Abstract
Biofilms contribute to virulence of Staphylococcus aureus. Formation of biofilms is multifactorial, involving polysaccharide, protein, and DNA components, which are controlled by various regulators. Here we report that deletion of the rsp gene resulted in an increase in biofilm formation in strain MW2, suggesting that Rsp is a repressor of biofilm formation. Using SDS-PAGE, we found that Rsp profoundly affected cell surface and secreted proteins. The rsp gene was transcribed monocistronically, and the transcripts were most abundant at the exponential growth phase. Microarray analyses revealed that Rsp represses 75 genes, including 9 genes encoding cell wall-anchored proteins, and activates 22 genes, including 5 genes encoding secreted proteases. Among these genes, fnbA, fnbB, sasG, and spa (which encode cell wall-anchored proteins) and splABCD (which encode secreted proteases) have been implicated in biofilm formation. To deconvolute Rsp's contribution to biofilm formation, we analyzed deletion mutants of these genes either in the wild-type or in the rsp mutant background. We found that fnbA deletion in the rsp mutant restored biofilm formation to the wild-type level, indicating that FnbA plays a major role in Rsp regulation of biofilm formation. Further studies revealed that Rsp inhibited biofilm formation at the stage of primary attachment through repressing fnbA. Rsp belongs to the AraC/XylS family of regulatory proteins. We expressed the putative Rsp DNA binding domain (RspDBD) in Escherichia coli and showed that RspDBD was able to specifically bind to a short DNA fragment containing the fnbA promoter, suggesting that Rsp represses fnbA expression by direct DNA binding.
INTRODUCTION
As a human pathogen, Staphylococcus aureus is capable of forming biofilms that are critical for establishing infections such as endocarditis, osteomyelitis, and indwelling device-related infections. Bacteria within a biofilm are difficult to eradicate, as they are protected from antibiotics and components of the host immune system (7, 44). Biofilm forms in distinct steps, including initial attachment, maturation, and detachment (42, 43). Studies have shown that polysaccharide intercellular adhesin (PIA) is the major component affecting the maturation of S. aureus biofilms (17). PIA is composed of poly-N-acetylglucosamine (PNAG), whose synthesis requires 4 gene products encoded in the icaADBC operon, which is repressed by the IcaR repressor encoded upstream of the operon in opposite orientation (8, 24). However, PIA-independent biofilm formation has been reported in various strains (2, 11, 40, 47, 55), suggesting that biofilm formation in S. aureus does not depend on one component. Indeed, recent studies have shown that DNA and protein components also contribute to biofilm formation in S. aureus (6, 9, 23, 37, 40, 46, 49, 56). In addition, extracellular proteases have been indicated to affect biofilm formation (4).
Several surface-anchored proteins have been shown to promote S. aureus biofilm formation, including fibronectin binding proteins FnbA and FnbB (40), surface proteins SasC (49) and SasG (6, 15), IgG-binding protein Spa (37), and biofilm-associated protein Bap produced by bovine strains (9, 55). Interestingly, SasG-promoted biofilm formation required proteolytic cleavage of the SasG protein (15). In addition, secreted proteins Emp and Eap have also been shown to be involved in biofilm formation in association with PIA under low-iron conditions (25).
Biofilm formation is subjected to complex regulation in S. aureus. Several regulatory genes have been shown to affect biofilm formation (38, 43). We have previously reported that Rbf, a member of the AraC/XylS family of transcriptional regulators, promotes biofilm formation in S. aureus by repressing icaR expression (10, 32). Besides rbf, the S. aureus chromosome contains several additional genes that encode homologs of the AraC/XylS family of transcriptional regulators. Here we report that one of these genes, MW2301, also regulates biofilm formation in strain MW2 by affecting production of surface proteins. Accordingly, we named this gene rsp.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. S. aureus MW2, a community-acquired methicillin-resistant S. aureus (CA-MRSA) strain originally isolated from a child with fatal septicemia and septic arthritis (5), was provided by the Network on Antimicrobial Resistance in S. aureus. S aureus strains were routinely cultivated with tryptic soy broth (TSB) or tryptic soy agar (Difco Laboratories, Detroit, MI) unless specified. S aureus RN4220 (28) was used as a recipient for electroporation by the procedure of Kraemer and Iandolo (27). Phage 52A and 80α were used for plasmid and chromosomal DNA transduction between S. aureus strains. Escherichia coli strains DH5α and XL1-Blue were used for plasmid construction and maintenance (48). E. coli was cultivated in Luria-Bertani broth or agar. Antibiotics were added to the culture medium when necessary at final concentrations of 5 to 10 μg/ml for chloramphenicol, 10 μg/ml for erythromycin, and 100 μg/ml for penicillin.
Table 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Bacterial strains | ||
| S. aureus | ||
| RN4220 | Restriction-negative laboratory strain | 28 |
| MW2 | Wild-type clinical isolate | 5 |
| CYL11710 | MW2 Δrsp | This study |
| CYL11801 | MW2 Δrsp (pML3871) | This study |
| CYL11804 | MW2 Δrsp (pML100) | This study |
| CYL11805 | MW2 (pML100) | This study |
| CYL12228 | MW2 Δrsp ΔfnbA | This study |
| CYL12230 | MW2 Δrsp ΔfnbB | This study |
| CYL12264 | MW2 Δrsp ΔfnbA (pML100) | This study |
| CYL12265 | MW2 Δrsp ΔfnbA (pML12255) | This study |
| CYL12266 | MW2 Δrsp ΔfnbB (pML100) | This study |
| CYL12267 | MW2 Δrsp ΔfnbB (pML12256) | This study |
| CYL12278 | MW2 Δspl::erm | This study |
| CYL12317 | MW2 Δrsp ΔsasG | This study |
| CYL12319 | MW2 ΔscpA | This study |
| CYL12336 | MW2 Δrsp Δspa | This study |
| E. coli | ||
| DH5α | Host strain for plasmids | Invitrogen |
| XL1-Blue | Host strain for plasmids | Stratagene |
| SoluBL21 | Host strain for recombinant protein isolation | Genlantis |
| Plasmids | ||
| pLI50 | E. coli-S. aureus shuttle vector | 31 |
| pKOR1 | Vector for allele replacement | 1 |
| pML100 | Shuttle vector, derived from pLI50 and pKOR1 | This study |
| pML3871 | Rsp expression plasmid derived from pML100 | This study |
| pML12255 | FnbA expression plasmid derived from pML100 | This study |
| pML12256 | FnbB expression plasmid derived from pML100 | This study |
| pCL3972 | Rsp DNA binding domain-expressing plasmid | This study |
Plasmid and strain construction.
The pKOR1 system (1) was used to construct the MW2 rsp deletion mutant (CYL11710) as described before (34) using PCR primers MW2301-1, MW2301-2, MW2301-3, and MW2301-4. Similarly, primers FnbA1, FnbA2, FnbA3, and FnbA4 were used in construction of the fnbA deletion; primers FnbB1, FnbB2, FnbB3, and FnbB4 were used in construction of the fnbB deletion; primers sasG-1, sasG-2, sasG-3, and sasG-4 were used to construct the sasG deletion; primers scpA-1, scpA-2, scpA-3, and scpA-4 were used to construct the scpA deletion; primers spa-6, spa-7 spa-11, and spa-12 were used to construct the spa deletion. The mutations were confirmed by PCR analyses. The primers are listed in Table 2. The spl deletion mutant, CYL12278, was constructed by chromosomal transduction from KB600 (45), which carries the Δspl::erm allele, to MW2 and verified by PCR.
Table 2.
Oligonucleotide primers used for plasmid and strain construction
| Primer purpose and name | Sequence |
|---|---|
| Plasmid and strain construction | |
| MW2301-1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTGCGAATATTTTGTAACAGGGCTA |
| MW2301-2 | ATCTTTACGACTCGCGGCCGCTTCGATTATATTTTCGCCGTGGTT |
| MW2301-3 | GAAAATATAATCGAAGCGGCCGCGAGTCGTAAAGATAATAACTACCA |
| MW2301-4 | GGGGACCACTTTGTACAAGAAAGCTGGGTTCGATTTTGATAGGTGTATCTAATGA |
| MW2301-8 | GAATTCCACAAGAGGACTGTTAGTATTTAGC |
| MW2301-12 | GGATCCCCATGTTTTACATTTAAATGATAGATTTC |
| MW2301-14 | AGATCGAAGTCAGCGTGCCAAAGT |
| MW2301-15 | AGCTAATGCCACCGCCATATCGTA |
| FnbA5′1 | ACAAGGGTTTATGATGACTTGAATACAATTTATAGG |
| FnbA5′2 | CGTACCTAAGATTGTTTTTCACTATAATATCTCCC |
| FnbA-1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTGGTGCAGGTAGTGTGATTTTG |
| FnbA-2 | TCCGCCGAACAACATAGCGGCCGCCCGCTATTGTCTTGAGATTGTGTT |
| FnbA-3 | TCAAGACAATAGCGGGCGGCCGCTATGTTGTTCGGCGGATTATTCAGC |
| FnbA-4 | GGGGACCACTTTGTACAAGAAAGCTGGGTGATGGTTGCTCAGTTGATGTC |
| FnbA-6F | GTCGACGCATTTAAAGGGAGATATTATAGTGAAAAACAATCTTAGG |
| FnbA-7R | GAGCTCCAATGAAGCAATCAGAAAACACTCT |
| FnbB-1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTATGTTGTTCGGCGGATTATTCAGC |
| FnbB-2 | TCCGAACAGTATACCGCGGCCGCGATGGTTGCTCAGTTGATGTC |
| FnbB-3 | AACTGAGCAACCATCGCGGCCGCGGTATACTGTTCGGAGGATTG |
| FnbB-4 | GGGGACCACTTTGTACAAGAAAGCTGGGTCATACGTTCAATCGCATCTGTT |
| FnbB-6F | GTCGACTACATTTAAGGGAGAATATTATAGTGAAAAG |
| FnbB-7R | GAATTCGAACGCCTTCATAGTGTCATTAAGTATGTA |
| sasG-1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTGTGTTTATAGCTAGCACCACAAACT |
| sasG-2 | CATTAATCCAGCAATTCCAATTATACTGTGAGCCAATTAAAATAGATGCTG |
| sasG-3 | CAGCATCTATTTTAATTGGCTCACAGTATAATTGGAATTGCTGGATTAATG |
| sasG-4 | GGGGACCACTTTGTACAAGAAAGCTGGGTCTATCGATTAACGCATTACTTGTTGCT |
| scpA-1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCCACTTCTAACCATGCTTCATAG |
| scpA-2 | GATGAATACCATTGATAATGGTCGCCATTGCTCAGCGTTTGCAATAGGGGTAACACT |
| scpA-3 | AGTGTTACCCCTATTGCAAACGCTGAGCAATGGCGACCATTATCAATGGTATTCATC |
| scpA-4 | GGGGACCACTTTGTACAAGAAAGCTGGGTCTGGACTAAAGGCATTCGTCGTAG |
| spa-6 | TCTAGAGGATCCATACCCCCTGTATGTATTTGT |
| spa-7 | CCCGGGGATCCAAACAATACACAACGATAGA |
| spa-11 | GGGGACAAGTTTGTACAAAAAAGCAGGCTCATCAGCAAGAAAACACACT |
| spa-12 | GGGGACCACTTTGTACAAGAAAGCTGGGTCTGCCTTTGTATTAGTATGG |
| Rsp1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTGTTCATAGACTGTGTAAACTATATTC |
| Rsp2 | GGGGACCACTTTGTACAAGAAAGCTGGGTTTATTAAATCTTGGAAACGAGTGAACGGAAC |
| Real-time RT-PCR | |
| SGfnbAF2 | GAATATTTGCAAGGGTCAGATCAGGTTAATTTTAGAACTG |
| SGfnbAR2 | CTGTGTGGTAATCAATGTCAAGCGGTGTATTG |
| fnbBF1 | GTAGAGGAAAGTGGGAGTTCAG |
| fnbBR1 | TGTGTTGATTGTGATGGTTGC |
| SGhlb1 | AAACACCTGTACTCGGCCGTTCTCAATCAG |
| SGhlb2 | ACTTACAATCGCTACGCCACCATCTTCTGC |
| ureA1 | TTGCACGTCGTGGTAAAGCA |
| ureA2 | AACTCTGCAACGGTCTTACC |
| SGmap1 | CCGCAGCTAAGCCATTAGA |
| SGmap2 | AGCGTTCACAGTGATTGCATA |
| spaF2 | GCCTTAAAGACGATCCTTCAGTGAGCAAAG |
| spaR2 | CCAGGTTTAACGACATGTACTCCGTTGC |
| SGgyrB3 | GGAATCGGTGGCGACTTTGATCTAGCGAAA |
| SGgyrB4 | CGCTCCATCCACATCGGCATCAGTCATAAT |
Plasmid pML100 was constructed by replacing the 153-bp ClaI-HindIII fragment of pLI50 with the 1,015-bp ClaI-HindIII fragment containing the pxyl/tetO promoter from pKOR1. Plasmids pML3871, pML12255, and pML12256 carrying the MW2 rsp, fnbA, and fnbB genes, respectively, were constructed by PCR amplification from MW2 chromosomal DNA using primer pairs MW2301-12/MW2301-14, FnbA-6F/FnbA-7R, and FnbB-6F/FnbB-7R, respectively. Plasmid pCL3972 was constructed by cloning the 314-bp DNA fragment (using primers Rsp1 and Rsp2) encoding the 104-amino-acid (aa) Rsp DNA binding domain into the Gateway Nova pET53-DEST expression system (Novagen, Madison, WI), which resulted in 6 histidine residues at the N terminus. All clones were validated by restriction mapping and sequencing of the inserts.
SDS-PAGE analysis of surface-associated and extracellular proteins.
Overnight S. aureus cultures were diluted (1:100) in 10 ml TSB with 10 μg/ml of chloramphenicol and incubated at 37°C with shaking at 225 rpm until reaching an optical density at 660 nm (OD660) of ∼1.7 (∼4 h). The cultures were centrifuged to separate the cell pellets and supernatants. Noncovalently bound cell surface-associated proteins were extracted as described previously (53) by resuspending the pellets with 4% SDS (adjusted to an OD660 of 100) at room temperature for 1 h followed by centrifugation to collect supernatants, and 30-μl samples were analyzed by 10% SDS-PAGE as described previously (29). Extracellular proteins were concentrated by precipitation of culture supernatants (1.35 ml) with cold trichloroacetic acid (TCA) (final concentration of 10%), washed with cold ethanol, and analyzed by 10% SDS-PAGE. Protein bands were revealed by staining the gel with Coomassie blue G-250.
Zymographic analysis.
For profiling S. aureus autolysis, cultures were grown in TSB for 3 h (OD660 of ∼1.7) and harvested by centrifugation. Surface-associated proteins were prepared with 4% SDS as described above. Culture supernatants were filter sterilized and concentrated 40-fold with Amicon Ultra-4 concentrators with a 5,000-molecular-weight cutoff (Millipore, Bedford, MA). A modified method of Sugai et al. (53) was used for bacteriolytic enzyme profiling analysis. Samples (20 μl) were mixed with 2× SDS sample buffer (final dithiothreitol [DTT] concentration of 12.5 μM), incubated at room temperature for 30 min, and loaded onto 10% SDS-PAGE gels containing heat-killed S. aureus RN4220 (about 2 mg per ml of gel solution). After electrophoresis, gels were first washed 3 times each with 250 ml deionized water for 30 min with slow shaking at room temperature and then washed with 250 ml buffer A (50 mM Tris-HCl [pH 7.6], 200 mM NaCl, 5 mM CaCl2) at room temperature for 30 min. The gels were incubated overnight with fresh buffer A at 37°C without shaking and then scanned with dark background.
For profiling extracellular proteases, filter-sterilized cultural supernatants (16 h) were concentrated 90-fold with Amicon Ultra-4 concentrators. Twenty microliters of concentrated samples was mixed with 2× SDS sample buffer as described above. Samples were loaded onto 10% SDS-PAGE gels containing gelatin (1 mg gelatin per ml of gel solution). After electrophoresis, gels were first washed twice with renaturing buffer (2.5% Triton X-100) for 30 min, washed once with buffer B (0.2 M Tris-Cl [pH 7.6], 5 mM CaCl2, 1 mM DTT), and then incubated overnight with fresh buffer B at 37°C as described by Beenken et al. (3). Gels were then washed with deionized water 3 times (5 min each) and stained with Coomassie blue G-250. The clear bands on the blue background gels indicate proteolytic activity.
Biofilm assay.
Biofilm assays using microtiter plates were carried out in TSB with 0.5% glucose and 1% NaCl using 96-well microtiter plates (Costar 3595; Corning, NY) as previously described (34) with the following modifications. The cultured microtiter plates were washed with sterile water after 24 h of incubation at 37°C. Each sample was loaded to microtiter wells in quadruplicate, and each experiment was repeated at least 3 times independently. Flow cell biofilm assays were carried out with a three-channel flow cell apparatus (Stovall Life Sciences, Inc., Greensboro, NC) as described previously (34). Two independent experiments were performed.
Northern blot analysis.
RNA was isolated as described by Groicher et al. (18) with some modifications. Overnight S. aureus cultures were diluted in fresh TSB to an OD660 of 0.05 and incubated at 37°C with shaking at 225 rpm to the desired OD660. Cultures were then mixed with an equal volume of an ice-cold 1:1 mixture of ethanol-acetone and kept at −20°C until all the samples were collected. Each sample containing about 2 × 109 CFU was centrifuged; the cell pellet was washed 2 times with TNE buffer (50 mM Tris [pH 7.6], 150 mM NaCl, 5 mM EDTA) and suspended in 50 μl of TNE buffer with 2.5 M NaCl. The cell walls were digested with 0.8 μg/μl of lysostaphin at 37°C for 5 min and immediately lysed with 1 ml RNAzol-RT (Molecular Research Center, Inc., Cincinnati, OH) and isolated according to the manufacturer's instructions. In some cases, the MICROExpress kit from Ambion (Austin, TX) was used to enrich mRNA from isolated total RNA by removing the rRNA.
A 751-bp rsp-specific DNA probe was synthesized with the PCR Dig probe synthesis kit using primers MW2301-14 and MW2301-15. RNA samples were denatured and separated in a 2% formaldehyde-1% agarose gel. Northern hybridization was carried out using Dig Easy Hyb (Roche Applied Science, Indianapolis, IN) according to the manufacturer's instructions. Hybridization was carried out at 50°C overnight.
Microarray profiling.
Overnight S. aureus cultures were diluted in fresh TSB to an OD660 of 0.05 and incubated at 37°C with shaking at 225 rpm until reaching an OD660 of 1.9 to 2.0. The cultures were mixed with an equal volume of an ice-cold 1:1 mixture of ethanol-acetone and kept frozen at −80°C. RNA was isolated as described previously (33). Three RNA samples from each strain prepared independently were used for microarray analysis as described previously (2). Data were analyzed using Gene-spring GX software version 7.3.1 (Agilent, Santa Clara, CA). Genes exhibiting ≥2-fold changes in expression, which were statistically significant as determined by Student's t test (P ≤ 0.05) and detectable above background levels based on Affymetrix algorithms, were considered to be differentially expressed under the conditions indicated. Confirmation of microarray data on selected genes by real-time reverse transcription-PCR (RT-PCR) was carried out as described previously (33) using primer pairs SGhlb1 and SGhlb2, ureA1 and ureA2, fnbBF1 and fnbBR1, SGmap1 and SGmap2, SGfnbAF2 and SGfnbAR2, spaF2 and spaR2, and SGgyrB3 and SGgyrB4 (Table 2).
Attachment assay.
The primary attachment assay was performed as described by Tchouaffi-Nana et al. (54) with some modifications. Overnight S. aureus cultures were diluted with TSB to an OD660 of 0.05 and incubated at 37°C with shaking at 225 rpm for 1.5 h (OD660 of ∼0.35) or 2.5 h (OD660 of ∼1.0). The cultures were adjusted to an OD660 of 0.05, 0.1, or 0.3 with TSB and loaded in quadruplicate to 24-well microtiter plates (TPP 92024; Switzerland) at 400 μl per well. The plates were incubated at different temperatures (37°C, 30°C, or 4°C) for 1 h for temperature-dependent studies or for different time periods (15, 30, or 60 min) at 37°C for time course studies. The plates were then washed 3 times with sterile water, air dried, fixed with ethanol, and stained with 0.4% crystal violet as described previously (34). The relative density of the crystal violet stain of the adherent S. aureus was determined with AlphaView image analysis tool (Alpha Innotech Corp.). Each sample was loaded to microtiter wells in quadruplicate, and each experiment was repeated at least 3 times independently.
Purification of Rsp DNA binding domain fused to histidine tag.
Expression of histidine-tagged Rsp DNA binding domain was done in SoluBL21(pCL3972) (Genlantis, San Diego, CA) grown in M9 medium containing 0.3% glycerol as the carbon source, 100 μg/ml penicillin, and 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), for 20 h at 23°C. Bacterial cells were harvested by centrifugation, subjected to osmotic shock (35), and then stored at −80°C. The cells were thawed, incubated with 400 μg/ml lysozyme (Sigma), and sonicated, and the resulting lysate was clarified by centrifugation. At this stage, the majority of the recombinant protein was found in the insoluble pellet. Soluble portion was purified from the clarified lysate by metal affinity chromatography using reagents purchased from EMD Chemicals, San Diego, CA. Following chromatography, the protein was dialyzed against buffer containing 25 mM Tris-Cl (pH 7.5), 10 mM NaCl, 1 mM EDTA, and 0.5 mM Tris(2-carboxyethyl)-phosphine (Sigma); concentrated by packing the dialysis bag in Sephadex (Sigma); and then aliquoted and stored at −80°C. As a mock control, parallel preparations were done using E. coli cells without rsp sequence.
Electrophoretic mobility shift assays (EMSAs).
A 222-bp DNA fragment containing the putative promoter region of fnbA was generated by PCR amplification of MW2 DNA using the oligonucleotide primers FnbA5′1 and FnbA5′2. The DNA fragment was end labeled with digoxigenin-dUTP using reagents purchased from Roche Applied Sciences (Indianapolis, IN). Binding reactions were performed in 20 μl of 20 mM HEPES (pH 7.6), 1 mM EDTA, 10 mM ammonium sulfate,1 mM DTT, 0.2% Tween 20, and 30 mM KCl containing 200 ng poly(dI-dC) and 0.7 ng of labeled DNA fragment. Reaction mixtures were incubated for 15 min at room temperature and then electrophoresed through 6.0% polyacrylamide gels buffered with 1/2× Tris-buffered EDTA (TBE). The DNA fragments were then electroblotted onto a nylon membrane (Applied Biosystems, Austin, TX). The digoxigenin-labeled DNA was detected using reagents purchased from Roche Applied Sciences.
Statistical analysis.
Data from the biofilm assay and attachment assay were analyzed by the GraphPad Prism program (San Diego, CA) using a paired Student t test for comparing two samples.
RESULTS
Phenotypic characterization of rsp.
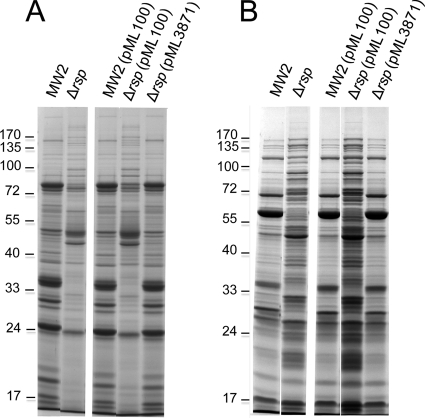
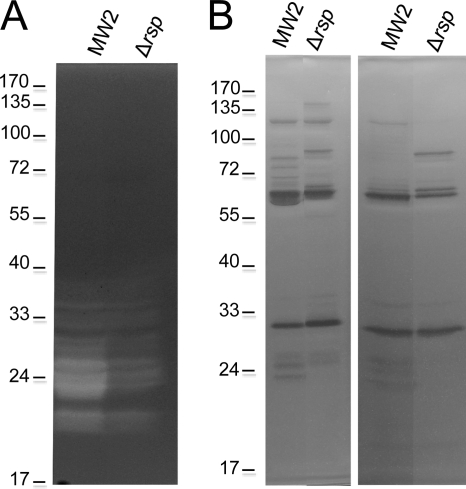
The S. aureus genome encodes 6 regulators containing the 99-residue consensus DNA binding domain with two helix-turn-helix (HTH) motifs that defines the AraC/XylS family of transcriptional regulators (14). We have previously shown that one of these regulators, Rbf, is involved in biofilm regulation by affecting PIA production (10, 32). The biological functions of other S. aureus AraC-like regulators have not been studied. The AraC family proteins are involved in sugar metabolism, stress responses, and pathogenesis (14). To further study AraC-like regulators in S. aureus, we deleted the rsp gene (MW2301) from the MW2 strain by allele replacement. To characterize the effect of the mutation, we first employed SDS-PAGE analyses as a preliminary indication of whether Rsp affects protein production. As shown in Fig. 1A, the mutation in rps had a strong negative effect on the production of extracellular proteins. This effect could be complemented back to the wild-type phenotype with a plasmid-borne rsp gene (pML3871), suggesting that the effect is specific to the rsp gene. It should be noted here that no inducer was needed for the complementation, suggesting that the pxyl/tetO promoter is leaky. We then characterized surface-associated proteins released by 4% SDS extraction. In the wild-type MW2 strain, many low-intensity surface-associated protein bands with one highly prominent protein band at about 60 kDa (Fig. 1B) were detected. In the mutant strain, the gel pattern was very different from that of the wild type (the 60-kDa protein is no longer dominant) and many new strong bands were found. The mutant complemented with pML3871 showed almost the same pattern and intensity of the protein bands as did the wild type. Finally, we compared the protein profiles of total cell lysates between the wild-type and the mutant strains and found no obvious difference (data not shown). These results suggest that Rsp has a profound effect on the expression of S. aureus surface-associated proteins and extracellular proteins.
Fig. 1.
Effect of Rsp on S. aureus MW2 secreted (A) and noncovalent surface-associated (B) proteins. Plasmid pML3871 (i.e., pML100-rsp) was used for complementation of the Δrsp mutation. Cultures were grown in TSB with 10 μg per ml of chloramphenicol as needed to an OD660 of about 1.7. (A) TCA-precipitated culture supernatants were applied to SDS-PAGE analysis. (B) Surface proteins released with 4% SDS were applied to SDS-PAGE analysis.
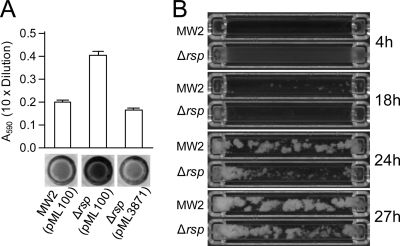
To study the biological effect of Rsp, we performed biofilm assays using 96-well polystyrene microtiter plates. As shown in Fig. 2A, we found that the rsp mutant produced more biofilm than did the wild-type MW2 and that the phenotype could be complemented by pML3871. These data suggest that Rsp represses biofilm formation in MW2. To further demonstrate the effect of Rsp on biofilm formation, we employed the flow cell method. Under our experimental condition, wild-type strain MW2 started to form biofilm at 14.2 ± 1.2 h and reached the peak biofilm abundance at 23.7 ± 0.5 h (Fig. 2B). Interestingly, in the mutant strain, the effect of Δrsp was biphasic. At the beginning, a homogeneous thin film of bacteria formed and peaked at 4.2 ± 0.2 h. The film then quickly cleared at 5.6 ± 0 h. The phenomenon was reproducible and was not detected in the MW2 wild-type strain. Then at 18.2 ± 0.2 h, the mutant started to form biofilm, which reached a maximal amount of biofilm comparable to that of the wild type at 26.8 ± 0.7 h. Taken together, the flow cell data suggest that Rsp may regulate biofilm under the flow condition at multiple stages, possibly through multiple factors.
Fig. 2.
Effect of Rsp on MW2 biofilm formation. (A) Biofilm formation was assayed in 96-well microtiter plates for MW2 (pML100) and MW2 Δrsp mutant with pML100 or pML3871 (i.e., pML100-rsp). Biofilm medium was supplemented with 5 μg per ml of chloramphenicol. The graph represents the average absorbance of the eluted crystal violet-stained wells from 3 independent experiments. A representative well stained with crystal violet of each strain is shown below the graph. (B) Biofilm formation in flow cells. Selected pictures taken every 20 min are shown with specific time points indicated.
Northern hybridization analysis for rsp expression.
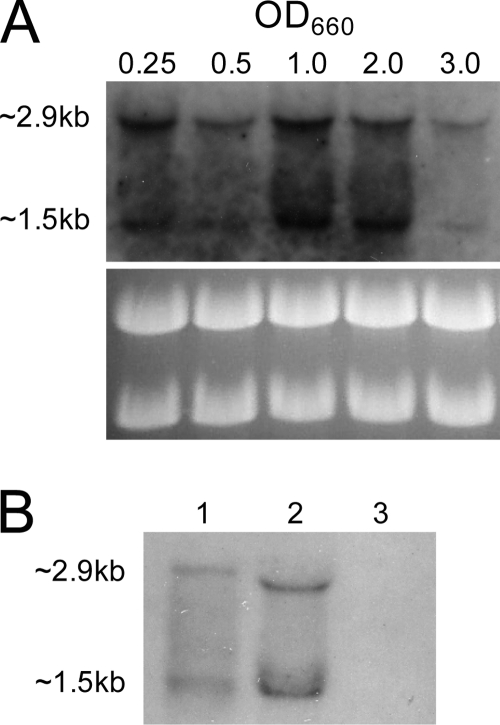
To study the expression of rsp in MW2, total RNAs were isolated from both the wild-type and Δrsp mutant strains and subjected to Northern hybridization using an rsp-specific probe. As shown in Fig. 3A, the probe hybridized to two transcripts, 2.9 kb and 1.5 kb, in the wild type. The rsp mRNA was detected from early exponential phase at an OD660 of 0.25, was slightly reduced at an OD660 of 0.5, and then peaked at mid- to late-exponential-phase growth at an OD660 of 1.0 to 2.0 and was markedly reduced at stationary phase at an OD660 of 3.0. When RNA samples were prepared from 8-h and 16-h MW2 cultures (OD660s of 3.7 and 4.0, respectively), rsp was not detectable by Northern blotting (data not shown), indicating that rsp is mainly expressed during exponential growth phase. The experiments were repeated two times using independently isolated RNA samples, and the results were reproducible. Because the 2.9-kb and 1.5-kb bands were very close to 23S and 16S rRNA bands, it is possible that the bands could be due to nonspecific hybridization with the rRNAs. Two lines of evidence were obtained to rule out such a possibility. First, we included a Δrsp mutant and showed that there was no detectable band compared to the wild-type strain (Fig. 3B). Second, we enriched the mRNA by removing the rRNAs and showed that the 2.9-kb and the 1.5-kb bands could still be detected by Northern blotting (Fig. 3B). It should be noted here that the enrichment process apparently resulted in some loss of rsp mRNA, as the intensity is weaker in lane 1 than in lane 2 of Fig. 3B, even though more total RNA was used for the enrichment in lane 1. The enrichment also altered the gel pattern, which could be due to reduced interference from large quantities of rRNAs. Since the coding region of the rsp gene is 2,106 nucleotides in length, our results suggest that rsp is most likely transcribed as a 2.9-kb monocistronic transcript. The 1.5-kb fragment detected in the Northern blots may be the processing or degradation product from the primary transcript.
Fig. 3.
Northern blot analysis of rsp transcription. (A) Total RNAs were prepared from MW2 cultures grown to different OD660s as indicated above the figure. Total RNA of 5 μg from each sample was applied to the formaldehyde gel and hybridized with a specific rsp (751-bp) probe. Ethidium bromide-stained ribosome bands are shown below as the loading control. (B) Lane 1, 5 μg of total RNA prepared from MW2 with rRNA removed. Lane 2, 3 μg of total RNA from MW2. Lane 3, total RNA of 3 μg from MW2 Δrsp. Cultures were grown to an OD660 of 1.0.
Identification of Rsp-regulated genes by microarray.
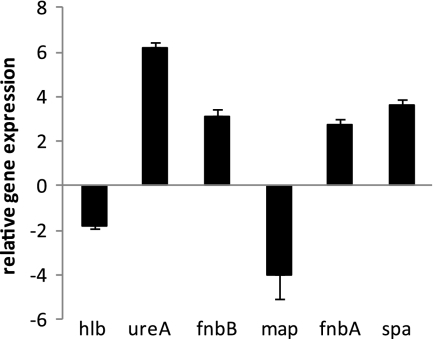
To understand how Rsp affects biofilm formation at the transcriptional level, we employed microarray analysis to identify genes differentially regulated by Rsp. Since the rsp gene was most highly expressed in the mid- to late log phase, we harvested RNA at an OD660 of about 2.0 from the wild-type MW2 and the isogenic Δrps mutant. We found that Rsp affected 22 genes positively and 75 genes negatively by at least 2-fold (Tables 3 and 4). Of the 22 Rsp-upregulated genes, half (11 genes) encode secreted proteins, including 5 proteases and 2 cell surface-associated proteins. Among the 75 downregulated genes, 17 are involved in metabolism, 12 are transporter genes, and 9 encode surface-anchored proteins. To confirm microarray results, we performed real-time RT-PCR on selected genes using gyrB expression for normalization. The results (Fig. 4) matched well with the microarray results.
Table 3.
Genes upregulated by rspa
| Open reading frame category and name | Fold change | Gene | Description |
|---|---|---|---|
| Secreted proteins | |||
| MW1755 | 6.7 | splA | Serine protease SplA |
| MW1754 | 9.0 | splB | Serine protease SplB |
| MW1753 | 6.0 | splC | Serine protease SplC |
| MW1752 | 6.0 | splF | Serine protease SplD, putative |
| MW1850 | 5.4 | scpA | Cysteine protease precursor SspB |
| MW1767 | 2.9 | lukD | Leukotoxin |
| MW1940 | 2.8 | hlb | Phospholipase C |
| MW0297 | 2.0 | geh | Glycerol ester hydrolase |
| MW0345 | 2.3 | Staphylococcal enterotoxin, putative | |
| MW0767 | 2.8 | empBP | Secretory extracellular matrix and plasma binding protein |
| MW1880 | 2.5 | map | Map protein, authentic frameshift |
| Regulation | |||
| MW2002 | 2.0 | kdpD | Sensor histidine kinase KdpD |
| MW2003 | 2.3 | kdpE | DNA binding response regulator KdpE |
| Metabolism | |||
| MW1675 | 2.7 | fhs | Formate-tetrahydrofolate ligase |
| MW0070 | 3.6 | plc | 1-Phosphatidylinositol phosphodiesterase |
| Other | |||
| MW2536 | 2.3 | nrdG | Anaerobic ribonucleoside triphosphate reductase activating protein |
| MW1609 | 2.6 | AbrB protein, putative | |
| MW0768 | 2.8 | Hypothetical protein | |
| MW0758 | 2.6 | Hypothetical protein | |
| MW0906 | 3.7 | Conserved hypothetical protein, similar to competence transcription factor | |
| COL-SA1713 | 2.9 | Hypothetical protein | |
| MW1851 | 4.6 | Hypothetical protein |
Upregulation indicates an increased expression level in the wild type compared to the rsp deletion mutant.
Table 4.
Genes downregulated by rspa
| Open reading frame category and name | Fold change | Gene | Description |
|---|---|---|---|
| Surface protein | |||
| MW0764 | 2.1 | clfA | Probably clumping factor A |
| MW2421 | 3.2 | fnbA | Fibronectin binding protein A |
| MW2420 | 3.9 | fnbB | Fibronectin binding protein B |
| MW1037 | 4.0 | Fibrinogen binding-related protein | |
| MW0517 | 3.9 | sdrD | Ser-Asp-rich fibrinogen-binding, bone sialoprotein-binding protein |
| MW0518 | 3.8 | sdrE | SdrE protein |
| MW0084 | 2.3 | spa | Immunoglobulin G-binding protein A precursor |
| MW2416 | 2.9 | sasG | Cell wall surface anchor family protein |
| MW2575 | 2.3 | Cell wall surface anchor family protein | |
| Metabolism | |||
| MW2444 | 2.4 | ddh | d-specific d-2-hydroxyacid dehydrogenase |
| MW2198 | 2.3 | fdhD | Formate dehydrogenase accessory protein FdhD |
| MW2443 | 3.0 | frp | NAD(P)H-flavin oxidoreductase |
| MW0358 | 3.4 | NAD(P)H-flavin oxidoreductase, putative | |
| MW2541 | 2.6 | gpxA | Glutathione peroxidase |
| MW2594 | 2.1 | hisH | Imidazole glycerol phosphate synthase, glutamine amidotransferase subunit |
| MW0415 | 3.1 | metB | Cystathionine gamma-synthase |
| MW0414 | 2.9 | Cysteine synthase/cystathionine beta-synthase family protein | |
| MW2206 | 4.8 | ureA | Urease, gamma subunit |
| MW2207 | 7.1 | ureB | Urease, beta subunit |
| MW2208 | 4.8 | ureC | Urease, alpha subunit |
| MW2212 | 3.1 | ureD | Urease accessory protein UreD |
| MW2209 | 4.2 | ureE | Urease accessory protein UreE |
| MW2210 | 2.9 | ureF | Urease accessory protein UreF |
| MW2211 | 2.7 | ureG | Urease accessory protein UreG |
| MW2157 | 2.0 | rplE | Ribosomal protein L5 |
| MW2154 | 2.1 | rplF | Ribosomal protein L6 |
| Regulation | |||
| MW0935 | 2.4 | Transcriptional regulator, MarR family | |
| MW2128 | 4.2 | Transcriptional regulator, MerR family | |
| Transporter | |||
| MW1236 | 2.4 | opuD | Osmoprotectant transporter, BCCT family |
| MW0528 | 3.7 | proP | Osmoprotectant proline transporter |
| MW0114 | 2.6 | Phosphonate ABC transporter, permease protein | |
| MW0115 | 2.2 | Phosphonate ABC transporter, permease protein | |
| MW0147 | 4.1 | ABC transporter, ATP-binding protein, authentic frameshift | |
| MW0148 | 3.5 | ABC transporter, permease protein | |
| MW0253 | 4.3 | ABC transporter, ATP-binding protein | |
| MW2370 | 2.0 | Amino acid ABC transporter, amino acid-binding protein | |
| MW2261 | 3.9 | ABC transporter, ATP-binding protein | |
| MW0218 | 2.1 | Phosphotransferase system, IIBC components | |
| MW0958 | 2.2 | Cobalt transport family protein | |
| MW2374 | 2.3 | Amino acid permease | |
| Other functions | |||
| MW2333 | 4.1 | fmhA | FmhA protein, methicillin resistance protein |
| MW0657 | 2.0 | norA | Multidrug resistance protein |
| MW0760 | 2.5 | sel | Enterotoxin L |
| MW0081 | 8.9 | Antigen, 67 kDa | |
| MW0748 | 3.1 | Pathogenicity island protein | |
| MW0924 | 2.3 | Acetyltransferase, GNAT family | |
| MW1425 | 2.8 | DNA polymerase, bacteriophage type | |
| MW2057 | 2.0 | Peptidase, M20/M25/M40 family | |
| MW2127 | 2.6 | Oxidoreductase, aldo-/ketoreductase family | |
| Unknown genes | |||
| MW0145 | 2.1 | Conserved hypothetical protein | |
| MW0149 | 2.8 | Conserved hypothetical protein | |
| MW0254 | 4.0 | Hypothetical protein | |
| MW0255 | 4.4 | Hypothetical protein | |
| MW0256 | 3.8 | Hypothetical protein | |
| MW0260 | 2.1 | Hypothetical protein | |
| MW0261 | 2.0 | Conserved hypothetical protein | |
| MW0262 | 2.1 | Conserved hypothetical protein | |
| MW0264 | 2.5 | Hypothetical protein | |
| MW0265 | 2.5 | Hypothetical protein | |
| MW0266 | 2.4 | Conserved hypothetical protein | |
| MW0267 | 2.4 | Hypothetical protein | |
| COL-SA1165 | 2.8 | Hypothetical protein | |
| MW1410 | 2.1 | Conserved hypothetical protein | |
| MW1405 | 2.3 | Conserved hypothetical protein | |
| MW1406 | 3.0 | Conserved hypothetical protein | |
| MW1408 | 2.1 | Conserved hypothetical protein | |
| MW1427 | 2.1 | Conserved hypothetical protein | |
| MW1969 | 2.5 | Conserved hypothetical protein | |
| MW2260 | 3.6 | Conserved hypothetical protein | |
| MW2263 | 2.3 | Hypothetical protein | |
| MW2560 | 2.2 | Conserved hypothetical protein | |
| MW2573 | 2.1 | Conserved hypothetical protein | |
| MW2600 | 2.1 | Hypothetical protein |
Downregulation indicates an increased expression level in the rsp mutant strain compared to the wild type.
Fig. 4.
Confirmation of microarray results of selected genes by real-time RT-PCR. RNAs were isolated from MW2 and MW2 Δrsp mutant at an OD660 of 2.0. Expression levels are expressed relative to that of the wild type. Data represent the means with standard errors from at least two independent experiments.
Effect of Rsp on proteolytic activity and bacteriolytic activity.
The microarray analyses above indicate that Rsp affects the expression of several proteases. To confirm these results, we concentrated culture supernatants and analyzed the effect of rsp on S. aureus proteolytic activities using gelatin-containing SDS-PAGE. As shown in Fig. 5A, the proteolytic activity in the MW2 Δrsp strain decreased compared to that of the wild-type strain. When casein was used as the substrate, we detected very weak proteolytic activity in MW2 but not in the Δrsp mutant (data not shown).
Fig. 5.
Zymographic analysis. (A) Effect of Rsp on proteolytic activity of concentrated culture supernatants in 10% SDS-PAGE gels containing gelatin. (B) Effect of Rsp on bacteriolytic activity of 4% SDS-released surface proteins (left panel) and concentrated culture supernatants (right panel) in 10% SDS-PAGE gels containing heat-killed S. aureus RN4220.
It has been shown elsewhere that the activity of Atl murein hydrolases could be modulated by extracellular proteases (21, 22, 41, 52). Furthermore, cell wall-associated Aaa autolysin (20) is likely to be vulnerable to proteolytic cleavage. Since we found that Rsp affected several proteases, we speculate that it may also affect autolysin through proteases, although no autolysin gene was found to be affected by Rsp in the microarray experiments. Accordingly, we tested whether deletion of rsp had an effect on bacteriolytic activity by using heat-killed S. aureus RN4220 as a substrate. As shown in Fig. 5B, deletion of rsp altered the autolysin zymographic pattern compared to that of the wild type, suggesting that Rsp affects autolysins most likely at the posttranscriptional level.
Regulation of biofim formation by Rsp through FnbA.
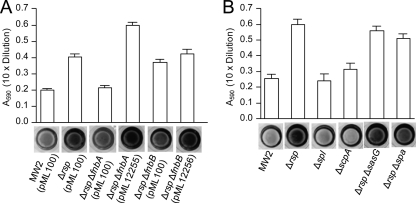
S. aureus possesses a total of 23 surface-anchored proteins (51), which are collectively termed microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) (13). The microarray analyses above (Table 4) showed that Rsp affected 9 such proteins, of which FnbA, FnbB, Spa, and SasG have been shown to promote biofilm formation (6, 15, 37, 40). We therefore hypothesized that Rsp affects biofilm by repressing one or more of these MSCRAMMs. To test this possibility, we deleted fnbA, fnbB, spa, and sasG individually from the chromosome of the MW2 Δrsp mutant and assayed their effect on biofilm. The results in Fig. 6 showed that, in comparison to biofilm formation in the MW2 Δrsp mutant, the deletion of fnbB or sasG had no significant effect whereas the deletion of fnbA or spa resulted in a significant reduction. The fnbA deletion reduced the biofilm formation of the Δrsp mutant (P < 0.0001) to the level similar to that of the wild-type MW2 while the spa deletion had only a slight effect, although the difference was statistically significant (P = 0.0310). Importantly, the reduction of biofilm in the Δrsp ΔfnbA double mutant could be complemented by the wild-type fnbA gene, though the complemented strain produced more biofilm than did the MW2 Δrsp mutant, which was most likely due to a multicopy vector effect. These results suggest that Rsp represses biofilm in MW2 primarily by downregulating the fnbA gene.
Fig. 6.
Biofilm assay in 96-well microtiter plates. Graphs represent the average absorbance of the eluted crystal violet dye from the stained wells, of which a representative well is shown below the graph. (A) Effect of ΔfnbA and ΔfnbB deletions on biofilm in the MW2 Δrsp background. Biofilm medium was supplemented with 5 μg/ml of chloramphenicol. Plasmids pML12255 and pML12256 carry MW2 fnbA and fnbB genes in pML100, respectively. Error bars indicate standard errors from 4 independent experiments. Student t test: MW2 Δrsp (pML100) versus MW2 Δrsp ΔfnbA (pML100), P < 0.0001; MW2 Δrsp (pML100) versus MW2 Δrsp ΔfnbB (pML100), P = 0.8308. (B) Effect of Δspl and ΔscpA in MW2 background and effect of ΔsasG and Δspa in the MW2 Δrsp background. Error bars indicate standard errors from 3 independent experiments. Student t test: MW2 Δrsp versus MW2 Δrsp Δspa, P = 0.031.
In the MW2 strain, the Spl proteases are encoded in the splABCD operon. All spl genes were upregulated by Rsp in the microarray analyses (Table 3). Spl proteases have been shown to affect biofilm in strain SH1000 (30); therefore, it is possible that Rsp could repress biofilm by activating the spl genes. To test this possibility, we deleted the spl operon in MW2. Our results (Fig. 6B) revealed that there was no effect of the spl deletion on biofilm formation. Besides the spl genes, scpA, which encodes the cysteine protease staphopain A, was also found to be activated by Rsp. However, deletion of scpA has no effect on biofilm formation (Fig. 6B). These results suggest that the Spl and ScpA proteases in MW2 do not play a critical role in biofilm formation and therefore are not critical factors mediating biofilm formation regulated by Rsp.
Effect of Rsp on attachment.
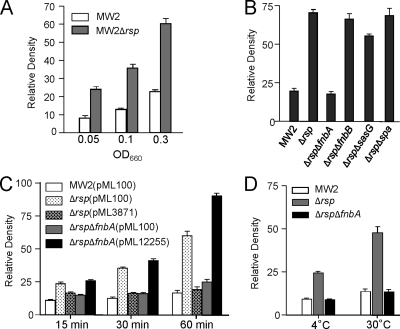
To determine whether Rsp affects biofilm formation at the initial attachment step, we compared the Δrsp mutant and the wild type in primary attachment assays. As shown in Fig. 7A, the mutant strain had about a 3-fold increase in attachment when the cultures were diluted to an OD660 of 0.05, 0.1, or 0.3 from early-log-phase cultures at an OD660 of ∼0.35 (Fig. 7A) or from mid-log-phase cultures at an OD660 of ∼1.0 (data not shown). In addition, the deletion of spa or fnbB had no significant effect whereas the deletion of fnbA reduced attachment of the Δrsp mutant to a level comparable to that of the wild type and deletion of sasG had only a slight effect but one that is statistically significant (P < 0.0001) (Fig. 7B). These results suggest that FnbA is the major factor contributing to the increased level of primary attachment in Δrsp mutant. To further ensure that FnbA affected biofilm formation at the primary attachment step, we performed additional experiments with shorter incubation times at 15 min and 30 min. The results (Fig. 7C) showed that even with a 15-min incubation time, the Δrsp ΔfnbA double mutant had a significant reduction in attachment (P < 0.0001) compared to that of the Δrsp mutant. Complementation experiments showed that the phenotype of the Δrsp mutant could be restored to that of the wild type by pML3871. In addition, the phenotype of the Δrsp ΔfnbA double mutant can be restored by pML12255 to that of the Δrsp mutant, though at the 60-min incubation time, MW2 Δrsp ΔfnbA (pML12255) had a higher attachment rate, which was likely due to a multiple-copy vector effect. We also performed the experiments at lower incubation temperatures. As shown in Fig. 7D, a significant effect (P < 0.0001) was also observed when the incubation temperature was at 30°C or 4°C. It should be noted here that deletion in splABCD or scpA had no effect on primary attachment (data not shown).
Fig. 7.
Attachment assay. Graphs represent average relative densities of 3 independent experiments. (A) MW2 and Δrsp mutant were grown to early growth phase and adjusted to an OD660 of 0.05, 0.1, or 0.3 with TSB and incubated at 37°C for 1 h. (B) Effect of ΔfnbA, ΔfnbB, ΔsasG, and Δspa on initial attachment in MW2 Δrsp background. Early-log-phase cultures were diluted to an OD660 of 0.3 and incubated at 37°C for 1 h. (C) Effect of incubation time on initial attachment. Same culture conditions as in panel B but with 10 μg/ml chloramphenicol and with different incubation time periods as indicated. Complementation tests using pML3871 (i.e., pML100-rsp) and pML12255 (i.e., pML100-fnbA) are also included. (D) Effect of temperature on initial attachment. Same culture conditions as in panel B but with lower incubation temperatures as indicated.
Binding of Rsp DNA binding domain to FnbA promoter.
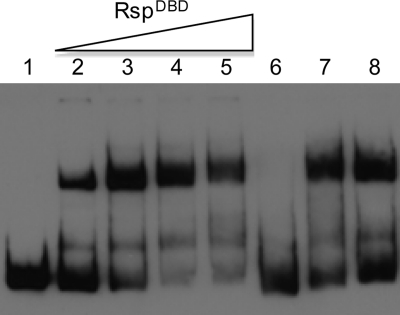
Rsp possesses two tandem HTH motifs in the predicted DNA binding domain. To determine whether Rsp binds its target gene through the DNA binding domain, we purified the putative Rsp DNA binding domain (RspDBD) fused to a His tag as described in Materials and Methods. Initial attempts revealed that the fusion protein is highly insoluble. However, upon moving the plasmid to a strain designed to express low-solubility recombinant proteins, we were able to purify a small amount for EMSA experiments. As shown in Fig. 8, we found that the RspDBD was able to bind the fnbA promoter region at a concentration of 15 nM. The shifted band was competed out with unlabeled DNA fragments of fnbA promoter in 50× excess. In addition, DNA fragments containing the agr promoter region (containing both P2 and P3 promoters) or the icaA promoter did not affect the binding, suggesting that the binding to the fnbA promoter is specific. Furthermore, mock-purified protein preparation showed no shifted band (data not shown).
Fig. 8.
Binding of Rsp DNA binding domain (RspDBD) to the fnbA promoter region. Electrophoretic mobility shift assays were performed in the absence (lane 1) or presence (lanes 2 to 8) of RspDBD protein. Lanes 2, 3, 4, and 5 contain 15, 30, 60, and 90 nM RspDBD, respectively. Lane 6 contains 90 nM RspDBD and a 50 molar excess of the unlabeled fnbA promoter fragment. Lanes 7 and 8 contain 90 nM RspDBD and a 50 molar excess of a nonspecific DNA fragment (lane 7, agr promoter fragment; lane 8, icaA promoter fragment).
DISCUSSION
Regulation of biofilm formation in S. aureus is very complex due in part to the fact that biofilm formation involves multiple components, which include polysaccharide, proteins, and extracellular DNA. In this study, we found that an AraC/XylS family regulator, Rsp, repressed biofilm formation. Our SDS-PAGE analyses revealed that Rsp had a profound effect on cell surface and secreted proteins in strain MW2, a community-acquired methicillin-resistant strain. Subsequent microarray analyses identified a number of genes encoding cell wall-anchored proteins that are repressed by Rsp, including FnbA, FnbB, SasG, and Spa, which have been previously shown to affect biofilm formation in S. aureus. By deleting individual genes encoding the four proteins in the Δrsp mutant background, we found that Rsp repressed biofilm formation primarily by downregulating FnbA and, to a much lesser extent, Spa. In addition, a number of genes encoding proteases were upregulated by Rsp, which could have an impact on biofilm by affecting cell surface and/or secreted proteins at the posttranslational level. However, deletion of these genes did not result in a detectable difference in biofilm formation.
The FnbA protein is anchored on the staphylococcal cell surface through the LPXTG motif and is capable of binding to fibronectin, fibrinogen, and elastin (50). FnbA has been shown to promote biofilm formation on polystyrene surfaces without coating with host plasma in methicillin-resistant S. aureus strains (39, 40), suggesting that binding to matrix proteins is not required. Spa (protein A) is known for binding to the Fc region of IgGs but can also bind to several host factors (12, 16, 19). FnbA and Spa have been shown to promote biofilm in the cell accumulation phase (37, 39, 40). Thus, we were surprised to find that Rsp affected biofilm formation at the initial attachment phase through FnbA. Because of the discrepancy with the earlier reports, we conducted additional experiments to ensure that FnbA affected the attachment phase of biofilm formation. We showed that the effect on attachment by FnbA in the Δrsp mutant could be detected even when the incubation time was as short as 15 min or the temperature as low as 4°C (Fig. 7C and D). Under these conditions, bacterial growth is minimal and therefore attachment of the bacterial cells to microtiter wells should not be affected by bacterial growth and thus should reflect only the attachment capability. Based on these results, we confirmed that FnbA was involved in mediating initial attachment for biofilm formation. It is likely that different procedures may lead to the discrepancy between our present study and the earlier reports. It is also possible that FnbA is highly expressed in the Δrsp mutant so that the effect on attachment could be readily observed in our study.
Our microarray data also showed that at least 5 other LPXTG proteins were also repressed by Rsp (Table 4). These surface proteins have not been shown to directly affect biofilm in vitro. However, from their abilities to bind to various matrix proteins, it is tempting to speculate that they are likely to promote biofilm formation in vivo in which bacteria are in contact with the extracellular matrix of the host tissue (13). Rsp, therefore, could potentially be involved in biofilm formation in vivo through these LPXTG proteins that it regulates, a topic which remains to be studied.
S. aureus produces several proteases, and some of these have been shown to inhibit biofilm formation. Double deletion of aur, which encodes metalloprotease aureolysin, and the spl operon, which encodes Spl serine proteases, has been shown to restore biofilm in sigB mutants, but the mechanism is unknown (30). Recently, Marti et al. (36) have shown that deletion of aur or sspA (which encodes V8 serine protease) in sigB deletion strains promotes robust biofilm formation by targeting a surface LPXTG protein, Bap. Indeed, degradation of FnbA, FnbB, and Spa surface-anchored proteins by proteases has been observed previously (26). In our microarray study, we found that all 4 genes in the spl operon in MW2 were upregulated by Rsp, raising the possibility that Rsp could repress biofilm formation through activation of Spl proteases. However, deletion of the spl operon did not increase biofilm formation in the wild-type MW2 background, indicating that Spl serine proteases may not play a role in Rsp-mediated repression of biofilm formation in MW2. The scpA gene, which encodes a cysteine protease, staphopain A, was also activated by Rsp. The role of ScpA in biofilm is unknown; however, our results showed that deletion of scpA in MW2 did not affect initial attachment or overall biofilm formation.
It is interesting that the biofilm formed by the rsp mutant exhibited two phases of bacterial growth in the flow cell, first with a smooth thin film, which dispersed within a few hours, and then later with robust biofilm formation in the chamber (Fig. 2B). It is likely that the thin film in the first few hours is due to the increased primary attachment phenotype of the mutant. However, it is difficult to comprehend why the thin biofilm dispersed after the initial attachment. It is also difficult to understand why it took a longer time for the mutant to form biofilm than the wild type did, especially considering that the mutant produced more biofilm than the wild type did under a static microtiter plate condition. Since Rsp affects many genes as revealed by microarray analyses, it is possible that one or more factors controlled by Rsp affect biofilm development in a temporal fashion, which may be required to resist shear force from the flow of the medium. It is also possible that differences in surface property between the microtiter plate and the flow cell contribute to the difference.
A number of regulators, including two-component systems and transcriptional regulators, have been shown to affect biofilm formation in S. aureus (38, 43). We had previously reported an AraC-like regulator, Rbf, which activates PIA production via repressing IcaR (10). Rsp is one of the 6 AraC-like regulators in S. aureus, which contains a DNA binding motif similar to that of Rbf. It also shares a low degree of overall protein homology with Rbf. However, we showed here that Rsp regulated biofilm through repressing surface protein FnbA at the primary attachment phase rather than affecting PIA. AraC family proteins typically sense a ligand that affects their regulatory activities. Our previous study suggests that functional domains of Rbf may recognize signals in response to concentration changes of NaCl and/or glucose (32). Rsp is a large protein (701 aa) similar in size to Rbf (716 aa), and both are much larger than most AraC family proteins, which are about 250 to 300 aa (14). It is most likely that Rsp also responds to certain environmental cues. The large size of the protein indicates that the protein may possess multiple domains. Here, we showed that the predicted DNA binding domain containing two adjacent HTH motifs was indeed capable of binding to the promoter of fnbA by EMSA experiments, suggesting that Rsp regulates its target genes by direct DNA binding at the promoter region.
ACKNOWLEDGMENTS
We thank Mark Smeltzer for helpful discussions.
This work was supported by grants AI067857 (C.Y.L.) and AI73780 (P.M.D.) from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 29 July 2011.
REFERENCES
- 1. Bae T., Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63 [DOI] [PubMed] [Google Scholar]
- 2. Beenken K. E., Blevins J. S., Smeltzer M. S. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 71:4206–4211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beenken K., et al. 2010. Epistatic relationships between sarA and agr in Staphylococcus aureus biofilm formation. PLoS One 5:e10790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boles B. R., Horswill A. R. 2008. agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4:e1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Centers for Disease Control and Prevention 1999. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus: Minnesota and North Dakota, 1997–1999. MMWR Morb. Mortal. Wkly. Rep. 48:707–710 [PubMed] [Google Scholar]
- 6. Corrigan R. M., Rigby D., Handley P., Foster T. J. 2007. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 153:2435–2446 [DOI] [PubMed] [Google Scholar]
- 7. Costerton J. W., Stewart P. S., Greenberg E. P. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1321 [DOI] [PubMed] [Google Scholar]
- 8. Cramton S. E., Gerke C., Schnell N. F., Nichols W. W., Gotz F. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427–5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cucarella C., et al. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183:2888–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cue D., et al. 2009. Rbf promotes biofilm formation by Staphylococcus aureus via repression of icaR, a negative regulator of icaADBC. J. Bacteriol. 191:6363–6373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fitzpatrick F., Humphreys H., O'Gara J. P. 2005. Evidence for icaADBC-independent biofilm development mechanism in methicillin-resistant Staphylococcus aureus clinical isolates. J. Clin. Microbiol. 43:1973–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Forsgren A., Sjoquist J. 1966. “Protein A” from S. aureus. I. Pseudo-immune reaction with human gamma-globulin. J. Immunol. 97:822–827 [PubMed] [Google Scholar]
- 13. Foster T. J., Hook M. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6:484–488 [DOI] [PubMed] [Google Scholar]
- 14. Gallegos M. T., Schleif R., Bairoch A., Hofmann K., Ramos J. L. 1997. AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Geoghegan J. A., et al. 2010. Role of surface protein SasG in biofilm formation by Staphylococcus aureus. J. Bacteriol. 192:5663–5673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gomez M. I., et al. 2004. Staphylococcus aureus protein A induces airway epithelial inflammatory responses by activating TNFR1. Nat. Med. 10:842–848 [DOI] [PubMed] [Google Scholar]
- 17. Götz F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367–1378 [DOI] [PubMed] [Google Scholar]
- 18. Groicher K. H., Firek B. A., Fujimoto D. F., Bayles K. W. 2000. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 182:1794–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hartleib J., et al. 2000. Protein A is the von Willebrand factor binding protein on Staphylococcus aureus. Blood 96:2149–2156 [PubMed] [Google Scholar]
- 20. Heilmann C., Hartleib J., Hussain M. S., Peters G. 2005. The multifunctional Staphylococcus aureus autolysin aaa mediates adherence to immobilized fibrinogen and fibronectin. Infect. Immun. 73:4793–4802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heilmann C., Hussain M., Peters G., Gotz F. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24:1013–1024 [DOI] [PubMed] [Google Scholar]
- 22. Houston P., et al. 2011. Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect. Immun. 79:1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Izano E. A., Amarante M. A., Kher W. B., Kaplan J. B. 2008. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 74:470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jefferson K. K., Cramton S. E., Gotz F., Pier G. B. 2003. Identification of a 5-nucleotide sequence that controls expression of the ica locus in Staphylococcus aureus and characterization of the DNA-binding properties of IcaR. Mol. Microbiol. 48:889–899 [DOI] [PubMed] [Google Scholar]
- 25. Johnson M., Cockayne A., Morrissey J. A. 2008. Iron-regulated biofilm formation in Staphylococcus aureus Newman requires ica and the secreted protein Emp. Infect. Immun. 76:1756–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Karlsson A., Saravia-Otten P., Tegmark K., Morfeldt E., Arvidson S. 2001. Decreased amounts of cell wall-associated protein A and fibronectin-binding proteins in Staphylococcus aureus sarA mutants due to up-regulation of extracellular proteases. Infect. Immun. 69:4742–4748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kraemer G. R., Iandolo J. J. 1990. High-frequency transformation of Staphylococcus aureus by electroporations. Curr. Microbiol. 21:373–376 [Google Scholar]
- 28. Kreiswirth B., et al. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712 [DOI] [PubMed] [Google Scholar]
- 29. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 30. Lauderdale K. J., Boles B. R., Cheung A. L., Horswill A. R. 2009. Interconnections between sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect. Immun. 77:1623–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee C. Y., Buranen S. L., Ye Z. H. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103:101–105 [DOI] [PubMed] [Google Scholar]
- 32. Lim Y., Jana M., Luong T. T., Lee C. Y. 2004. Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J. Bacteriol. 186:722–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Luong T. T., Dunman P. M., Murphy E., Projan S. J., Lee C. Y. 2006. Transcription profiling of the mgrA regulon in Staphylococcus aureus. J. Bacteriol. 188:1899–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Luong T. T., Lei M. G., Lee C. Y. 2009. Staphylococcus aureus Rbf activates biofilm formation in vitro and promotes virulence in a murine foreign-body infection model. Infect. Immun. 77:335–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Magnusdottir A., Johansson I., Dahlgren L. G., Nordlund P., Berglund H. 2009. Enabling IMAC purification of low abundance recombinant proteins from E. coli lysates. Nat. Methods 6:477–478 [DOI] [PubMed] [Google Scholar]
- 36. Marti M., et al. 2010. Extracellular proteases inhibit protein-dependent biofilm formation in Staphylococcus aureus. Microbes Infect. 12:55–64 [DOI] [PubMed] [Google Scholar]
- 37. Merino N., et al. 2009. Protein A-mediated multicellular behavior in Staphylococcus aureus. J. Bacteriol. 191:832–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. O'Gara J. P. 2007. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 270:179–188 [DOI] [PubMed] [Google Scholar]
- 39. O'Neill E., et al. 2007. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J. Clin. Microbiol. 45:1379–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. O'Neill E., et al. 2008. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 190:3835–3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oshida T., et al. 1995. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-l-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. U. S. A. 92:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. O'Toole G., Kaplan H. B., Kolter R. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49–79 [DOI] [PubMed] [Google Scholar]
- 43. Otto M. 2008. Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 322:207–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Otto M. 2006. Bacterial evasion of antimicrobial peptides by biofilm formation. Curr. Top. Microbiol. Immunol. 306:251–258 [DOI] [PubMed] [Google Scholar]
- 45. Reed S. B., et al. 2001. Molecular characterization of a novel Staphylococcus aureus serine protease operon. Infect. Immun. 69:1521–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rice K. C., et al. 2007. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 104:8113–8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rohde H., et al. 2005. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol. 55:1883–1895 [DOI] [PubMed] [Google Scholar]
- 48. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 49. Schroeder K., et al. 2009. Molecular characterization of a novel Staphylococcus aureus surface protein (SasC) involved in cell aggregation and biofilm accumulation. PLoS One 4:e7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Signas C., et al. 1989. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proc. Natl. Acad. Sci. U. S. A. 86:699–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stranger-Jones Y. K., Bae T., Schneewind O. 2006. Vaccine assembly from surface proteins of Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 103:16942–16947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sugai M., et al. 1989. Purification of a 51 kDa endo-beta-N-acetylglucosaminidase from Staphylococcus aureus. FEMS Microbiol. Lett. 52:267–272 [DOI] [PubMed] [Google Scholar]
- 53. Sugai M., Akiyama T., Komatsuzawa H., Miyake Y., Suginaka H. 1990. Characterization of sodium dodecyl sulfate-stable Staphylococcus aureus bacteriolytic enzymes by polyacrylamide gel electrophoresis. J. Bacteriol. 172:6494–6498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tchouaffi-Nana F., et al. 2010. Nitazoxanide inhibits biofilm formation by Staphylococcus epidermidis by blocking accumulation on surfaces. Antimicrob. Agents Chemother. 54:2767–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tormo M. A., Knecht E., Gotz F., Lasa I., Penades J. R. 2005. Bap-dependent biofilm formation by pathogenic species of Staphylococcus: evidence of horizontal gene transfer? Microbiology 151:2465–2475 [DOI] [PubMed] [Google Scholar]
- 56. Vergara-Irigaray M., et al. 2009. Relevant role of fibronectin-binding proteins in Staphylococcus aureus biofilm-associated foreign-body infections. Infect. Immun. 77:3978–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]