviations for three independent experiments.
DISCUSSION
The spontaneous generation of genetic variation in bacteria can be induced using three natural methods (2): small local changes in the nucleotide sequence of the genome, intragenomic rearrangement of genomic sequences, and acquisition of DNA sequences from the environment. Previous studies showed that the generation of the WIG− variant was dependent on endogenous DNA rearrangement of gtfB and gtfC using homologous recombination (40). Here we found that in mixed cultures of WIG+ and WIG− cells, the reversion from WIG− to WIG+ was enabled with exogenous DNA uptake, indicating that extracellular DNA recombination contributes to the expansion of mutants in a population. In general, natural genetic transformation in bacteria uses active uptake of extracellular DNA and heritable incorporation of this genetic information (9). Most of the natural transformations in streptococci are regulated by a ComDE two-component regulatory system and the extracellular CSP level. Here we found that endogenous and exogenous DNA recombination generates WIG− cells and that both were induced by the same signal transducer, the so-called com system. The RecA-induced DNA recombination in pneumococci is regulated by ComX (23, 24, 34), and ComY has been shown to be essential for binding DNA to the cell surface and is also dependent on ComX (27). We speculated that the generation of the ancestral WIG− variant and expansion of the mutant through the uptake of extracellular DNA may be temporally coincident in the population. This coordinated mechanism may be required for the release of extracellular DNA from the cell. In S. pneumoniae a competence-specific recA transcript was shown to induce lytA, which is located immediately downstream of recA and encodes an autolysin (23, 28). Conversely, the downstream sequence of recA in S. mutans is different from that in S. pneumoniae, where disruption of S. mutans SMU2083 and SMU2084, located downstream of the recA gene, did not affect the amount of extracellular DNA uptake (data not shown). Recent studies indicate that S. mutans can control the expression of many of its bacteriocins through the com system (14, 17, 46). The coordination of bacteriocin production and competence suggests that S. mutans may generate DNA for uptake using lysis of neighboring species (17). Perry et al. (34) indicate that there also is a relationship between competence-regulated bacteriocin production and cell lysis and discuss the dissemination of a fitness-enhancing gene in the oral biofilm. We propose that coordination among endogenous DNA recombination, exogenous DNA recombination, and cell lysis through the com system contributes to the production of heterogeneity in the clonal population, where this complex regulatory system may not be limited to in vitro conditions. S. mutans is organized on biofilms, where the cells are closely packed together and multiple species of microorganisms coexist. A previous study indicates that the S. mutans WIG− variant has a stronger presence within biofilm cells than among planktonic cells (3). Our data are in agreement with this (Fig. 2). Oral streptococci such as S. gordonii and S. salivarius contain WIG homologs sharing high homology with that of S. mutans. Future research will examine interspecies interaction through uptake of WIG.
In S. mutans, no binding motif for ComE is present in the promoter region of ComX, suggesting a missing link between both regulators and ComX. Recent studies reported that competence in S. mutans is controlled by two different quorum-sensing systems, ComDE and ComRS (20, 25). They proposed a model where the ComRS system is the proximal regulator of comX and ComDE is an upstream regulator that may be connected to the ComRS system. Signaling information from competence regulators such as ComDE is integrated through the control of ComRS activity, though their effects may not direct and could be relayed through one or more undiscovered components (25). The disruption of SMU61, encoding comR, in the FSM-11 strain completely inhibited the appearance of WIG− cells (data not shown). This result supports at least our conclusion that the appearance of WIG− cells is regulated by competence. However, the detailed mechanisms underlying the integration of the ComRS and ComDE systems will need further investigation.
E. coli and other bacteria have a highly effective response to DNA damage, the SOS response, that minimizes the lethal and mutagenic consequences of damage (12). Cell DNA damage blocks the replication fork, generating a single-strand DNA region to which RecA binds to form a nucleoprotein filament. These complexes stimulate the self-cleavage of the LexA repressor and lead to the induction of the SOS genes, including recA. However, S. pneumoniae has a natural competence system similar to that of S. mutans and lacks an SOS-like induction system, instead using the competence regulatory cascade to coordinate responses to stress (36). We did not find a lexA-like sequence in the genome database of S. mutans. Additionally, acidic and oxidative stresses and mitomycin C induce the SOS response in E. coli, resulting in accelerated natural competency (Fig. 1B). Wen et al. (45) indicated that acid and oxidative stresses are linked to the development of natural competence. This suggests that the development of natural competence in S. mutans maybe a substitute for the SOS response. However, the parallel is only partial because competence was not induced by nalidixic acid and ampicillin, which induce the SOS response in E. coli, and kanamycin and streptomycin induce heat shock protein expression (41), suggesting that stress signals are processed differently in the two species.
The number of WIG− cell among planktonic cells increased during prolonged cultivation periods (Fig. 2), suggesting that the former were derived from the biofilm environment. The biofilm biomass contents after 16 h and 32 h of cultivation were similar. In addition, the biomass in the outflow solution also remained unchanged using both cultivation periods (data not shown). This indicates that after 16 h the biofilm had attained a steady state where the intact mature biofilm does not disperse after prolonged culture. Because WIG is associated with the biofilm architecture (15, 39), the prevalence of WIG− cells in biofilms may change the structure, resulting in the release of WIG− cells. Plaque pH can be lower than 5.0 and remain so for some time; this low pH leads to demineralization of the enamel (42). Our data show that an acidic pH promotes the presence of WIG− cells (Fig. 1A). In addition, the growth rate of WIG− cells with sucrose supplementation was greater than that of the wild type (31), suggesting that carbohydrate fermentation by the mutant contributes to an early shift to a more acidic environment. Therefore, the WIG− variant may contribute to further demineralization and development of caries. An improved understanding of the behavior of the variant in situ would help to determine its role in natural ecosystems.
Other than sucrose-dependent adherence, S. mutans is diverse in its antigenic polysaccharides that determine serotype, and it also varies in sugar fermentation, bacteriocin type, capacity to produce acid, interaction with salivary components, and release of surface proteins (44). In natural ecosystems, population heterogeneity may mitigate the impact of environmental perturbation because the presence of diverse members extends the range of conditions under which the organism can persist (7, 25). Diversity can also enhance population stability and productivity because of cooperative interactions among community members (4, 5, 7, 26). Therefore, one consequence of the appearance of the S. mutans WIG− variant would be to enhance fitness in the human oral environment. Our data may help in understanding how S. mutans can adapt to the oral environment and ultimately help explain the evolution of S. mutans. The mechanism of the appearance of the WIG− variant observed in this work may serve as a useful model for studying population heterogeneity among species.
ACKNOWLEDGMENTS
We thank Justin Merritt and Fengxia Qi (University of Oklahoma Health Sciences Center) for providing the pFW5 plasmid and Ryoma Nakao, Saori Yoneda, and Soichi Furukawa for their advice and valuable discussions.
This work was supported in-part by Grants-in-Aid for the Development of Scientific Research (no. 21390506) and a Research Fellowship of the Japan Society for the Promotion of Science (no. 10J10410, JSPS Research Fellow) from the Ministry of Education, Science and Culture of Japan. This study was also supported in part by the “Strategic Research Base Development” Program for Private Universities from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (MEXT), 2010-2014 (S1001024).
Footnotes
Published ahead of print on 29 July 2011.
REFERENCES
- 1. Aoki H., Shiroza T., Hayakawa M., Sato S., Kuramitsu H. K. 1986. Cloning of a Streptococcus mutans glucosyltransferase gene coding for insoluble glucan synthesis. Infect. Immun. 53:587–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arber W. 2000. Genetic variation: molecular mechanisms and impact on microbial evolution. FEMS Microbiol. Rev. 24:1–7 [DOI] [PubMed] [Google Scholar]
- 3. Banas J. A., et al. 2007. Evidence that accumulation of mutants in a biofilm reflects natural selection rather than stress-induced adaptive mutation. Appl. Environ. Microbiol. 73:357–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boles B. R., Thoendel M., Singh P. K. 2004. Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl. Acad. Sci. U. S. A. 101:16630–16635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boles B. R., Singh P. K. 2008. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc. Natl. Acad. Sci. U. S. A. 105:12503–12508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carlsson J. 1997. Bacterial metabolism in dental biofilms. Adv. Dent. Res. 11:75–80 [DOI] [PubMed] [Google Scholar]
- 7. Carr M. H., Anderson T. W., Hixon M. A. 2002. Biodiversity, population regulation, and the stability of coral-reef fish communities. Proc. Natl. Acad. Sci. U. S. A. 99:11241–11245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Costerton J. W., Lewandowski Z., Caldwell D. E., Korber D. R., Lappin-Scott H. M. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711–745 [DOI] [PubMed] [Google Scholar]
- 9. Cvitkovitch D. G. 2001. Genetic competence and transformation in oral streptococci. Crit. Rev. Oral Biol. Med. 12:217–243 [DOI] [PubMed] [Google Scholar]
- 10. Dunny G. M., Lee L. N., LeBlanc D. J. 1991. Improved electroporation and cloning vector system for gram-positive bacteria. Appl. Environ. Microbiol. 57:1194–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elkins J. G., Hassett D. J., Stewart P. S., Schweizer H. P., McDermott T. R. 1999. Protective role of catalase in Pseudomonas aeruginosa biofilm resistance to hydrogen peroxide. Appl. Environ. Microbiol. 65:4594–4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Foster P. L. 2005. Stress responses and genetic variation in bacteria. Mutat. Res. 569:3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fux C. A., Wilson S., Stoodley P. 2004. Detachment characteristics and oxacillin resistance of Staphyloccocus aureus biofilm emboli in an in vitro catheter infection model. J. Bacteriol. 186:4486–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hale J. D., Ting Y. T., Jack R W., Tagg J. R., Heng N. C. 2005. Bacteriocin (mutacin) production by Streptococcus mutans genome sequence reference strain UA159: elucidation of the antimicrobial repertoire by genetic dissection. Appl. Environ. Microbiol. 71:7613–7617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hazlett K. R., Mazurkiewicz J. E., Banas J. A. 1999. Inactivation of the gbpA gene of Streptococcus mutans alters structural and functional aspects of plaque biofilm which are compensated by recombination of the gtfB and gtfC genes. Infect. Immun. 67:3909–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoiby N., et al. 2001. Pseudomonas aeruginosa and the in vitro and in vivo biofilm mode of growth. Microb. Infect. 3:23–35 [DOI] [PubMed] [Google Scholar]
- 17. Kreth J., Merritt J., Shi W., Qi F. 2005. Co-ordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighbouring species. Mol. Microbiol. 57:392–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kuramitsu H. K., He X., Lux R., Anderson M. H., Shi W. 2007. Interspecies interactions within oral microbial communities. Microbiol. Mol. Biol. Rev. 71:653–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee M. S., Morrison D. A. 1999. Identification of anew regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181:5004–5016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lemme A., Gröbe L., Reck M., Tomasch J., Wagner-Döbler I. 2011. Subpopulation-specific transcriptome analysis of competence-stimulating-peptide-induced Streptococcus mutans. 193:1863–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Y. H., Lau P. C., Lee J. H., Ellen R P., Cvitkovitch D. G. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183:897–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Love P. E., Yasbin R. E. 1986. Induction of the Bacillus subtilis SOS-like response by Escherichia coli RecA protein. Proc. Natl. Acad. Sci. U. S. A. 83:5204–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martin B., Garcia P., Castanie M. P., Claverys J. P. 1995. The recA gene of Streptococcus pneumoniae is part of a competence-induced operon and controls lysogenic induction. Mol. Microbiol. 15:367–379 [DOI] [PubMed] [Google Scholar]
- 24. Martin B., Quentin Y., Fichant G., Claverys J. P. 2006. Independent evolution of competence regulatory cascades in streptococci? Trends Microbiol. 14:339–345 [DOI] [PubMed] [Google Scholar]
- 25. Mashburn-Warren L., Morrison D. A., Federle M. J. 2010. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol. Microbiol. 78:589–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McCann K. S. 2000. The diversity-stability debate. Nature 405:228–233 [DOI] [PubMed] [Google Scholar]
- 27. Merritt J., Qi F., Shi W. 2005. A unique nine-gene comY operon in Streptococcus mutans. Microbiology 151:157–166 [DOI] [PubMed] [Google Scholar]
- 28. Mortier-Barriere I., de Saizieu A., Claverys J. P., Martin B. 1998. Competence-specific induction of recA is required for full recombination proficiency during transformation in Streptococcus pneumoniae. Mol. Microbiol. 27:159–170 [DOI] [PubMed] [Google Scholar]
- 29. Motegi M., et al. 2006. Assessment of genes associated with Streptococcus mutans biofilm morphology. Appl. Environ. Microbiol. 72:6277–6287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Narisawa N., Haruta S., Arai H., Ishii M., Igarashi Y. 2008. Coexistence of antibiotic-producing and antibiotic-sensitive bacteria in biofilms is mediated by resistant bacteria. Appl. Environ. Microbiol. 74:3887–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Narisawa N. 2010. Development, characterization and ecological implications of a smooth colony variant of biofilm-forming cariogenic Streptococcus mutans. J. Oral Biosci. 52:245–251 [Google Scholar]
- 32. Nomura R., et al. 2006. Isolation and characterization of Streptococcus mutans in heart valve and dental plaque specimens from a patient with infective endocarditis. J. Med. Microbiol. 55:1135–1140 [DOI] [PubMed] [Google Scholar]
- 33. Olson M. E., Ceri H., Morck D. W., Buret A. G., Read R. R. 2002. Biofilm bacteria: formation and comparative susceptibility to antibiotics. Can. J. Vet. Res. 66:86–92 [PMC free article] [PubMed] [Google Scholar]
- 34. Perry J. A., Jones M. B., Peterson S. N., Cvitkovitch D. G., Levesque C. M. 2009. Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Mol. Microbiol. 72:905–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Podbielski A., Spellerberg B., Woischnik M., Pohl B., Liitticken R. 1996. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS). Gene 177:137–147 [DOI] [PubMed] [Google Scholar]
- 36. Prudhomme M., Attaiech L., Sanchez G., Martin B., Claverys J. P. 2006. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313:89–92 [DOI] [PubMed] [Google Scholar]
- 37. Sauer K., Camper A. K., Ehrlich G. D., Costerton J. W., Davies D. G. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shiroza T., Kuramitsu H. K. 1993. Construction of a model secretion system for oral streptococci. Infect. Immun. 61:3745–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thurnheer T., van der Ploeg J. R., Giertsen E., Guggenheim B. 2006. Effects of Streptococcus mutans gtfC deficiency on mixed oral biofilms in vitro. Caries Res. 40:163–171 [DOI] [PubMed] [Google Scholar]
- 40. Ueda S., Kuramitsu H. K. 1988. Molecular basis for the spontaneous generation of colonization-defective mutants of Streptococcus mutans. Mol. Microbiol. 2:135–140 [DOI] [PubMed] [Google Scholar]
- 41. VanBogelen R. A., Neidhardt F. C. 1990. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 87:5589–5593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Van Houte J., Lopman J., Kent R. 1996. The final pH of bacteria comprising the predominant flora on sound and carious human root and enamel surfaces. J. Dent. Res. 75:1008–1014 [DOI] [PubMed] [Google Scholar]
- 43. Vieira J., Messing J. 1987. Production of single-stranded plasmid DNA. Methods Enzymol. 153:3–11 [DOI] [PubMed] [Google Scholar]
- 44. Waterhouse J. C., Russell R. R. 2006. Dispensable genes and foreign DNA in Streptococcus mutans. Microbiology 152:1777–1788 [DOI] [PubMed] [Google Scholar]
- 45. Wen Z. T., Suntharaligham P., Cvitkovitch D. G., Burne R. A. 2005. Trigger factor in Streptococcus mutans is involved in stress tolerance, competence development, and biofilm formation. Infect. Immun. 73:219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yonezawa H., et al. 2008. Differential expression of the Smb bacteriocin in Streptococcus mutans isolates. Antimicrob. Agents Chemother. 52:2742–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhu H., Qu F., Zhu L. H. 1993. Isolation of genomic DNAs from plants, fungi and bacteria with benzyl chloride. Nucleic Acids Res. 21:5279–5280 [DOI] [PMC free article] [PubMed] [Google Scholar]