Abstract
DNA is constantly exposed to chemical and environmental mutagens, causing lesions that can stall replication. In order to deal with DNA damage and other stresses, Escherichia coli utilizes the SOS response, which regulates the expression of at least 57 genes, including umuDC. The gene products of umuDC, UmuC and the cleaved form of UmuD, UmuD′, form the specialized E. coli Y-family DNA polymerase UmuD′2C, or polymerase V (Pol V). Y-family DNA polymerases are characterized by their specialized ability to copy damaged DNA in a process known as translesion synthesis (TLS) and by their low fidelity on undamaged DNA templates. Y-family polymerases exhibit various specificities for different types of DNA damage. Pol V carries out TLS to bypass abasic sites and thymine-thymine dimers resulting from UV radiation. Using alanine-scanning mutagenesis, we probed the roles of two active-site loops composed of residues 31 to 38 and 50 to 54 in Pol V activity by assaying the function of single-alanine variants in UV-induced mutagenesis and for their ability to confer resistance to UV radiation. We find that mutations of the N-terminal residues of loop 1, N32, N33, and D34, confer hypersensitivity to UV radiation and to 4-nitroquinoline-N-oxide and significantly reduce Pol V-dependent UV-induced mutagenesis. Furthermore, mutating residues 32, 33, or 34 diminishes Pol V-dependent inhibition of recombination, suggesting that these mutations may disrupt an interaction of UmuC with RecA, which could also contribute to the UV hypersensitivity of cells expressing these variants.
INTRODUCTION
Escherichia coli has five DNA polymerases that replicate DNA under different circumstances (22). The replicative polymerase in E. coli is DNA polymerase III (Pol III), a member of the C family. DNA polymerases IV and V are members of the Y family, which specialize in copying damaged DNA in a process known as translesion synthesis (TLS) (22, 47). Y-family DNA polymerases also copy undamaged DNA in an error-prone manner, possibly subjecting DNA to untargeted mutagenesis and potentially leading to antibiotic resistance or cancer (16, 17, 22, 52).
E. coli DNA polymerases IV (DinB) and V (UmuD′2C) are the products of the dinB and umuDC genes, respectively. Due to their potentially mutagenic nature, these proteins are highly regulated in a specific cellular response to DNA damage and other stresses called the SOS response (22, 55). The SOS response is initiated when single-stranded DNA (ssDNA) forms downstream from a lesion in DNA due to the inability of the replicative DNA polymerase to copy damaged DNA. RecA then coats the ssDNA to form a RecA-ssDNA nucleoprotein filament, which is the inducing signal for the SOS response. The RecA-ssDNA filament facilitates the autocleavage of LexA, the repressor of the SOS genes, allowing the expression of at least 57 genes (22, 66). While many genes are induced in the SOS response, expression of only the umuDC genes is required for SOS mutagenesis (71).
The umuDC genes in E. coli encode UmuD, a polymerase manager protein, and UmuC, the DNA polymerase subunit. The RecA-ssDNA nucleoprotein filament plays another role in regulation of SOS mutagenesis by facilitating the cleavage of the UmuD protein to form UmuD′ (12, 46, 64). Approximately 20 to 40 min after SOS induction, UmuD cleaves its N-terminal arms between C24 and G25 to form UmuD′2, the form that interacts with UmuC to form Pol V (UmuD′2C), which is active in SOS mutagenesis (57, 78, 81). UmuD′2 is required for UmuC to be active in translesion synthesis (57, 78).
Other protein interactions have also been found to be important for Pol V activity. The β clamp substantially increases processivity in both Pol III and Pol IV in E. coli but increases the processivity of Pol V only 3- to 5-fold (25, 35, 42, 80). The β clamp interacts directly with UmuC utilizing a canonical (4, 6, 20, 41, 73) β-binding motif, an interaction that is critical for UmuC to participate in TLS (4, 6, 73).
In addition to the roles of RecA in the induction of the SOS response and the cleavage of UmuD, RecA also plays a role in the activation of Pol V for TLS (23, 31, 58, 60, 61). While the exact mechanism of Pol V stimulation by RecA remains to be determined, it is understood that RecA is required for Pol V mutagenesis. It is thought that Pol V has a preference for binding to the end of RecA nucleoprotein filaments in order to target the Pol V complex to DNA lesions, as well as binding deep in the groove of the RecA-ssDNA filament (21).
Pol V bypasses the major lesions produced by UV radiation, which are thymine-thymine cyclobutane pyrimidine dimers (T-T CPDs) and thymine-thymine (6-4) photoproducts (77). Pol V generally accurately bypasses T-T CPDs by inserting two dA nucleotides opposite the lesion (75, 77). Pol V bypasses T-T (6-4) photoproducts inaccurately by adding dA across from the 5′ T and dG across from the 3′ T (3, 77). Pol V also bypasses abasic sites by inserting dA, following the “A” rule (36, 57, 72, 77, 78). Opposite C8-dG-acetylaminofluorine, Pol V inserts dA (3, 23, 24); Pol V bypasses N2-benzo[a]pyrene-dG with relative inaccuracy, while it bypasses N6-benzo[a]pyrene-dA fairly accurately (37, 63, 83).
In this work, we sought to identify residues of UmuC that contribute to lesion bypass and, more specifically, to UV-induced mutagenesis. We hypothesized that two loops of UmuC interact with template DNA as well as with the incoming nucleotide and therefore are likely to contribute to mutagenesis. We show here that the N-terminal residues of loop 1, residues 31 to 34, play a significant role in cell survival in response to UV radiation. We also show that loop 2 does not play as significant a role in conferring survival or in UV-induced mutagenesis. Furthermore, we show that the mutation of each of the three UmuC loop 1 residues 32 to 34 to alanine results in less inhibition of recombination than that in wild-type UmuC, which could indicate the disruption of an interaction between Pol V and RecA and therefore could explain the ability of these variants to confer UV hypersensitivity.
MATERIALS AND METHODS
Plasmids and strains.
Low-copy-number plasmid pGY9738 carries the synthetic umuD′C operon and encodes resistance to spectinomycin (60 μg/ml). Strains used (Table 1) were grown in Luria broth at 37°C unless otherwise noted. Competent cells were made by using the CaCl2 method (59). Transformations were performed as described previously (7). Variants of pGY9738 were made with QuikChange or QuikChange Lightning site-directed mutagenesis kits (Agilent Technologies). β-Binding sites are defined as β2 (313LTP315) and β1 (357QLNLF361). Constructs with mutated β-binding sites are designated β1 + 2, which indicates the presence of the mutations 313LTP315 → 313AAA315 and 357QLNLF361 → 357AAAAA361 (6). The presence of the mutations was confirmed by DNA sequencing (Massachusetts General Hospital DNA Core Facility, Cambridge, MA).
Table 1.
Strains and plasmids
| Strain or plasmid | Relevant genotype or description | Reference or source |
|---|---|---|
| Bacterial strain | ||
| AB1157 | argE3 umuDC+ | Laboratory stock |
| GW8017 | AB1157 ΔumuDC | 27 |
| PB102 | AB1157 ΔumuC ΔrecJ | P1(JW2860) → AB1157 ΔumuC (1) |
| AB1157 ΔumuC | 30 | |
| GW2771 | umuDC+ | 51 |
| GW2771 spq-2 | GW2771 spq-2 | 51 |
| GW2771 spq-2 dnaQ903 | dnaQ903::Tet | 65, 67 |
| Plasmid | ||
| pGB2 | Vector; pSC101 derived, Specr | 15 |
| pGY9738 | oC1umuD′C; pSC101 derived | 69 |
UV survival and mutagenesis assays.
UV survival and mutagenesis assays were performed as described previously (7, 65). Variants were exposed to 25 J/m2 254-nm UV radiation unless otherwise noted for mutagenesis assays. Each point shown represents the average of at least three trials, and the error bars indicate the standard deviation.
NFZ and 4-NQO survival assays.
Strains harboring alanine variants of the N-terminal loop 1 residues (residues 32 to 34) were exposed to increasing concentrations of nitrofurazone (NFZ; 5-nitro-2-furaldehyde semicarbazone; TCI America) and 4-nitroquinoline-N-oxide (4-NQO; Acros Organics), as described previously (7, 65). Stock solutions (10 mg/ml) were freshly prepared in N,N-dimethyl formamide (Fisher Scientific) and protected from light. Serial dilutions of overnight cultures were plated on LB agar plates containing 60 μg/ml spectinomycin and the indicated amount of either NFZ or 4-NQO, and the plates were incubated at 37°C for 20 to 24 h. Each point shown represents the average of at least three trials, and the error bars indicate the standard deviation.
Immunoblotting.
Western blotting was performed as described previously (65). Strains harboring variants of the N-terminal loop 1 residues 31 to 34 (alanine or conservative mutations) were exposed to 10 J/m2 UV radiation, while all others were exposed to 25 J/m2 UV radiation. Proteins were resolved by 14% SDS-PAGE and transferred to a polyvinylidene difluoride membrane. The membrane was blocked overnight in 5% milk in 1× Tris-buffered saline (TBS)–Tween buffer (20 mM Tris-HCl, pH 7.6, 137 mM NaCl, 1% Tween 20). The membrane was then probed with anti-UmuC (6) in 2.5% milk with 0.5× TBS-Tween buffer and washed for 2 min and then 3 times for 10 min each time with 1× TBS-Tween buffer. The membrane was probed with horseradish peroxidase-conjugated goat anti-rabbit antibody (Pierce) in 2.5% milk with 0.5× TBS-Tween buffer, developed with SuperSignal chemiluminescence reagent (Pierce), and exposed to film, which was subsequently developed with a Kodak photoprocessor.
Genetic transduction.
Strains used (Table 1) were grown in Luria broth at 37°C overnight. Cultures were centrifuged at 1,500 × g for 10 min and resuspended in 2.5 ml of a solution containing 10 mM MgSO4 and 5 mM CaCl2, and the suspension was transferred to glass test tubes. Recipient cells (100 μl) harboring each variant were incubated at 30°C for 30 min with 50 μl bacteriophage P1vir ΔyeaB (Kanr). As controls, 100 μl of recipient cells was incubated without P1vir ΔyeaB and 100 μl of P1vir ΔyeaB was incubated without recipient cells. After incubation, 100 μl of 1 M sodium citrate and 400 μl of Luria broth were added to each test tube, and cells recovered for 2 h at 30°C with no shaking. Reaction mixtures were centrifuged at 3,800 × g for 5 min and resuspended in 200 μl of Luria broth. A 100-μl aliquot of each reaction mixture was plated on selective medium (spectinomycin, 60 μg/ml; kanamycin, 30 μg/ml) and incubated at 30°C for 20 to 22 h. Phage titer was determined as described previously (45), except that 10-μl aliquots of serial dilutions of phage P1 were plated onto bacteria in top agar.
RESULTS
Identification of UmuC active-site loops.
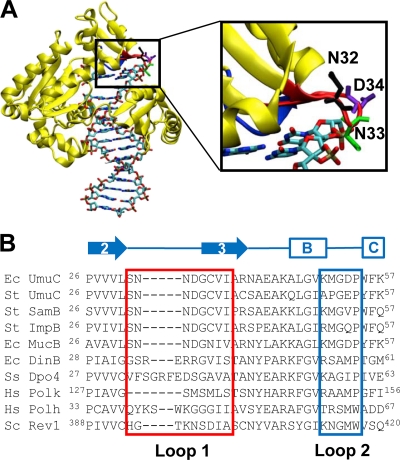
We identified UmuC loops 1 and 2 through modeling and homology searches (Fig. 1). The homology model of UmuC is based upon the crystal structure of Dpo4 since there is currently no crystal structure of UmuC (6). UmuC loops 1 and 2 are predicted to be near the active site of the polymerase and are hypothesized to play a role in DNA template binding and alignment, as well as in interactions with the incoming nucleotide. We defined loop 1 as residues 31 to 38 and loop 2 as residues 50 to 54. For comparison, the sequences of some other Y-family DNA polymerases are also shown, including E. coli DinB, Sulfolobus solfataricus Dpo4, human Pol κ, human Pol η, and Saccharomyces cerevisiae Rev1 (Fig. 1). Loop 1 is highly conserved among UmuC sequences but not within the entire Y family of polymerases, while loop 2 is more variable.
Fig. 1.
E. coli UmuC is a Y-family DNA polymerase that shares little homology with other members of the Y family. (A) Homology model of UmuC (6). UmuC loop 1 (residues 31 to 38) is shown in red, and loop 2 (residues 50 to 54) is shown in blue. UmuC residues N32 (black), N33 (green), and D34 (purple) are predicted to determine the gap size that dictates which lesions can fit into the active site (13, 14, 62). The backbone of UmuC is shown in yellow. DNA is rendered as sticks and colored by atom identity. The illustration was prepared using the VMD package (28). (B) Amino acid sequences of representative Y-family polymerases showing conserved residues aligned with loop 1 (residues 31 to 38, red box) and loop 2 (residues 50 to 54, blue box). Secondary structure based on the crystal structure of Dpo4 is shown above the alignment (11, 39). Ec, Escherichia coli; St, Salmonella enterica serovar Typhimurium; Hs, Homo sapiens; Ss, Sulfolobus solfataricus; Sc, Saccharomyces cerevisiae. The first five sequences are UmuC and its homologs, which share almost 100% homology in loop 1.
Variants in the N-terminal region of loop 1 cause hypersensitivity to UV radiation.
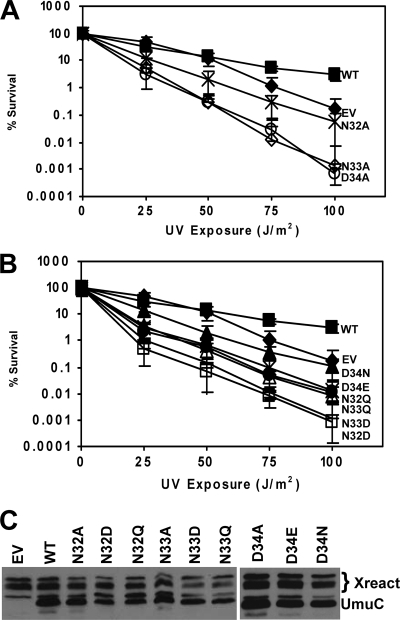
Alanine-scanning mutagenesis was used to determine the extent to which mutating the residues of loop 1 (S31 to I38) contributes to the ability of UmuC to facilitate UV-induced mutagenesis in vivo. We first characterized the alanine variants of UmuC N32, N33, and D34 for UV-induced mutagenesis, which is a prominent phenotype of UmuC. Variants of UmuC expressed in the GW8017 (ΔumuDC) strain do not normally cause sensitivity to UV. Moreover, cells in which the umuDC genes have been deleted are only modestly sensitive to UV (8, 50). However, UmuC with alanine mutations at residues 32 to 34 causes significantly greater sensitivity to UV radiation than wild-type UmuC (Fig. 2A). Strains harboring either UmuC N33A or D34A variants are substantially more sensitive to UV than strains without UmuC.
Fig. 2.
N-terminal loop 1 variants 32, 33, and 34 cause hypersensitivity to UV radiation in a strain that is not normally sensitive to UV. (A) Assays were performed with the pGY9738 plasmid and the following derivatives in GW8017: pGY9738 (umuD′C wild type [WT]; ■), pGB2 (empty vector [EV]; ◆), pGY9738-N32A (umuD′C N32A; ×), pGY9738-N33A (umuD′C N33A; ♢), and pGY9738-D34A (umuD′C D34A; ○). (B) Conservative mutations of N32, N33, and D34 conferred hypersensitivity to UV radiation. Assays were performed with the pGY9738 plasmid and the following derivatives in GW8017: pGY9738 (umuD′C, wild type; ■), pGB2 (empty vector; ◆), pGY9738-D34N (umuD′C D34N; ▴), pGY9738-D34E (umuD′C D34E; ×), pGY9738-N32Q (umuD′C N32Q; ●), pGY9738-N33Q (umuD′C N33Q; ▵), pGY9738-N33D (umuD′C N33D; +), and pGY9738-N32D (umuD′C N32D; □). (C) Immunoblot showing steady-state levels of UmuC variants expressed from plasmids encoding the umuD′C genes in GW8017. The wild-type plasmid was pGY9738, and the empty vector was pGB2. Xreact, cross-reaction.
We made conservative substitutions of N32, N33, and D34 by changing N32 or N33 to Gln or D34 to Glu. These substitutions increase the length of the side chain at these positions by one methylene (CH2) group while maintaining similar chemical characteristics of the side chain functional groups. We also mutated N32 or N33 to Asp or D34 to Asn to test the effects of altering the charge of the side chains on the ability of UmuC to contribute to UV survival. Each conservative substitution caused significantly greater UV sensitivity in a ΔumuDC strain than wild-type UmuC (Fig. 2B). Therefore, these residues may play an important role in the ability of UmuC to bypass UV-induced lesions in DNA, supporting the conjecture that these residues are involved in binding the DNA substrate, since altering these residues even in a conservative manner causes extreme sensitivity of the cells harboring these mutations to UV light.
We determined the steady-state expression levels of the UmuC variants to rule out the possibility that the decrease in survival of cells expressing these variants was due to a change in protein levels. Western blots showed that steady-state expression levels for each variant were similar to that of wild-type UmuC (Fig. 2C). Therefore, the effects noted above are not due to altered expression levels of the UmuC variants. It should be noted that LexA is able to repress umuDC expression in this context; SOS induction causes an approximately 2-fold increase in umuDC expression compared to levels in uninduced cells (71).
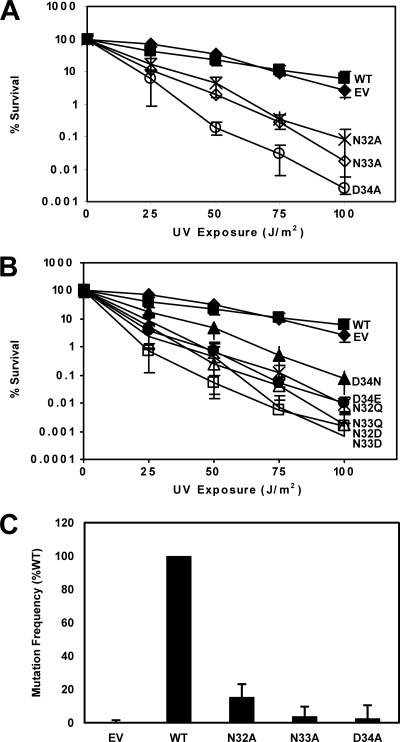
Given the extreme sensitivity to UV light conferred by UmuC variants with substitutions at residues 32, 33, and 34, we examined the effect of UV radiation on cells expressing these UmuC variants in the AB1157 strain (umuDC+) to determine whether these variants caused sensitivity to UV radiation even in a strain with a chromosomal copy of umuDC. UmuC variants with either alanine or more conservative substitutions at positions 32, 33, and 34 conferred extreme sensitivity to UV radiation in comparison to wild-type UmuC or to the empty vector (Fig. 3A and B). The argE3 reversion frequency of wild-type umuDC+ strains expressing UmuC variants containing single alanine substitutions at positions 32, 33, and 34 was determined by exposure to 5 J/m2 UV radiation. Cells expressing the UmuC variants showed a significant decrease in mutation frequency compared to cells expressing wild-type UmuC (Fig. 3C). We previously observed that plasmid-borne umuC can suppress the extreme sensitivity to UV that is characteristic of strains in which both umuC and recJ have been deleted (19, 48). RecJ helps to partially degrade DNA at replication forks that are stalled due to lesions to help aid in restart of DNA replication. Without RecJ, replication is delayed, and recovery of DNA synthesis relies on TLS specifically by Pol V to bypass UV-induced lesions (19). However, changing UmuC N32, N33, and D34 to alanine conferred sensitivity to UV radiation on the cells harboring plasmids expressing these variants and therefore failed to complement the ΔumuC ΔrecJ strain in comparison to cells expressing wild-type UmuC (Fig. 4A to C). Conservative mutations of N32, N33, and D34 also conferred hypersensitivity to UV radiation in this strain (Fig. 4A to C). Therefore, these UmuC variants fail to fulfill various cellular functions of UmuC, including induced mutagenesis and complementation of a ΔumuC ΔrecJ strain, in addition to conferring hypersensitivity on a wild-type strain.
Fig. 3.
N-terminal loop 1 variants 32, 33, and 34 cause hypersensitivity to UV radiation in a wild-type strain. (A) Assays were performed with the pGY9738 plasmid and the following derivatives in AB1157: pGY9738 (umuD′C wild type; ■), pGB2 (empty vector; ◆), pGY9738-N32A (umuD′C N32A; ×), pGY9738-N33A (umuD′C N33A; ♢), and pGY9738-D34A (umuD′C D34A; ○). (B) Conservative mutations of N32, N33, and D34 conferred hypersensitivity to UV radiation. Assays were performed with the pGY9738 plasmid and the following derivatives in AB1157: pGY9738 (umuD′C wild type; ■), pGB2 (empty vector; ◆), pGY9738-D34N (umuD′C D34N; ▴), pGY9738-D34E (umuD′C D34E; ×), pGY9738-N32Q (umuD′C N32Q; ●), pGY9738-N33Q (umuD′C N33Q; ▵), pGY9738-N32D (umuD′C N32D; □), and pGY9738-N33D (umuD′C N33D; +). (C) UV (5 J/m2)-induced mutation frequency of selected variants in plasmid pGY9738 (umuD′C) in strain AB1157. The wild-type plasmid was pGY9738, and the empty vector was pGB2. Frequencies were as follows: for the empty vector, induced mutant, 2.78 × 10−7; spontaneous mutants, 2.99 × 10−7; mutation frequency, −2.16 × 10−8 (set equal to 0); for the wild type, induced mutants, 9.53 × 10−6; spontaneous mutants, 3.40 × 10−6; mutation frequency, 6.13 × 10−6; for N32A, induced mutants, 1.73 × 10−6; spontaneous mutants, 7.89 × 10−7; mutation frequency, 9.50 × 10−7; for N33A, induced mutants, 8.20 × 10−7; spontaneous mutants, 5.83 × 10−7; mutation frequency, 2.37 × 10−7; and for D34A, induced mutants, 6.67 × 10−7; spontaneous mutants, 5.01 × 10−7; mutation frequency, 1.66 × 10−7.
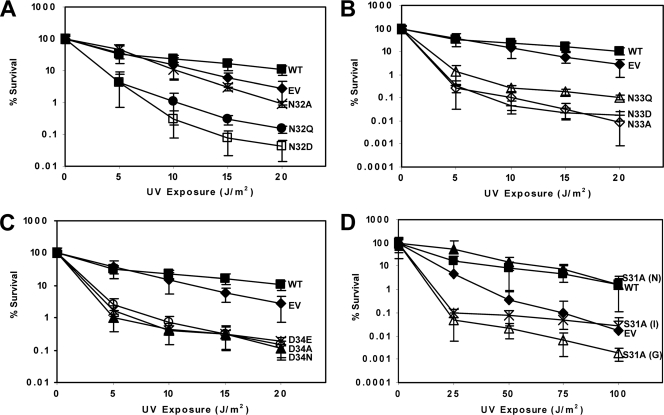
Fig. 4.
UmuC N-terminal loop 1 variants S31, N32, N33, and D34 confer hypersensitivity to UV radiation in a ΔumuC ΔrecJ strain. (A) Mutation of N-terminal loop 1 residue N32 causes sensitivity to UV radiation. Assays were performed with pGY9738 plasmid and the following derivatives in PB102 (AB1157 ΔumuC ΔrecJ): pGY9738 (umuD′C wild type; ■), pGB2 (empty vector; ◆), pGY9738-N32A (umuD′C N32A; ×), pGY9738-N32Q (umuD′C N32Q; ●), and pGY9738-N32D (umuD′C N32D; □). (B) Mutation of N-terminal loop 1 residue N33 causes hypersensitivity to UV radiation. Assays were performed with pGY9738 plasmid and the following derivatives in PB102: pGY9738 (umuD′C wild type; ■), pGB2 (empty vector; ◆), pGY9738-N33Q (umuD′C N33Q; ▵), pGY9738-N33D (umuD′C N33D; +), and pGY9738-N33A (umuD′C N33A; ♢). (C) Mutation of N-terminal loop 1 residue D34 causes hypersensitivity to UV radiation. Assays were performed with pGY9738 plasmid and the following derivatives in PB102: pGY9738 (umuD′C wild type; ■), pGB2 (empty vector; ◆), pGY9738-D34E (umuD′C D34E; ×), pGY9738-D34A (umuD′C D34A; ○), and pGY9738-D34N (umuD′C D34N; ▴). (D) N-terminal loop 1 variant S31A causes hypersensitivity to UV radiation and confers upon strains a growth defect (G), a nongrowth defect (N), or an intermediate growth defect (I). Assays were performed with the pGY9738 plasmid and the following derivatives in PB102: pGY9738-S31A (N) (umuD′C S31A [N]; ▴), pGY9738 (umuD′C wild type; ■), pGY9738-S31A (I) (umuD′C S31A [I]; ×), pGB2 (empty vector; ◆), and pGY9738-S31A (G) (umuD′C S31A [G]; ▵).
Cells expressing the S31A variant of UmuC exhibit a growth defect, observed in the AB1157, GW8017, and PB102 strains, that was not observed with other variants (data not shown). Cells of the PB102 strain harboring S31A are extremely sensitive to UV radiation (Fig. 4D). However, we sometimes observed these cells to be resistant to UV, which we attribute to the possible acquisition of suppressor mutations. Because of the apparent instability of strains harboring the S31A variant, we decided not to pursue further characterization of this variant.
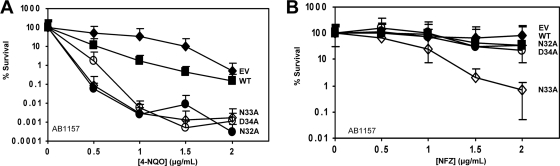
N-terminal loop 1 variants confer sensitivity to 4-NQO, and N33A causes sensitivity to NFZ.
We wanted to investigate whether the N-terminal loop 1 variants contribute to survival in the presence of other DNA-damaging agents. We assayed survival of the AB1157 (umuDC+) strain harboring low-copy-number plasmids expressing the UmuC N32A, N33A, and D34A variants in the presence of NFZ and 4-NQO. NFZ is thought to cause N2-furfuryl-dG adducts, and strains in which dinB, the gene encoding DNA polymerase IV, is deleted are very sensitive to NFZ (30). The major adduct formed from 4-NQO is at the N-2 position of guanine; 4-NQO also forms adducts at the C-8 position of dG as well as at the N-6 position of dA (22). UmuC variants N32A, N33A, and D34A conferred sensitivity to 4-NQO in the umuDC+ strain (Fig. 5A). UmuC variant N33A conferred sensitivity to NFZ, but N32A and D34A did not (Fig. 5B). It has been observed that NFZ is a relatively weak DNA-damaging agent (49), which could account for these differences. Nonetheless, mutation of the N-terminal loop 1 residues causes sensitivity to 4-NQO as well as, to some extent, to NFZ.
Fig. 5.
Cells harboring UmuC variants N32A, N33A, and D34A are sensitive to 4-NQO (A), while only cells harboring UmuC variant N33A are sensitive to NFZ (B). (A and B) Assays were performed with the pGY9738 plasmid and the following derivatives in AB1157: pGB2 (empty vector; ◆), pGY9738 (umuD′C wild type; ■), pGY9738-N32A (umuD′C N32A; ●), pGY9738-N33A (umuD′C N33A; ♢), and pGY9738-D34A (umuD′C D34A; ○).
Deletion of dnaQ or modification of the β-binding motifs of UmuC does not alter UV hypersensitivity of N-terminal loop variants.
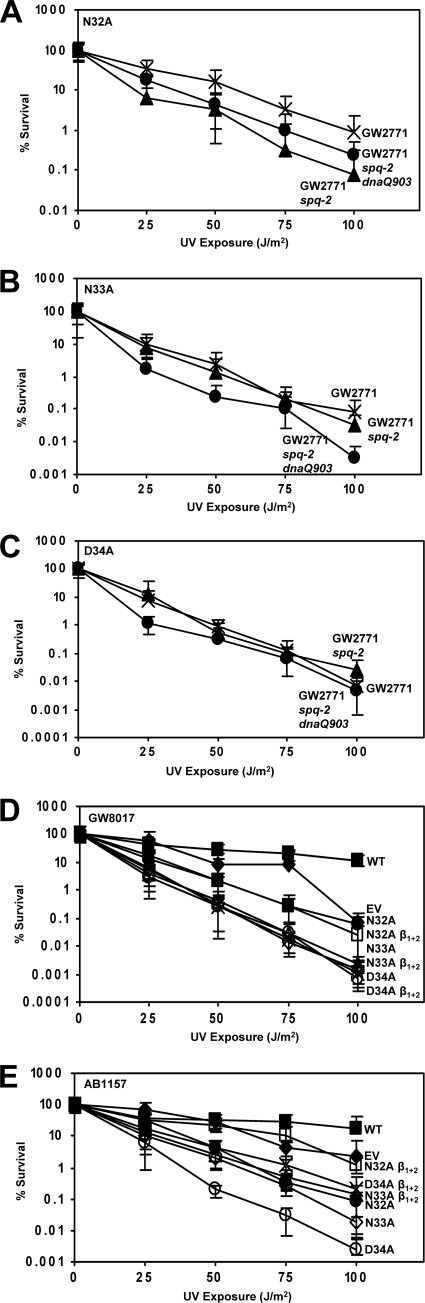
Disrupting dnaQ, which encodes the ε subunit of Pol III, disrupts the proofreading function of Pol III and has been shown to allow Pol III to bypass damaged DNA under some circumstances (40, 53, 79). To determine whether the proofreading subunit of DNA polymerase III plays a role in the hypersensitivity to UV conferred by N32, N33, and D34 variants, we expressed UmuC N32A, N33A, and D34A in GW2771, GW2771 spq-2, and GW2771 spq-2 dnaQ903 strains (Table 1). We compared the effects of the UmuC N-terminal loop variants in GW2771, a wild-type strain, to those in GW2771 spq-2, where spq-2 is an antimutator allele of dnaE (amino acid substitution V832G in DnaE) (38, 67) that suppresses the mutator effect of the disruption of dnaQ, and the isogenic strain in which dnaQ has been disrupted. Whereas N32A caused modest sensitivity to UV in these strains, as in other strains (Fig. 6A), variants N33A and D34A caused hypersensitivity to UV in all three strains (Fig. 6B and C). Therefore, deletion of dnaQ did not suppress the UV hypersensitivity caused by UmuC variants N33A and D34A.
Fig. 6.
Deletion of the proofreading subunit of Pol III (dnaQ) or mutation of β clamp-binding sites of UmuC does not suppress sensitivity to UV radiation caused by mutation of N32, N33, and D34. (A to C) The respective plasmids were assayed in the following strains: GW2771 (×), GW2771 spq-2 (▴), and GW2771 spq-2 dnaQ903 (●). N-terminal loop 1 variants N32A (A), N33A (B), and D34A (C) confer sensitivity to UV radiation, despite the deletion of dnaQ. Assays were performed with derivatives of pGY9738 in the listed strains. (D) The hypersensitivity to UV radiation conferred by variants N32A, N33A, and D34A in GW8017 was not suppressed by the mutation of the β-binding motifs of UmuC. (E) The hypersensitivity to UV radiation conferred by UmuC variants N32A, N33A, and D34A in AB1157 was modestly suppressed by the mutation of the β-binding motifs of UmuC. (D and E) Assays were performed with plasmid pGY9738 and derivatives in GW8017 (D) and AB1157 (E): pGB2 (empty vector; ◆), pGY9738 (umuD′C wild type; ■), pGY9738-N32A (umuD′C N32A; ●), pGY9738-N32Aβ1 + 2 (umuD′C N32A β1 + 2; □), pGY9738-N33A (umuD′C N33A; ♢), pGY9738-N33A β1 + 2 (umuD′C N33A β1 + 2; ▵), pGY9738-D34A (umuD′C D34A; ○), and pGY9738-D34A β1 + 2 (umuD′C D34A β1 + 2; ×).
Next, we tested the role of binding to the β processivity clamp in the ability of UmuC mutations N32A, N33A, and D34A to confer UV hypersensitivity. We combined the N32A, N33A, and D34A individual mutations with mutations of both of the known β-binding sites of UmuC to alanine. The mutations in the β-binding sites of UmuC are designated β1 + 2, indicating the multiple mutations 313LTP315 → 313AAA315 and 357QLNLF361 → 357AAAAA361. These mutations disrupt the binding of UmuC to the β clamp, and therefore, the resultant UmuC should not be recruited to the replication fork (6, 65). N32A, N33A, and D34A conferred hypersensitivity to UV radiation when combined with mutations in the β-binding motifs in the GW8017 strain (Fig. 6D), indicating that the UV hypersensitivity is caused by a mechanism that is at least partially independent of binding to the β clamp. When wild-type UmuC is present on the chromosome, as in the case of AB1157, the mutation of the β-binding motifs partially suppresses the hypersensitivity conferred by N33A and D34A and almost fully suppresses the moderate sensitivity conferred by N32A (Fig. 6E). Therefore, disruption of the β-binding motifs in this context appears to allow wild-type UmuC to confer resistance to DNA damage, presumably by allowing access of wild-type UmuC to DNA.
Variants in the N-terminal region of loop 1 partially suppress inhibition of RecA-mediated homologous recombination.
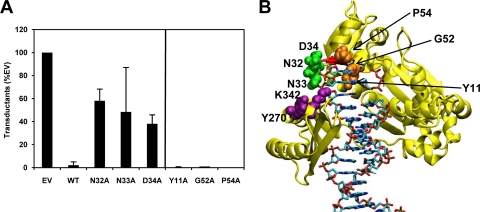
UmuD′ and UmuC inhibit RecA-mediated homologous recombination when present at elevated levels, such as in the SOS response, which may be important for regulating RecA and its involvement in cellular processes (10, 56, 68, 70, 74, 76). UmuC variants N32A, N33A, and D34A were expressed from the respective derivatives of plasmid pGY9738 in a ΔumuDC strain. Cells were exposed to bacteriophage P1vir ΔyeaB and plated on selective medium to measure transductants that result from RecA-mediated homologous recombination. We observed that the N32A, N33A, and D34A variants confer intermediate inhibition of homologous recombination compared to wild-type UmuD′C, which inhibits recombination (Fig. 7A). This suggests that these mutations may disrupt a direct interaction between RecA and UmuC, although other interpretations are possible, such as a disruption of an interaction between Pol V and RecA that is mediated by UmuD′2. Two residues, K342 and Y270, were previously shown (70) to enhance the inhibition of homologous recombination when mutated to glutamine and cysteine, respectively. These two residues are predicted to be in close proximity to N32, N33, and D34, offering further evidence that this region is important for interactions that modulate recombination (Fig. 7B).
Fig. 7.
UmuD′2C inhibits homologous recombination facilitated by RecA. (A) Variants in plasmid pGY9738 (umuD′C wild type) were expressed in strain GW8017 (ΔumuDC). Cells harboring the variants N32A, N33A, and D34A show intermediate inhibition of RecA-mediated homologous recombination, whereas cells harboring other variants (Y11A, G52A, P54A; to the right of the vertical line) inhibit homologous recombination to a similar extent as wild-type umuD′C. Transduction efficiency is measured in CFU/PFU as a percentage of that for the empty vector (1.31 × 10−5 CFU/PFU, normalized to 100.0%): wild type, 2.09%; N32A, 58.1%; N33A, 48.4%; D34A, 38.1%; Y11A, 0.358%, G52A, 0.956%; P54A, 0.000%. (B) Homology model of UmuC (6). The backbone of UmuC is shown in yellow. UmuC loop 1 (residues 31 to 38) is shown in red, and loop 2 (residues 50 to 54) is shown in blue. UmuC residues N32, N33, and D34 (green) are near previously studied residues K342 and Y270 (purple) in the UmuC model. Previously studied residue Y11, as well as residues G52 and P54 (orange), is shown for comparison. Cells harboring variants Y270C and K342Q show a significant decrease in RecA-mediated homologous recombination. The image was prepared using VMD (28).
In order to assess the specificity of this effect, we determined the ability of UmuC variants with other mutations to inhibit homologous recombination. Mutants with the UmuC loop 2 G52A and P54A mutations exhibited an inhibition of homologous recombination that is similar to that of the strain with wild-type UmuC (Fig. 7A). Moreover, the previously studied variant Y11A confers hypersensitivity to UV radiation (65), but we find that the UmuC Y11A variant inhibits homologous recombination to a similar extent as wild-type UmuC. G52, P54, and Y11 are predicted to be located in the interior of the protein and so are unlikely to be available for protein-protein interactions (Fig. 7B). These observations suggest that N32, N33, and D34 have a specific role in UmuC function both in UV resistance, presumably via DNA binding and TLS, and in modulating homologous recombination.
C-terminal region of loop 1 does not play a significant role in UV survival.
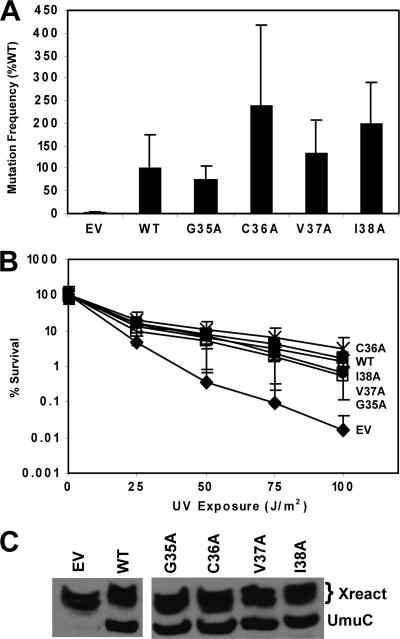
Cells expressing UmuC variants at the C terminus of loop 1 (G35A, C36A, V37A, or I38A) have a mutation frequency similar to or greater than that of cells expressing wild-type UmuC; therefore, these variants are proficient for UV-induced mutagenesis (Fig. 8A). Additionally, UmuC variants G35A, C36A, V37A, and I38A each complement the ΔumuC ΔrecJ strain as well as the strain with wild-type UmuC for UV resistance (Fig. 8B). For the cells harboring these variants, there is a good correlation between UV-induced mutagenesis and UV resistance in the ΔumuC ΔrecJ strain.
Fig. 8.
UmuC C-terminal loop 1 variants do not confer sensitivity to UV radiation and are proficient for mutagenesis. (A) UV (25 J/m2)-induced mutation frequency of selected variants in plasmid pGY9738 (umuD′C) in strain GW8017 (ΔumuDC). The wild-type plasmid was pGY9738, and the empty vector was pGB2. Frequencies were as follows: for the empty vector, induced mutants, 7.66 × 10−7; spontaneous mutants, 3.36 × 10−7; mutation frequency, 4.31 × 10−7; for the wild type, induced mutants, 1.85 × 10−5; spontaneous mutants, 6.48 × 10−6; mutation frequency, 1.21 × 10−5; for G35A, induced mutants, 1.73 × 10−5; spontaneous mutants, 7.58 × 10−6; mutation frequency, 9.68 × 10−6; for C36A, induced mutants, 3.25 × 10−5; spontaneous mutants, 2.30 × 10−6; mutation frequency, 3.03 × 10−5; for V37A, induced mutants, 2.24 × 10−5; spontaneous mutants; 5.41 × 10−6; mutation frequency, 1.70 × 10−5; and for I38A, induced mutants, 2.80 × 10−5; spontaneous mutants, 2.73 × 10−6; mutation frequency, 2.53 × 10−5. (B) C-terminal loop 1 variants do not confer sensitivity to UV radiation. Assays were performed with pGY9738 plasmid and the following derivatives in PB102: pGY9738-C36A (umuD′C C36A; ×), pGY9738 (umuD′C wild type; ■), pGY9738-I38A (umuD′C I38A; ▵), pGY9738-V37A (umuD′C V37A; ●), pGY9738-G35A (umuD′C G35A; □), and pGB2 (empty vector; ◆). (C) Immunoblot showing steady-state levels of UmuC expressed from variant umuDC plasmids (GW8017). The wild-type plasmid was pGY9738, and the empty vector was pGB2.
The steady-state levels of the loop 1 C-terminal UmuC variants G35A, C36A, V37A, and I38A after exposure to UV radiation were determined by using Western blotting (Fig. 8C). Each of these variants is present at levels similar to wild-type UmuC. Therefore, the expression levels of UmuC are not altered by the presence of these loop 1 mutations.
Variants in UmuC loop 2 contribute little to UV survival.
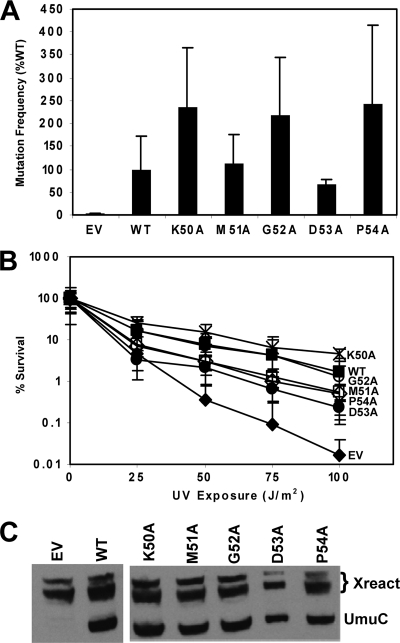
We also used alanine-scanning mutagenesis to determine the importance of residues in loop 2 (K50 to P54) in UV-induced mutagenesis. Cells expressing these UmuC variants have similar mutation frequencies as cells expressing wild-type UmuC (Fig. 9A). Each of the loop 2 variants confers UV resistance in a ΔumuC ΔrecJ strain that is similar to that in cells harboring wild-type UmuC (Fig. 9B). Of these variants, only cells expressing UmuC D53A are significantly more sensitive to UV radiation than cells expressing wild-type UmuC (Fig. 9B) and exhibit a lower mutation frequency than cells expressing wild-type UmuC (Fig. 9A). With the exception of D53A, mutating each residue of loop 2 to alanine did not have a significant effect on UmuC-dependent UV-induced mutagenesis or UV survival. A Western blot (Fig. 9C) shows that the steady-state expression level of each UmuC variant is similar to that of wild-type UmuC, indicating that the mutations constructed in UmuC do not alter its expression levels.
Fig. 9.
UmuC loop 2 variants do not confer sensitivity to UV radiation and are proficient for mutagenesis. (A) UV (25 J/m2)-induced mutation frequency of selected variants in plasmid pGY9738 (umuD′C) in strain GW8017 (ΔumuDC). The wild-type plasmid was pGY9738, and the empty vector was pGB2. Frequencies were as follows: for the empty vector, induced mutants, 7.66 × 10−7; spontaneous mutants, 3.36 × 10−7; mutation frequency, 4.31 × 10−7; for the wild type, induced mutants, 1.85 × 10−5; spontaneous mutants, 6.48 × 10−6; mutation frequency, 1.21 × 10−5; for K50A, induced mutants, 3.28 × 10−5; spontaneous mutants, 2.96 × 10−6; mutation frequency, 2.98 × 10−5; for M51A, induced mutants, 1.71 × 10−5; spontaneous mutants, 3.00 × 10−6; mutation frequency, 1.41 × 10−5; for G52A, induced mutants, 3.38 × 10−5; spontaneous mutants, 6.36 × 10−6; mutation frequency, 2.74 × 10−5; for D53A, induced mutants, 1.16 × 10−5; spontaneous mutants, 3.32 × 10−6; mutation frequency, 8.29 × 10−6; and for P54A, induced mutants, 3.77 × 10−5; spontaneous mutants, 7.22 × 10−6; mutation frequency, 3.05 × 10−5. (B) Loop 2 variants do not confer sensitivity to UV radiation. Assays were performed with pGY9738 plasmid and the following derivatives in PB102: pGY9738-K50A (umuD′C K50; ×), pGY9738 (umuD′C wild type; ■), pGY9738-G52A (umuD′C G52A; ○), pGY9738-M51A (umuD′C M51A; □), pGY9738-P54A (umuD′C P54A; ♢), pGY9738-D53A (umuD′C D53A; ●), and pGB2 (empty vector; ◆). (C) Immunoblot showing steady-state levels of UmuC expressed from variant umuDC plasmids (GW8017). The wild-type plasmid was pGY9738, and the empty vector was pGB2.
DISCUSSION
We identified loops 1 and 2 to be likely to harbor residues important for the function of UmuC in mutagenesis. Loop 1, residues 31 to 38, is located just after predicted β-sheet 2 in the fingers domain. Loop 2, residues 50 to 54, is located between predicted α helices B and C and is also in the fingers domain (11). Residue P54 of loop 2 was noted by Boudsocq et al. to be an important conserved residue for hydrophobic core formation (11). The residues in loop 1, however, are not conserved among the Y-family polymerases (11) but are conserved among UmuC homologs, including those found on plasmids (Fig. 1) (82). Considering that these residues are conserved among UmuC homologs but are not conserved among Y-family polymerases, we hypothesized that they may contribute to the specificity of UmuC in TLS. Loop 2 residues are more conserved among Y-family polymerases and less conserved among UmuC homologs (Fig. 1), suggesting that loop 2 may not play as important a role in lesion bypass specificity. We observed that the N-terminal part of loop 1, residues 32 to 34, plays a significant role in UmuC function. When each of these residues is mutated to alanine, cells harboring the variants are nonmutable (Fig. 3C). The ability of Pol V to inhibit homologous recombination is also disrupted by mutating residues 32 to 34 (Fig. 7A), so perturbation of the interaction between Pol V and RecA may partially explain the extreme sensitivity to UV radiation conferred by these variants. On the other hand, mutations of the residues in the C-terminal region of loop 1 and loop 2 do not have as significant an impact on UmuC-dependent mutagenesis (Fig. 8 and 9).
To date, there is no crystal structure of UmuC, the polymerase subunit of Pol V, so we rely on molecular modeling based on homology to other Y-family polymerases as well as comparisons to polymerases in other families. Y-family polymerases are notable for their lack of a specific α helix, the O helix, found in the active sites of A-family DNA polymerases, that strongly contributes to fidelity (29). This α helix is positioned such that it interacts with the incoming nucleotide as well as with the template strand base and contributes to the high fidelity of replicative DNA polymerases (2, 34, 54). Loops 1 and 2 in UmuC as defined here are in a similar location in UmuC as the O helix is in the A family of DNA polymerases, which suggests a role in fidelity or specificity (Fig. 1) (29). Chandani and colleagues describe the opening next to the active site of Y-family polymerases as a “chimney” (13, 14) with a cluster of amino acids (UmuC S31, N32, N33) that control the size of the opening and potentially dictate which adducts are bypassed by UmuC. We observed that UmuC variants S31A, N32A, N33A, and D34A fail to complement the UV sensitivity of the ΔumuC ΔrecJ strain as well as the less sensitive ΔumuDC strain. Conservative mutations of these residues also failed to complement these strains, suggesting that they are extremely important for UmuC function, perhaps in determining which lesions UmuC bypasses by controlling the size of the active-site opening. However, our observations suggest that the UV sensitivity conferred by mutations of the loop 1 N-terminal residues may not be entirely due to action at the replication fork, as concomitant mutation of the β-binding motifs suppresses their UV hypersensitivity only in the presence of wild-type UmuC and not in its absence.
UmuC I38 is a conserved residue that is located above the base of the incoming nucleotide, is next to the opening of the active site, and has been shown to be important for efficient bypass of N2-benzo[a]pyrene-dG (14, 62). The residue neighboring I38, UmuC M51, also contacts the incoming nucleotide in the model, giving rise to the suggestion that these residues may play a significant role in lesion bypass or, more specifically, deoxynucleoside triphosphate (dNTP) insertion. We find that cells (ΔumuC ΔrecJ) harboring plasmids expressing UmuC M51A were only modestly sensitive to UV radiation and were fully proficient for UV-induced mutagenesis (Fig. 9B), suggesting that M51 may not have a significant role in UV mutagenesis but may still have a role in dNTP insertion (14, 62). In our experiments, the I38A variant complemented the ΔumuC ΔrecJ strain and was fully proficient in UV-induced mutagenesis. The differences in these observations could be due to the fact that other studies probed UmuC bypass of adducts of a common metabolite of the carcinogen benzo-[a]-pyrene (14, 62), while here we are mainly concerned with photoproducts of UV radiation.
Mutating S31 to L results in a UmuC variant that is unable to complement a ΔumuDC strain in UV mutagenesis (5). S31 is located in the “flue” region of the UmuC protein (14) and predicted to be within 5 Å of the deoxyribose of the template base (5). We observed that strains expressing UmuC S31A had a growth defect and subsequently identified populations of strains expressing UmuC S31A with severe, intermediate, or no growth defects, indicating that this variant acquired suppressor mutations. Strains expressing UmuC S31L did not have this growth defect (5). The location and behavior of S31A as well as those of S31L suggest that this residue plays an important role in UmuC function and perhaps, more specifically, in template DNA alignment in the active site.
Human DNA Pol η, a functional homolog of UmuC, is encoded by the hRAD30A gene, and without it, humans develop the genetic disorder xeroderma pigmentosum variant (XPV), making them 100 times more susceptible to UV radiation-induced skin cancer from exposure to sunlight (18, 26, 32, 44). Human Pol η bypasses T-T CPDs formed from exposure to UV radiation accurately by inserting primarily adenine opposite each T of the CPD (33, 43). Motif II of human Pol η (residues 52 to 73) aligns with the loop 2 region in UmuC (Fig. 1). The S62G variant is more efficient in bypassing T-T CPDs, 8-oxo-dG, O4-methyl-dT, O6-methyl-dG, abasic sites, and etheno-dA lesions, as well as in copying undamaged DNA, than is wild-type Pol η (26). A crystal structure of human Pol η depicts S62 interacting with the 5′ base of a T-T CPD (9, 26). Human Pol η R61A, also in loop 2, exhibits decreased efficiency but improved fidelity opposite the T-T CPD lesion (9). We observed that several variants of UmuC loop 2 (residues 50 to 54) conferred increased mutagenesis on strains harboring those mutations, although these increases were not statistically significant compared to wild-type UmuC.
Human Pol η loop 1 residue Q38 hydrogen bonds to both thymines of a T-T CPD lesion (9). Mutating Gln to Ala decreases the efficiency of Pol η, causing stalling after the T-T CPD, perhaps due to incorrect base stacking and therefore misalignment with the incoming nucleotide (9). This interaction with the template base supports the idea that loop 1 residues interact with the nascent base pair in the active site (13, 14).
There are very few examples of variants of UmuC that confer hypersensitivity to UV radiation as seen here with the N-terminal loop 1 residues 32 to 34. A variant of UmuC with a mutation of the steric gate residue Y11 failed to complement a ΔumuDC strain for UV-induced mutagenesis, rendered these cells hypersensitive to UV radiation, and was dominant negative (65). Though the Y11A variant confers similar hypersensitivity to UV radiation as the mutations N32A, N33A, and D34A, its apparent mechanism of action is different from that of the variants reported here. Disruption of the dnaQ gene, which encodes the proofreading subunit of DNA Pol III, almost completely suppressed the UV hypersensitivity of cells expressing UmuC Y11A, while with the variants reported here, disruption of dnaQ had almost no effect on UV sensitivity. Moreover, mutating the β clamp-binding sites of UmuC did not suppress the UV hypersensitivity conferred by N33A and D34A in a ΔumuDC strain, in striking contrast to our observations with Y11A. By analogy to Dpo4, we hypothesized that the UmuC N-terminal loop 1 residues 31 to 34 interact with the DNA template, and mutating these residues possibly disrupts this interaction. This perturbed interaction with the DNA substrate may cause cells to confer hypersensitivity to UV radiation if the UmuC variants cannot bypass lesions caused by exposure to UV radiation. Because mutating the β clamp-binding sites of UmuC along with mutations at residues 32 to 34 did not suppress their hypersensitivity to UV radiation in a ΔumuDC strain, unlike the previously studied Y11A variant (65), there is likely an additional factor responsible for the observed UV hypersensitivity.
Even more intriguing is the possibility that a RecA interaction is interrupted by the mutation of N32, N33, or D34 to alanine. UmuC requires RecA for TLS, but the site of interaction on UmuC is not completely elucidated (31, 60, 61). Our data indicate that mutating N32, N33, or D34 to alanine confers intermediate inhibition of RecA-mediated homologous recombination compared to that of the empty vector and wild-type UmuC (Fig. 7A). A selection experiment for UmuD′ and UmuC variants that increased the inhibition of RecA-mediated homologous recombination identified (69) seven UmuC variants: F10L, Y270C, K277E, F287L, F287S, K342Q, and F351I. We mapped these residues on a model of UmuC (Fig. 7B) and observed that N32, N33, and D34 are in close proximity to Y270 and K342, two residues that when mutated were found to enhance the inhibition of RecA-mediated homologous recombination (69). All five residues (N32, N33, D34, Y270, K342) are located near the incoming template strand of ssDNA, consistent with the idea that inhibition of RecA-mediated homologous recombination by UmuD′2C occurs via a physical interaction of Pol V with RecA (10, 56, 68–70). Taken together, these observations suggest that a site of interaction between UmuC and RecA is the location on UmuC where the single-stranded DNA template enters UmuC. Our observations cannot rule out alternative models, however, including a scenario in which UmuD′2 mediates interactions with UmuC and RecA or in which competition for DNA substrates forms the basis of Pol V-induced inhibition of homologous recombination. Disruption of the interaction of Pol V with RecA could lead to the UV sensitivity that we observed, as that interaction is critical for TLS (23, 31, 58, 60, 61). In sum, we show here that the N terminus of loop 1 plays an essential role in cellular resistance to DNA-damaging agents and facilitates possible interactions with RecA that could be critical for Pol V-mediated UV mutagenesis.
ACKNOWLEDGMENTS
We are pleased to acknowledge generous financial support from a New Faculty Award from the Camille & Henry Dreyfus Foundation (to P.J.B.), the NSF (CAREER Award, MCB-0845033 to P.J.B.), and the NU Office of the Provost (to P.J.B.). P.J.B. is a Cottrell Scholar of the Research Corporation for Science Advancement.
We thank Jason Walsh, Danielle Kania, and Megan Troy for technical assistance. We also thank Mark D. Sutton (University of Buffalo) for helpful discussions.
Footnotes
Published ahead of print on 22 July 2011.
REFERENCES
- 1. Baba T., et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beard W. A., Wilson S. H. 2003. Structural insights into the origins of DNA polymerase fidelity. Structure 11:489–496 [DOI] [PubMed] [Google Scholar]
- 3. Becherel O. J., Fuchs R. P. 1999. SOS mutagenesis results from up-regulation of translesion synthesis. J. Mol. Biol. 294:299–306 [DOI] [PubMed] [Google Scholar]
- 4. Becherel O. J., Fuchs R. P. P., Wagner J. 2002. Pivotal role of the beta-clamp in translesion DNA synthesis and mutagenesis in E. coli cells. DNA Repair (Amst.) 1:703–708 [DOI] [PubMed] [Google Scholar]
- 5. Beuning P. J., et al. 2009. Characterization of novel alleles of the Escherichia coli umuDC genes identifies additional interaction sites of UmuC with the beta clamp. J. Bacteriol. 191:5910–5920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beuning P. J., Sawicka D., Barsky D., Walker G. C. 2006. Two processivity clamp interactions differentially alter the dual activities of UmuC. Mol. Microbiol. 59:460–474 [DOI] [PubMed] [Google Scholar]
- 7. Beuning P. J., Simon S. M., Godoy V. G., Jarosz D. F., Walker G. C. 2006. Characterization of Escherichia coli translesion synthesis polymerases and their accessory factors. Methods Enzymol. 408:318–340 [DOI] [PubMed] [Google Scholar]
- 8. Beuning P. J., Simon S. M., Zemla A., Barsky D., Walker G. C. 2006. A non-cleavable UmuD variant that acts as a UmuD′ mimic. J. Biol. Chem. 281:9633–9640 [DOI] [PubMed] [Google Scholar]
- 9. Biertumpfel C., et al. 2010. Structure and mechanism of human DNA polymerase eta. Nature 465:1044–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boudsocq F., Campbell M., Devoret R., Bailone A. 1997. Quantitation of the inhibition of Hfr × F− recombination by the mutagenesis complex UmuD′C. J. Mol. Biol. 270:201–211 [DOI] [PubMed] [Google Scholar]
- 11. Boudsocq F., Ling H., Yang W., Woodgate R. 2002. Structure-based interpretation of missense mutations in Y-family DNA polymerases and their implications for polymerase function and lesion bypass. DNA Repair (Amst.) 1:343–358 [DOI] [PubMed] [Google Scholar]
- 12. Burckhardt S. E., Woodgate R., Scheuermann R. H., Echols H. 1988. UmuD mutagenesis protein of Escherichia coli: overproduction, purification, and cleavage by RecA. Proc. Natl. Acad. Sci. U. S. A. 85:1811–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chandani S., Jacobs C., Loechler E. L. 2010. Architecture of y-family DNA polymerases relevant to translesion DNA synthesis as revealed in structural and molecular modeling studies. J. Nucleic Acids 2010:pii=784081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chandani S., Loechler E. L. 2009. Y-family DNA polymerases may use two different dNTP shapes for insertion: a hypothesis and its implications. J. Mol. Graph. Model. 27:759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Churchward G., Belin D., Nagamine Y. 1984. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene 31:165–171 [DOI] [PubMed] [Google Scholar]
- 16. Cirz R. T., et al. 2005. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 3:e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cirz R. T., et al. 2007. Complete and SOS-mediated response of Staphylococcus aureus to the antibiotic ciprofloxacin. J. Bacteriol. 189:531–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cleaver J. E. 1999. Stopping DNA replication in its tracks. Science 285:212–213 [DOI] [PubMed] [Google Scholar]
- 19. Courcelle C. T., Chow K. H., Casey A., Courcelle J. 2006. Nascent DNA processing by RecJ favors lesion repair over translesion synthesis at arrested replication forks in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 103:9154–9159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dalrymple B. P., Kongsuwan K., Wijffels G., Dixon N. E., Jennings P. A. 2001. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc. Natl. Acad. Sci. U. S. A. 98:11627–11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Frank E. G., et al. 2000. Visualization of two binding sites for the Escherichia coli UmuD′(2)C complex (DNA pol V) on RecA-ssDNA filaments. J. Mol. Biol. 297:585–597 [DOI] [PubMed] [Google Scholar]
- 22. Friedberg E. C., et al. 2006. DNA repair and mutagenesis, 2nd ed ASM Press, Washington, DC [Google Scholar]
- 23. Fujii S., Fuchs R. P. 2009. Biochemical basis for the essential genetic requirements of RecA and the beta-clamp in Pol V activation. Proc. Natl. Acad. Sci. U. S. A. 106:14825–14830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fujii S., Fuchs R. P. 2004. Defining the position of the switches between replicative and bypass DNA polymerases. EMBO J. 23:4342–4352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fujii S., Gasser V., Fuchs R. P. 2004. The biochemical requirements of DNA polymerase V-mediated translesion synthesis revisited. J. Mol. Biol. 341:405–417 [DOI] [PubMed] [Google Scholar]
- 26. Glick E., Vigna K. L., Loeb L. A. 2001. Mutations in human DNA polymerase eta motif II alter bypass of DNA lesions. EMBO J. 20:7303–7312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guzzo A., Lee M. H., Oda K., Walker G. C. 1996. Analysis of the region between amino acids 30 and 42 of intact UmuD by a monocysteine approach. J. Bacteriol. 178:7295–7303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Humphrey W., Dalke A., Schulten K. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38, 27–28 [DOI] [PubMed] [Google Scholar]
- 29. Jarosz D. F., Beuning P. J., Cohen S. E., Walker G. C. 2007. Y-family DNA polymerases in Escherichia coli. Trends Microbiol. 15:70–77 [DOI] [PubMed] [Google Scholar]
- 30. Jarosz D. F., Godoy V. G., Delaney J. C., Essigmann J. M., Walker G. C. 2006. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature 439:225–228 [DOI] [PubMed] [Google Scholar]
- 31. Jiang Q., Karata K., Woodgate R., Cox M. M., Goodman M. F. 2009. The active form of DNA polymerase V is UmuD′(2)C-RecA-ATP. Nature 460:359–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johnson R. E., Kondratick C. M., Prakash S., Prakash L. 1999. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science 285:263–265 [DOI] [PubMed] [Google Scholar]
- 33. Johnson R. E., Washington M. T., Prakash S., Prakash L. 2000. Fidelity of human DNA polymerase eta. J. Biol. Chem. 275:7447–7450 [DOI] [PubMed] [Google Scholar]
- 34. Johnson S. J., Taylor J. S., Beese L. S. 2003. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc. Natl. Acad. Sci. U. S. A. 100:3895–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kornberg A., Baker T. A. (ed.). 1992. DNA replication, 2nd ed W. H. Freeman & Company, New York, NY [Google Scholar]
- 36. Lawrence C. W., Borden A., Banerjee S. K., LeClerc J. E. 1990. Mutation frequency and spectrum resulting from a single abasic site in a single-stranded vector. Nucleic Acids Res. 18:2153–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lenne-Samuel N., Janel-Bintz R., Kolbanovskiy A., Geacintov N. E., Fuchs R. P. 2000. The processing of a benzo (a) pyrene adduct into a frameshift or a base substitution mutation requires a different set of genes in Escherichia coli. Mol. Microbiol. 38:299–307 [DOI] [PubMed] [Google Scholar]
- 38. Lifsics M. R., Lancy E. D., Jr., Maurer R. 1992. DNA replication defect in Salmonella typhimurium mutants lacking the editing (epsilon) subunit of DNA polymerase III. J. Bacteriol. 174:6965–6973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ling H., Boudsocq F., Woodgate R., Yang W. 2001. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell 107:91–102 [DOI] [PubMed] [Google Scholar]
- 40. Livneh Z. 1986. Replication of UV-irradiated single-stranded DNA by DNA polymerase III holoenzyme of Escherichia coli: evidence for bypass of pyrimidine photodimers. Proc. Natl. Acad. Sci. U. S. A. 83:4599–4603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lopez de Saro F. J., Georgescu R. E., Goodman M. F., O'Donnell M. 2003. Competitive processivity-clamp usage by DNA polymerases during DNA replication and repair. EMBO J. 22:6408–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maor-Shoshani A., Livneh Z. 2002. Analysis of the stimulation of DNA polymerase V of Escherichia coli by processivity proteins. Biochemistry 41:14438–14446 [DOI] [PubMed] [Google Scholar]
- 43. Masutani C., Kusumoto R., Iwai S., Hanaoka F. 2000. Mechanisms of accurate translesion synthesis by human DNA polymerase eta. EMBO J. 19:3100–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Masutani C., et al. 1999. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature 399:700–704 [DOI] [PubMed] [Google Scholar]
- 45. Miller J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 46. Nohmi T., Battista J. R., Dodson L. A., Walker G. C. 1988. RecA-mediated cleavage activates UmuD for mutagenesis: mechanistic relationship between transcriptional derepression and posttranslational activation. Proc. Natl. Acad. Sci. U. S. A. 85:1816–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ohmori H., et al. 2001. The Y-family of DNA polymerases. Mol. Cell 8:7–8 [DOI] [PubMed] [Google Scholar]
- 48. Ollivierre J. N., Sikora J. L., Beuning P. J. 2011. The dimeric SOS mutagenesis protein UmuD is active as a monomer. J. Biol. Chem. 286:3607–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ona K. R., Courcelle C. T., Courcelle J. 2009. Nucleotide excision repair is a predominant mechanism for processing nitrofurazone-induced DNA damage in Escherichia coli. J. Bacteriol. 191:4959–4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Opperman T., Murli S., Smith B. T., Walker G. C. 1999. A model for a umuDC-dependent prokaryotic DNA damage checkpoint. Proc. Natl. Acad. Sci. U. S. A. 96:9218–9223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Opperman T., Murli S., Walker G. C. 1996. The genetic requirements for UmuDC-mediated cold sensitivity are distinct from those for SOS mutagenesis. J. Bacteriol. 178:4400–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pages V., Fuchs R. P. 2002. How DNA lesions are turned into mutations within cells? Oncogene 21:8957–8966 [DOI] [PubMed] [Google Scholar]
- 53. Pages V., Janel-Bintz R., Fuchs R. P. 2005. Pol III proofreading activity prevents lesion bypass as evidenced by its molecular signature within E. coli cells. J. Mol. Biol. 352:501–509 [DOI] [PubMed] [Google Scholar]
- 54. Patel P. H., Suzuki M., Adman E., Shinkai A., Loeb L. A. 2001. Prokaryotic DNA polymerase I: evolution, structure, and “base flipping” mechanism for nucleotide selection. J. Mol. Biol. 308:823–837 [DOI] [PubMed] [Google Scholar]
- 55. Radman M. 1974. Phenomenology of an inducible mutagenic DNA repair pathway in Escherichia coli: SOS repair hypothesis, p. xv, 289 In Prakash L., et al. (ed.), Molecular and environmental aspects of mutagenesis. Thomas, Springfield, IL [Google Scholar]
- 56. Rehrauer W. M., Bruck I., Woodgate R., Goodman M. F., Kowalczykowski S. C. 1998. Modulation of RecA nucleoprotein function by the mutagenic UmuD′C protein complex. J. Biol. Chem. 273:32384–32387 [DOI] [PubMed] [Google Scholar]
- 57. Reuven N. B., Arad G., Maor-Shoshani A., Livneh Z. 1999. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD′, RecA, and SSB and is specialized for translesion replication. J. Biol. Chem. 274:31763–31766 [DOI] [PubMed] [Google Scholar]
- 58. Reuven N. B., Arad G., Stasiak A. Z., Stasiak A., Livneh Z. 2001. Lesion bypass by the Escherichia coli DNA polymerase V requires assembly of a RecA nucleoprotein filament. J. Biol. Chem. 276:5511–5517 [DOI] [PubMed] [Google Scholar]
- 59. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 60. Schlacher K., Cox M. M., Woodgate R., Goodman M. F. 2006. RecA acts in trans to allow replication of damaged DNA by DNA polymerase V. Nature 442:883–887 [DOI] [PubMed] [Google Scholar]
- 61. Schlacher K., et al. 2005. DNA polymerase V and RecA protein, a minimal mutasome. Mol. Cell 17:561–572 [DOI] [PubMed] [Google Scholar]
- 62. Seo K. Y., et al. 2009. Amino acid architecture that influences dNTP insertion efficiency in Y-family DNA polymerase V of E. coli. J. Mol. Biol. 392:270–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shen X., et al. 2002. Efficiency and accuracy of SOS-induced DNA polymerases replicating benzo[a]pyrene-7,8-diol 9,10-epoxide A and G adducts. J. Biol. Chem. 277:5265–5274 [DOI] [PubMed] [Google Scholar]
- 64. Shinagawa H., Iwasaki H., Kato T., Nakata A. 1988. RecA protein-dependent cleavage of UmuD protein and SOS mutagenesis. Proc. Natl. Acad. Sci. U. S. A. 85:1806–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shurtleff B. W., Ollivierre J. N., Tehrani M., Walker G. C., Beuning P. J. 2009. Steric gate variants of UmuC confer UV hypersensitivity on Escherichia coli. J. Bacteriol. 191:4815–4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Simmons L. A., Foti J. J., Cohen S. E., Walker G. C. 2008. The SOS regulatory network. In Bock A., et al. (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC [Google Scholar]
- 67. Slater S. C., Lifsics M. R., O'Donnell M., Maurer R. 1994. holE, the gene coding for the theta subunit of DNA polymerase III of Escherichia coli: characterization of a holE mutant and comparison with a dnaQ (epsilon-subunit) mutant. J. Bacteriol. 176:815–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sommer S., Bailone A., Devoret R. 1993. The appearance of the UmuD′C protein complex in Escherichia coli switches repair from homologous recombination to SOS mutagenesis. Mol. Microbiol. 10:963–971 [DOI] [PubMed] [Google Scholar]
- 69. Sommer S., Boudsocq F., Devoret R., Bailone A. 1998. Specific RecA amino acid changes affect RecA-UmuD′C interaction. Mol. Microbiol. 28:281–291 [DOI] [PubMed] [Google Scholar]
- 70. Sommer S., Coste G., Bailone A. 2000. Specific amino acid changes enhance the anti-recombination activity of the UmuD′C complex. Mol. Microbiol. 35:1443–1453 [DOI] [PubMed] [Google Scholar]
- 71. Sommer S., Knezevic J., Bailone A., Devoret R. 1993. Induction of only one SOS operon, umuDC, is required for SOS mutagenesis in Escherichia coli. Mol. Gen. Genet. 239:137–144 [DOI] [PubMed] [Google Scholar]
- 72. Strauss B. S. 1991. The ‘A rule’ of mutagen specificity: a consequence of DNA polymerase bypass of non-instructional lesions? Bioessays 13:79–84 [DOI] [PubMed] [Google Scholar]
- 73. Sutton M. D., Duzen J. M., Maul R. W. 2005. Mutant forms of the Escherichia coli beta sliding clamp that distinguish between its roles in replication and DNA polymerase V-dependent translesion DNA synthesis. Mol. Microbiol. 55:1751–1766 [DOI] [PubMed] [Google Scholar]
- 74. Sutton M. D., Kim M., Walker G. C. 2001. Genetic and biochemical characterization of a novel umuD mutation: insights into a mechanism for UmuD self-cleavage. J. Bacteriol. 183:347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Szekeres E. S., Jr., Woodgate R., Lawrence C. W. 1996. Substitution of mucAB or rumAB for umuDC alters the relative frequencies of the two classes of mutations induced by a site-specific T-T cyclobutane dimer and the efficiency of translesion DNA synthesis. J. Bacteriol. 178:2559–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Szpilewska H., Bertrand P., Bailone A., Dutreix M. 1995. In vitro inhibition of RecA-mediated homologous pairing by UmuD′C proteins. Biochimie 77:848–853 [DOI] [PubMed] [Google Scholar]
- 77. Tang M., et al. 2000. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature 404:1014–1018 [DOI] [PubMed] [Google Scholar]
- 78. Tang M., et al. 1999. UmuD′(2)C is an error-prone DNA polymerase, Escherichia coli pol V. Proc. Natl. Acad. Sci. U. S. A. 96:8919–8924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Vandewiele D., Borden A., O'Grady P. I., Woodgate R., Lawrence C. W. 1998. Efficient translesion replication in the absence of Escherichia coli Umu proteins and 3′-5′ exonuclease proofreading function. Proc. Natl. Acad. Sci. U. S. A. 95:15519–15524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wagner J., Fujii S., Gruz P., Nohmi T., Fuchs R. P. 2000. The beta clamp targets DNA polymerase IV to DNA and strongly increases its processivity. EMBO Rep. 1:484–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Woodgate R., Rajagopalan M., Lu C., Echols H. 1989. UmuC mutagenesis protein of Escherichia coli: purification and interaction with UmuD and UmuD′. Proc. Natl. Acad. Sci. U. S. A. 86:7301–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Woodgate R., Sedgwick S. G. 1992. Mutagenesis induced by bacterial UmuDC proteins and their plasmid homologues. Mol. Microbiol. 6:2213–2218 [DOI] [PubMed] [Google Scholar]
- 83. Yin J., Seo K. Y., Loechler E. L. 2004. A role for DNA polymerase V in G → T mutations from the major benzo[a]pyrene N2-dG adduct when studied in a 5′-TGT sequence in E. coli. DNA Repair (Amst.) 3:323–334 [DOI] [PubMed] [Google Scholar]