Abstract
Mycobacterium tuberculosis, the etiological agent of tuberculosis, remains a significant cause of morbidity and mortality throughout the world despite a vaccine and cost-effective antibiotics. The success of this organism can be attributed, in part, to its ability to adapt to potentially harmful stress within the host and establish, maintain, and reactivate from long-term persistent infection within granulomatous structures. The DosRS-DosT/DevRS-Rv2027c, and MprAB two-component signal transduction systems have previously been implicated in aspects of persistent infection by M. tuberculosis and are known to be responsive to conditions likely to be found within the granuloma. Here, we describe initial characterization of a locus (Rv0081-Rv0088) encoding components of a predicted formate hydrogenylase enzyme complex that is directly regulated by DosR/DevR and MprA, and the product of the first gene in this operon, Rv0081. In particular, we demonstrate that Rv0081 negatively regulates its own expression and that of downstream genes by binding an inverted repeat element in its upstream region. In contrast, DosR/DevR and MprA positively regulate Rv0081 expression by binding to recognition sequences that either partially or completely overlap that recognized by Rv0081, respectively. Expression of Rv0081 initiates from two promoter elements; one promoter located downstream of the DosR/DevR binding site but overlapping the sequence recognized by both Rv0081 and MprA and another promoter downstream of the DosR/DevR, Rv0081, and MprA binding sites. Interestingly, Rv0081 represses Rv0081 and downstream determinants following activation of DosRS-DosT/DevRS-Rv2027c by nitric oxide, suggesting that expression of this locus is complex and subject to multiple levels of regulation. Based on this and other published information, a model is proposed detailing Rv0081-Rv0088 expression by these transcription factors within particular growth environments.
INTRODUCTION
Tuberculosis (TB) continues to be a major health burden worldwide, even though this disease has been recognized since antiquity. The etiological agent, Mycobacterium tuberculosis, is currently estimated to latently infect about one-third of the world's population. In 2009, 1.7 million individuals succumbed to TB, and there were 9 million new cases of active disease reported, many resulting from the reactivation of M. tuberculosis from persistent infection (2). While current antitubercular drugs are largely effective at killing actively replicating bacilli, these antibiotics are much less effective against persistent M. tuberculosis, necessitating the use of prolonged antibiotic treatment regimens (3). The emergence of multidrug-resistant (MDR) or extensively drug-resistant (XDR) strains of M. tuberculosis has further compromised established treatment strategies. Understanding the mechanisms by which M. tuberculosis establishes, maintains, or reactivates from latency is tantamount for development of new therapeutics or vaccine candidates to reduce the global TB burden and treat infections with MDR and XDR strains.
M. tuberculosis infects and survives within alveolar macrophages following respiratory infection of susceptible hosts. Most often, infection triggers host immune responses that culminate in the production of granulomas, which act to limit M. tuberculosis replication and subsequent dissemination (47). It is thought that persistence of M. tuberculosis within the host is initiated following recognition of certain environmental stimuli within the granuloma, including nutrient limitation, oxygen limitation, reactive oxygen and nitrogen intermediates, acidic pH, and cell wall/cell membrane-perturbing compounds. To respond to and combat such stress, M. tuberculosis encodes a variety of regulatory factors, including extracytoplasmic function (ECF) sigma factors, two-component signal transduction systems (TCSS), serine/threonine protein kinases (STPK), and other transcription factors (18).
Several TCSS have been implicated in aspects of persistent infection by M. tuberculosis, including MprAB and DosRS-DosT (also termed DevRS-Rv2027c). MprAB has been shown to control expression of a regulon in M. tuberculosis comprising ∼200 genes (28, 43). Activation by MprAB occurs in response to detergents, alkaline pH, nutrient limitation, and other stimuli (6, 28, 34, 43). mprAB is also activated in vivo during both acute and persistent infection states (60), within an artificial granuloma model system (34), and is required for virulence of M. tuberculosis in the low-dose murine model of TB (69). Recently, MprAB was shown to directly participate in a positive feedback loop with ECF sigma factor SigE, indirectly influencing activation of other stress-responsive networks, including those controlled by STPK PknB, transcription factor ClgR, and stringent starvation factor RelA (5, 22, 23, 28, 40, 41, 59, 68).
Similarly, DosRS-DosT/DevRS-Rv2027c upregulates itself and a well-defined regulon of 48 genes in M. tuberculosis following exposure to hypoxia, nitric oxide (NO), carbon monoxide (CO), and the cytochrome c reductant ascorbate (9, 30, 35, 45, 61, 66). dosRS (devRS) is induced during growth of M. tuberculosis in monocytes/macrophages (20, 27, 56), in the artificial granuloma model system (34), and in mice (56, 60, 66). This system is also required for virulence of M. tuberculosis in several animal model systems, including mice, guinea pigs, and rabbits (19, 39), although this remains controversial (44, 50). Duplication of the dosRS (devRS) region and point mutations in dosT (Rv2027c) in M. tuberculosis W/Beijing family members have been noted (21, 24); however, the contribution of these changes to the enhanced virulence attributed to these strains has not yet been established. Importantly, DosR/DevR is essential for survival of M. tuberculosis during anaerobic dormancy and is required for metabolic processes that occur during entrance into and throughout the dormant state in vitro (36).
Comparisons between the MprAB and DosRS-DosT/DevRS-Rv2027c regulons have indicated that a subset of determinants may be coregulated by these transcription factors (28, 43, 45). One of these determinants, Rv0081, is a member of the ArsR/SmtB family of metal-dependent transcriptional regulators (11). Rv0081 is the first gene in a larger locus comprising seven additional, closely spaced or overlapping determinants exhibiting homology to components of a formate hydrogenlyase (FHL) complex. In Escherichia coli, these genes are involved in the conversion of formate to CO2 and H2 under conditions of anaerobic respiration in the absence of an external terminal electron acceptor (37). Currently, nothing is known about the function of Rv0081 in the context of M. tuberculosis gene regulation, physiology, or pathogenesis. Incubation of this protein with whole blood isolates from latently infected individuals from certain countries of Africa stimulates significant levels of IFN-γ, suggesting that this protein may be antigenic (8). Here, an initial characterization of Rv0081 and its promoter region was carried out. We show that Rv0081 is a transcriptional regulator of itself and other genes comprising the Rv0081-Rv0088 locus and that it directly binds an inverted repeat element in its own promoter region. Importantly, we also demonstrate that Rv0081 is directly regulated by response regulators MprA and DosR/DevR, that the binding sites for these transcription factors completely or partially overlap that recognized by Rv0081, respectively, and that Rv0081 and downstream genes are cotranscribed in an operon and are upregulated in response to NO. Thus, Rv0081-Rv0088 is regulated by multiple transcription factors in M. tuberculosis and may participate in activities controlled by the MprAB and DosRS-DosT/DevRS-Rv2027c TCSS.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Strains and plasmids used in this study are described in Table 1. Escherichia coli Top10 (Invitrogen, Carlsbad, CA) and DH5α were used for cloning. E. coli BL21(DE3)/pLysS (Novagen, La Jolla, CA) was used to express recombinant proteins. E. coli strains were grown with aeration at 37°C in Luria-Bertani (LB) broth or LB agar (Thermo Fisher Scientific, Waltham, MA). When required, medium was supplemented with 25 μg/ml chloramphenicol (Sigma, St. Louis, MO), 150 μg/ml hygromycin B (AG Scientific, San Diego, CA), 100 μg/ml ampicillin (Thermo Fisher Scientific, Waltham, MA), and/or 50 μg/ml kanamycin sulfate (Thermo Fisher Scientific, Waltham, MA). Mycobacterium strains used in this study were all derivatives of Mycobacterium tuberculosis H37Rv (ATCC 27294) or Mycobacterium smegmatis mc2155 (ATCC 700084) (American Type Culture Collection, Manassas, VA). Mycobacteria were grown at 37°C in 7H9 broth or 7H10 agar medium (Difco, Franklin Lakes, NJ) supplemented with 0.5% glycerol, 10% albumin-dextrose-catalase or oleic acid-albumin-dextrose-catalase (Difco, Franklin Lakes, NJ), and 0.05% Tween 80. For β-galactosidase or quantitative reverse transcription-PCR (qRT-PCR) expression assays, mycobacteria were grown in Sauton's medium (1). When required, mycobacteria medium was supplemented with 25 μg/ml kanamycin sulfate and 50 μg/ml hygromycin B.
Table 1.
Strains, plasmids, and bacteriophages used in this study
| Strain, plasmid, or bacteriophage | Genotype or description | Application | Reference or source |
|---|---|---|---|
| Strains | |||
| E. coli DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | Lab collection | |
| E. coli TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu)7697 galU galK rpsL (StrR) endA1 nupG | Invitrogen | |
| E. coli HB101 | supE44 hsdS20(rB− mB−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 leuB6 thi-1 | Lab collection | |
| E. coli BL21(DE3)/pLysS | F−ompT hsdSB(rB− mB−) gal dcm (DE3)/pLysE; Camr | Novagen | |
| M. smegmatis mc2155 | Laboratory strain | ATCC 700084 | |
| M. tuberculosis H37Rv | Laboratory strain | ATCC 27294 | |
| TB193 | H37Rv ΔRv0081 | This study | |
| TB222 | H37Rv ΔRv0081 carrying pTZ1165 | This study | |
| TB228 | H37Rv ΔRv0081 carrying pTZ1111 and pTZ1161 | This study | |
| TB229 | H37Rv ΔRv0081 carrying pTZ1111 and pTZ1165 | This study | |
| Plasmids | |||
| pCR2.1-TOPO | 3.9-kb plasmid for cloning PCR products; Ampr Kanr | PCR cloning vector | Invitrogen |
| pET15b | Inducible His-tagged protein expression plasmid; Ampr | Overexpression | Lab collection |
| pJEM15 | E. coli-Mycobacterium shuttle vector for transcriptional fusion with lacZ; ampr | Reporter assay | 62 |
| pMV361 | Integrative protein expression vector in mycobacterial cell under hsp60 promoter; Kanr | Protein expression | 58 |
| pTZ229 | pET15b containing mprA coding sequence; Ampr | Overexpression | 70 |
| pTZ763 | pYUB854 containing dosR upstream and downstream regions; Hygr | dosR deletion | This study |
| pTZ765 | pET15b containing dosR coding sequence; Ampr | Overexpression | This study |
| pTZ1064 | pYUB854 containing mprA upstream and mprB downstream regions; Hygr | mprAB deletion | This study |
| pTZ1070 | pET15b containing Rv0081 coding sequence; Ampr | Overexpression | This study |
| pTZ1110 | pYUB854 containing Rv0081 upstream and downstream regions; Hygr | Rv0081 deletion | This study |
| pTZ1111 | pJEM15 containing Rv0081 upstream sequence; Kanr | Reporter assay | This study |
| pTZ1161 | pMV361 derivative with Kanr replaced by Hygr; Hygr | Protein expression | This study |
| pTZ1165 | pTZ1161 containing Rv0081 coding sequence; Hygr | Protein expression | This study |
| pYUB854 | 3.9-kb cosmid for generating gene deletions in Mycobacterium; carries Hygr cassette flanked by multiple cloning sites and λpac site; Hygr | Mycobacterium gene deletion vector | 4 |
| pYUB870 | 8.9-kb plasmid for unmarking mutations in Mycobacterium; carries tnpR and sacB cassettes; Kanr | Mycobacterium gene deletion unmarking vector | 4 |
| Mycobacteriophages | |||
| phAE87 | Derivative of conditionally replicating mycobacteriophage PH101(Ts) | 4 | |
| phTZ763 | phAE87 derivative containing pTZ763 | dosR deletion | This study |
| phTZ1064 | phAE87 derivative containing pTZ1064 | mprAB deletion | This study |
| phTZ1110 | phAE87 derivative containing pTZ1110 | Rv0081 deletion | This study |
DNA manipulations.
Restriction enzyme digestion, cloning, subcloning, and DNA electrophoresis were performed according to standard techniques (51). Oligonucleotides and primers were synthesized by Eurofins MWG Operon (Huntsville, AL) and are listed in Table S1 of the supplemental material. PCR was performed using High Fidelity Platinum PCR supermix or Taq polymerase (Invitrogen, Carlsbad, CA). All amplified products were cloned into pCR2.1-TOPO (Invitrogen, Carlsbad, CA) and sequenced to confirm the absence of mutations. Cloned inserts were then removed by restriction enzyme digestion, resolved on a 1% agarose gel, recovered by gel purification, and subcloned into the appropriate vector. Ligations were performed using the Quick Ligation kit (New England BioLabs, Beverly, MA) or T4 DNA ligase (Invitrogen, Carlsbad, CA). When necessary, plasmid DNA was treated with Antarctic phosphatase (New England BioLabs, Beverly, MA) to prevent religation of vector ends. Electroporation of plasmid DNA into mycobacteria was conducted as previously described (33). Plasmid DNA was prepared using the QIAprep Spin miniprep kit (Qiagen, Venlo, Netherlands) as recommended by the manufacturer. DNA fragments were purified using either the QIAquick gel extraction kit or QIAquick PCR purification kit (Qiagen, Venlo, Netherlands). DNA sequencing was performed with an ABI Prism BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems, Carlsbad, CA) using an automated long capillary method (ABI PRISM 3100 genetic analyzer; Applied Biosystems, Carlsbad, CA).
Production and purification of recombinant proteins in E. coli.
Overproduction and purification of 6×His-MprA has been previously described (70). A similar strategy was used to generate 6×His-DosR/DevR and 6×His-Rv0081. Briefly, coding sequences for dosR (devR; Rv3133c) and Rv0081 were generated by PCR. Cloned inserts were then subcloned into a pET15b derivative, allowing N-terminal fusion of dosR (devR) (pTZ765) and Rv0081 (pTZ1070) to a 6×His epitope tag. E. coli BL21(DE3)/pLysS strains containing overexpression constructs were grown overnight on selective LB agar medium containing kanamycin and chloramphenicol, suspended in LB selective broth, and grown to mid-exponential phase. Protein overproduction was induced by the addition of 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside; Invitrogen, Carlsbad, CA) for 3 h at 30°C. Induced cells were suspended in lysis buffer (20 mM Tris [pH 7.9], 500 mM NaCl, 5 mM imidazol, and 6 μg of DNase/ml), passed 3 times through a French press, and centrifuged at 25,000 × g for 30 min. Cellular supernatants were passed over a nickel nitrilotriacetic acid-agarose column (Qiagen, Valencia, CA), and collected fractions were pooled. Purified protein fractions were dialyzed overnight against dialysis buffer (20 mM Tris [pH 7.9], 150 mM NaCl, 20% glycerol). Purified proteins were stored at −80°C.
FPLC of His-Rv0081.
His-Rv0081 protein was purified and dialyzed as described above. Rv0081 was separated on an AKTAexplorer 10 S fast protein liquid chromatograph (FPLC; GE Healthcare Life Science, Piscataway, NJ) using a Superose 6 HR10/30 column with 20 mM Tris [pH 7.9], 150 mM NaCl, and 20% glycerol as running buffer. The molecular mass of Rv0081 was calculated from a standard curve generated with protein standards, including cytochrome c (12.33 kDa), ovalbumin (44.3 kDa), and ferritin (450 kDa). FPLC was performed at a flow rate of 0.2 ml per minute for 250 min, and 0.2-ml fractions were collected and subjected to SDS-PAGE to confirm the oligomeric state of Rv0081.
EMSAs.
For electrophoretic mobility shift assays (EMSAs), DNA from regions upstream of Rv0079, Rv0081, Rv0082, and hycD in M. tuberculosis H37Rv were amplified by PCR and labeled with [γ-32P]ATP (MP Biomedical, Solon, OH) using T4 polynucleotide kinase (Invitrogen, Carlsbad, CA). Radiolabeled probes were incubated with recombinant protein in a reaction buffer [20 mM KCl, 5% glycerol, 10 ng of salmon sperm DNA per ml, 25 mM Tris-HCl (pH 8.0), 6 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol, and 4.0 μg of poly(dI-dC)] for 30 min at room temperature. For some experiments, excess cold DNA from the same region or from a different DNA region was included in reaction mixtures to demonstrate binding specificity. Following incubation, binding reaction mixtures were loaded onto 5% nondenaturing polyacrylamide gels and electrophoresed at 130 V for about 3 h at 4°C. Gels were dried and exposed to X-ray films overnight.
β-Galactosidase assays.
The Rv0081-lacZ reporter was generated by amplifying the region upstream of Rv0081 (from −229 to +29 relative to the Rv0081 translational start site) from M. tuberculosis H37Rv and subcloning into pJEM15 (62), resulting in vector pTZ1111. This plasmid was subsequently introduced into Mycobacterium strains expressing Rv0081 from an integrative vector under the control of the heterologous hsp60 promoter (pTZ1165). This plasmid was constructed by amplifying the Rv0081 coding sequence from H37Rv and subcloning downstream of the hsp60 promoter in pTZ1161, a hygromycin-resistant derivative of pMV361 (58). Resulting strains were grown in Sauton's medium to mid-log phase (optical density at 600 nm [OD600], 0.5), washed three times in 1× phosphate-buffered saline (PBS), suspended in lysis buffer (300 mM Tris [pH 6.8], 100 mM NaCl, and 5 mM EDTA), and mechanically lysed using a bead beater (BioSpec Products, Bartlesville, OK). Cell debris was separated from the whole-cell lysate by centrifugation. For lysates prepared from M. tuberculosis, supernatants were clarified by passage through 0.22-μm low-protein-binding syringe filters (Corning, Lowell, MA). β-Galactosidase assays and quantification were carried out as previously described (42), with the modification that enzymatic activity was normalized to equivalent amounts of total protein.
Extraction of DNA and RNA.
Mycobacterial genomic DNA was isolated as described previously (33). For isolation of RNA, M. tuberculosis was grown in 7H9 broth to mid-log phase (OD600, 0.5). For some assays, bacterial cultures were exposed to 100 μM diethylenetriamine (DETA)/NO (Sigma-Aldrich, St. Louis, MO) for 40 min to induce activation of DosRST/DevRS and its regulon (66). RNA was fixed by treating cultures with RNAlater (Ambion, Austin, TX) for at least 40 min at room temperature prior to processing. Bacteria were collected by centrifugation, washed twice with 1× PBS, and then mechanically disrupted in the presence of RNA-Bee (Tel-Test, Friendswood, TX) by bead beating. RNA was then extracted with chloroform, precipitated with isopropanol, and stored at −80°C until use. Resulting RNA was resuspended in diethylpyrocarbonate (DEPC)-treated water, treated with Turbo DNase (Ambion, Austin, TX) to remove contaminating genomic DNA, and purified using an RNeasy miniprep column (Qiagen, Valencia, CA).
RT-PCR and 5′-RACE.
For RT-PCR, cDNA from M. tuberculosis strains was prepared by incubating 500 ng total RNA with random M. tuberculosis decamers (25) in a reverse transcription reaction mixture containing SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA). To test for the presence of DNA contamination, identical reactions lacking SuperScript III were also run. For cotranscription studies, reaction mixtures included primer sets that spanned intergenic regions. Amplification conditions consisted of 30 rounds of denaturation at 95°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for between 30 s and 2 min. Reactions were run on an MJ Research PTC-200 thermocycler (Bio-Rad, Hercules, CA). For qRT-PCR, reaction mixtures included primer sets that amplified 100- to 150-bp fragments from specific coding sequences. Amplification conditions consisted of 40 rounds of denaturation at 95°C for 30 s and annealing/extension at 60°C for 15 s using iQ SYBR green Supermix (Bio-Rad, Hercules, CA). Reactions were run on an iCycler iQ real-time PCR detection system (Bio-Rad, Hercules, CA). Single PCR products were confirmed by performing a postamplification melt curve analysis for each reaction. Transcript levels of target genes were normalized to the ribosomal gene rrs. For 5′ random amplification of cDNA ends (RACE), the FirstChoice RLM-RACE kit (Ambion, Austin, TX) was used essentially as recommended by the manufacturer. Briefly, 3 to 5 μg of total RNA was ligated to 300 ng of 5′-RACE adaptor by using 5 units of T4 RNA ligase (Ambion, Austin, TX) for 1 h at 37°C. RNA was then reverse transcribed with TB-specific decamers and Moloney murine leukemia virus (M-MLV) reverse transcriptase (Ambion, Austin, TX) for 1 h at 50°C. PCR was then performed using a primer nested within the gene of interest and a primer specific to the 5′ RACE adaptor (5′ RACE outer primer). Resulting PCR products were cloned into pCR2.1-TOPO and sequenced using the M13rev primer. When possible, between 10 and 20 clones were sequenced for each primer set.
Construction of unmarked deletion mutations in M. tuberculosis.
mprAB, dosR (devR), and Rv0081 were deleted from M. tuberculosis H37Rv by specialized phage transduction as described previously (4). Briefly, 1.0-kb regions upstream and downstream of target genes were PCR amplified from H37Rv genomic DNA and subcloned into pYUB854 flanking the Hygr cassette. For mprAB, the generated deletion removed 1,890 bp, extending from +101 downstream of the mprA translational start site through +1301 downstream of the mprB translational start site. The generated deletion in dosR (devR) removed the entire coding sequence for this gene. This deletion also removed the dosS (devS) translational start site, which overlaps with the dosR (devR) coding sequence by 4 nucleotides. For Rv0081, the generated deletion removed 303 bp extending from +28 to +330 relative to the Rv0081 translational start site. Following packaging and transduction into M. tuberculosis, hygromycin-resistant clones were screened for loss of mprAB, dosR (devR), or Rv0081 by PCR. Resulting ΔmprAB::Hygr, ΔdosR::Hygr, and ΔRv0081::Hygr strains were electroporated with pYUB870 to unmark the mutation. Following counterselection on medium containing 10% sucrose, resulting sucrose-resistant, kanamycin-sensitive, and hygromycin-sensitive colonies were screened for unmarking of the mprAB, dosR, and Rv0081 mutations by PCR. The Rv0081 mutation was complemented by cloning the Rv0081 coding sequence behind a heterologous hsp60 promoter carried on a derivative of pMV361 (58).
MEME analysis.
Predicted binding sites for Rv0081, MprA, and DosR were determined using MEME (Multiple Em for Motif Elicitation) (http://meme.sdsc.edu/meme4_6_1/cgi-bin/meme.cgi).
Statistical analyses.
All statistical analyses were conducted using Student's t test or a one-way analysis of variance. Values were determined to be statistically significant at a P level of <0.05.
RESULTS
Rv0081 autogenously regulates its own expression through direct binding of the Rv0081 upstream region.
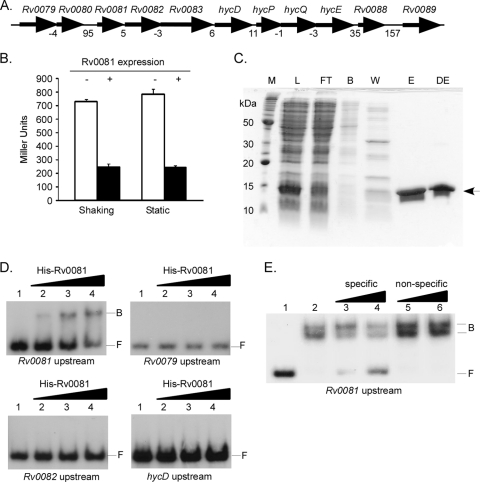
In M. tuberculosis, Rv0081 is the first gene in an eight-gene locus (Rv0081-Rv0088) predicted to encode components of a formate hydrogenlyase enzyme complex (Fig. 1A). Bioinformatic analyses indicate that Rv0081 is a metal-dependent transcriptional regulator of the Smt/ArsR family (11). To determine whether Rv0081 regulates its own expression, a reporter plasmid carrying the Rv0081 promoter fused to lacZ was constructed (pTZ1111) and introduced into M. smegmatis strains expressing M. tuberculosis Rv0081 from the heterologous hsp60 promoter carried on an integrated vector. Strains carrying pTZ1111 and expressing hsp60-Rv0081 produced ∼3-fold less β-galactosidase when cultures were shaking or incubated statically than did strains carrying pTZ1111 and the integration vector-only control (Fig. 1B). To determine whether regulation of the Rv0081 upstream region by Rv0081 was direct or indirect, His-Rv0081 was overproduced in E. coli, purified by column chromatography (Fig. 1C), and utilized in EMSAs. Binding by His-Rv0081 was observed following incubation with DNA from the Rv0081 upstream region (Fig. 1D). In contrast, His-Rv0081 binding was not observed following incubation with regions upstream of Rv0079, Rv0082, or hycD (Fig. 1D). Binding by His-Rv0081 to its own upstream region was concentration dependent (Fig. 1D) and sequence specific (Fig. 1E), as binding to radiolabeled probe from the Rv0081 upstream region was reduced in the presence of excess unlabeled DNA from the same region (Fig. 1E, lanes 3 and 4) but not with unlabeled DNA from the hycD upstream region (Fig. 1E, lanes 5 and 6). Thus, Rv0081 is a negative regulator and directly binds its own upstream region to repress transcription.
Fig. 1.
Autogenous regulation of the Rv0081 promoter by Rv0081. (A) Genomic organization of the Rv0079-Rv0089 locus. Numbers below genes indicate the intergenic distance. (B) Rv0081 promoter-lacZ reporter assays were conducted in wild-type M. smegmatis carrying vector only (white bars) or vector expressing hsp60-Rv0081 (black bars). Cultures were grown with shaking or were grown statically, and β-galactosidase activity was quantified. Data are presented in Miller units. (C) His-Rv0081 was overproduced and purified using nickel nitrilotriacetic acid-agarose column chromatography. Enriched His-Rv0081 is depicted with an arrow. M, marker; L, lysate; FT, flowthrough; B, binding buffer; W, wash buffer; E, eluate; DE, dialyzed eluate. (D) Binding by His-Rv0081 was assessed using electrophoretic mobility shift assays to regions upstream of Rv0081, Rv0079, Rv0082, or hycD. Reaction mixtures contained 1.0 ng radiolabeled probe alone (lane 1) or probe with 10 ng (lane 2), 40 ng (lane 3), or 80 ng (lane 4) of His-Rv0081. (E) Binding by Rv0081 to its own upstream region is sequence specific. Radiolabeled probe (1.0 ng) from the Rv0081 upstream region was incubated alone (lane 1) or in the presence of 100 ng of His-Rv0081 (lanes 2 to 6). Reaction mixtures also contained 150-fold and 300-fold excess cold DNA from the Rv0081 upstream region (lanes 3 and 4) or nonspecific cold DNA from the hycD upstream region (lanes 5 and 6). B, bound; F, free.
Rv0081 binds an inverted repeat element in the Rv0081 upstream region.
SmtB/ArsR family members often bind inverted repeat (IR) elements comprised of two 12-bp IR subunits separated by a 2-bp spacer (10). The Rv0081 upstream region contains a partially conserved 12-2-12 IR element (Fig. 2A) which has homology to IR elements recognized by other SmtB/ArsR family members (see Fig. S1 in the supplemental material). To determine whether Rv0081 recognized this IR element, EMSAs were repeated using DNA probes that either contained or lacked IR subunits (Fig. 2A). Probes carrying both IR subunits were bound by His-Rv0081 in a concentration-dependent manner (Fig. 2B, primer pairs1/2 and 1/3). Interestingly, two protein-DNA complexes were observed with each probe, suggesting that Rv0081 may bind at multiple locations on each DNA fragment or bind one site and form higher-order complexes. In contrast, the DNA probe lacking IR subunit 2 and the 2-bp spacer was not bound by His-Rv0081 (Fig. 2B, primers 1 and 4). To confirm that Rv0081 bound specifically to each IR subunit, EMSAs were repeated using three 59-bp DNA probes that were either wild type for both IR subunits (probe 1), mutant for IR subunit 1 but wild type for IR subunit 2 (probe 2), or wild type for IR subunit 1 and mutant for IR subunit 2 (probe 3) (Fig. 2C). As observed previously, incubation of His-Rv0081 with probe containing wild-type IR subunits resulted in formation of two protein-DNA complexes (Fig. 2D, probe 1). In contrast, a single protein-DNA complex (corresponding to the lower molecular mass form seen with probe 1) was observed following incubation of His-Rv0081 with the probe containing a mutant IR subunit 1 but wild-type IR subunit 2 (Fig. 2D, probe 2). His-Rv0081 was unable to bind the probe with a wild-type IR subunit 1 but mutant IR subunit 2 (Fig. 2D, probe 3). These results indicate that Rv0081 binds preferentially to IR subunit 2, which may then recruit a second Rv0081 molecule to IR subunit 1. Finally, to determine whether Rv0081 was monomeric or formed higher-order complexes in solution, purified His-Rv0081 was subjected to FPLC. Purified His-Rv0081 eluted as a single protein peak (see Fig. S2 in the supplemental material). Compared to protein standards, including cytochrome c, ovalbumin, and ferritin, the estimated molecular mass of His-Rv0081 was determined to be 14.08 kDa. This is in good agreement with the predicted molecular mass for monomeric His-Rv0081 (13.15 kDa). Thus, Rv0081 is monomeric in solution and binds DNA cooperatively.
Fig. 2.
Rv0081 binds a loosely conserved 12-bp IR element located in its own upstream region. (A) The Rv0080-Rv0081 intergenic region. Bold letters indicate the end of Rv0080 or beginning of the Rv0081 coding sequence. Arrows indicate locations of oligonucleotides used to generate DNA probes. Shaded regions show the location of the 12-bp inverted repeat subunits. (B) Rv0080-Rv0081 intergenic probes were incubated with His-Rv0081 and analyzed in EMSAs. Generated probes carried the entire Rv0080-Rv0081 intergenic region (primers 1 and 2), a region extending from the end of Rv0080 into the Rv0080-Rv0081 intergenic region, including both IR subunits (primers 1 and 3), or a region extending from the end of Rv0080 into the Rv0080-Rv0081 intergenic region, including the first IR subunit (primers 1 and 4). Two nanograms of each radiolabeled probe was incubated alone (lane 1) or with 100 ng (lane 2) or 800 ng (lane 3) of His-Rv0081. B, bound; F, free. (C) Complete nucleotide sequence of the 59-bp DNA probes from the Rv0081 upstream region containing wild-type or mutant IR subunit sequences. Sequences were wild type for both IR subunits (probe 1), mutant for IR subunit 1 but wild type for IR subunit 2 (probe 2), or wild type for IR subunit 1 but mutant for IR subunit 2 (probe 3). (D) Wild-type or mutant IR probes were incubated with His-Rv0081 and analyzed in EMSAs. Two nanograms of each DNA probe was incubated alone (lane 1) or with 100 ng (lane 2), 400 ng (lane 3), or 1,200 ng of His-Rv0081.
Response regulators MprA and DosR/DevR also bind the Rv0081 upstream region and regulate its expression in M. tuberculosis.
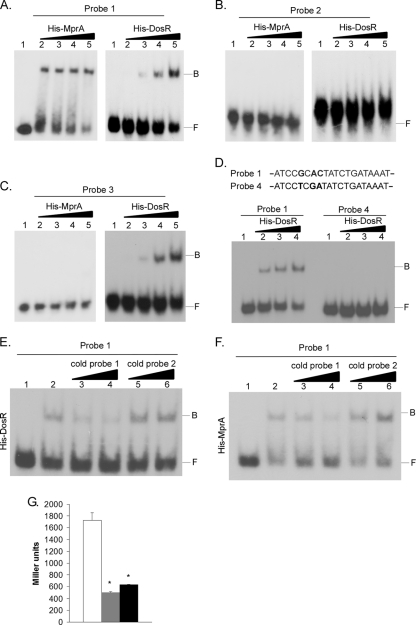
The Rv0081 upstream region contains sequences that resemble known bindings sites for response regulator MprA (see Fig. S3 in the supplemental material) (28, 29, 43). DosR/DevR may also directly bind to the Rv0081 upstream region, as expression of this gene is decreased in a dosR mutant of M. tuberculosis (45), and the upstream region of Rv0081 contains sequences that are loosely conserved with known DosR/DevR recognition motifs (see Fig. S4 in the supplemental material) (15–17, 26, 45). To determine whether MprA and/or DosR/DevR bound sequences in this region (Fig. 2A), DNA probes generated to the Rv0081 upstream region were utilized in EMSAs with His-MprA (70) or His-DosR/DevR. Both MprA and DosR/DevR bound a 258-bp probe that spanned the intergenic region between Rv0080 and Rv0081 (data not shown). To determine whether the MprA and/or DosR/DevR binding site(s) overlapped that of Rv0081, EMSAs were repeated using the three 59-bp probes containing wild-type or mutant IR subunits (Fig. 2C). Both His-MprA and His-DosR/DevR bound to probe 1 in a concentration-dependent manner (Fig. 3A). In contrast, His-MprA and His-DosR/DevR failed to bind probe 2, even at the largest amounts of protein added (Fig. 3B). Finally, binding by His-DosR/DevR but not His-MprA was observed with probe 3 (Fig. 3C). Thus, MprA and DosR/DevR bind to the Rv0081 upstream region, and this binding partially or completely overlaps regions recognized by Rv0081. DosR/DevR has previously been shown to bind DNA nonspecifically in EMSAs (15). Therefore, specificity of DosR/DevR binding to this region was investigated by EMSA using a variant of the wild-type 59-bp DNA probe, in which three highly conserved residues within the 20-bp predicted consensus motif were mutated (probe 4) (see Fig. S4; position 5G→T, 7A→G, 8C→A). While His-DosR/DevR bound to wild-type probe (probe 1), as observed previously, His-DosR/DevR failed to bind probe 4 (Fig. 3D), demonstrating that the three mutated nucleotides are specifically required for His-DosR/DevR interaction. Specificity of DosR-DevR binding was also confirmed in competition EMSAs. Addition of excess cold DNA from probe 1 but not probe 2 effectively reduced binding by His-DosR/DevR to radiolabeled DNA from probe 1 (Fig. 3E). Competition assays were also performed with His-MprA to demonstrate specificity of this binding interaction. Addition of excess cold DNA from probe 1 but not probe 2 also reduced binding by His-MprA to radiolabeled DNA from probe 1 (Fig. 3F). When taken together, these results indicate that binding by MprA and DosR/DevR to the Rv0081 upstream region is both concentration dependent and sequence specific. Finally, to determine whether MprA and DosR/DevR positively or negatively regulates transcription of Rv0081, Rv0081 promoter-lacZ reporter assays were performed in wild-type, ΔmprAB, or ΔdosR mutant strains of M. tuberculosis. β-Galactosidase production in both the ΔmprAB and ΔdosR mutants was ∼3- to 4-fold lower than that observed in wild-type H37Rv (Fig. 3G). Thus, MprA and DosR/DevR directly bind to the Rv0081 upstream region and positively regulate its expression.
Fig. 3.
MprA and DosR bind to the Rv0081 upstream region and are positive regulators of Rv0081 expression. EMSAs were conducted using the 59-bp DNA probes that were wild type for both IR subunits (A), mutant for IR subunit 1 but wild type for IR subunit 2 (B), or wild type for IR subunit 1 but mutant for IR subunit 2 (C). Seven nanograms of each DNA probe was incubated alone (lane 1) or with increasing amounts of His-MprA or His-DosR (lanes 2 to 5). Binding reaction mixtures contained 200 ng (lane 2), 400 ng (lane 3), 800 ng (lane 4), or 1,600 ng (lane 5) of unphosphorylated His-MprA, or 600 ng (lane 2), 1,200 ng (lane 3), 2,400 ng (lane 4), or 4,800 ng (lane 5) of unphosphorylated His-DosR. B, bound; F, free. (D) Specificity of DosR binding was evaluated in an EMSA using a variant of probe 1 in which three conserved nucleotides within the 20-bp consensus were mutated (probe 4). Three nanograms of probe 1 or probe 4 was incubated alone (lane 1) or with 300 ng (lane 2), 600 ng (lane 3), or 1,000 ng (lane 4) of unphosphorylated His-DosR. Bold residues indicate mutated nucleotides. (E) DosR binding competition assays. Radiolabeled probe 1 (3.0 ng) was incubated alone (lane 1) or with 600 ng of unphosphorylated His-DosR (lanes 2 to 6). Reaction mixtures also contained 200-fold (lane 3) or 400-fold (lane 4) excess cold DNA from probe 1 or 200-fold (lane 5) or 400-fold (lane 6) nonspecific cold DNA from probe 2 (lanes 5 and 6). (F) MprA binding competition assays. Radiolabeled probe 1 (3.0 ng) was incubated alone (lane 1) or with 200 ng of unphosphorylated His-MprA (lanes 2 to 6). Reaction mixtures also contained 200-fold (lane 3) or 400-fold (lane 4) excess cold DNA from probe 1 or 200-fold (lane 5) or 400-fold (lane 6) nonspecific cold DNA from probe 2 (lanes 5 and 6). (G) Rv0081 promoter-lacZ reporter assays were conducted in wild-type M. tuberculosis (white bars), M. tuberculosis ΔdosR (gray bars), or M. tuberculosis ΔmprAB (black bars). Cultures were grown with shaking in Sauton's medium, and β-galactosidase activity was quantified. Data are presented in Miller units. Asterisks denote statistical significance (P < 0.05).
Identification of TSPs within the Rv0081 upstream region.
Gene expression studies indicated that Rv0081 expression was negatively regulated by Rv0081 and positively regulated by both DosR/DevR and MprA. To delineate promoters within the Rv0081 upstream region, transcriptional start points (TSPs) were identified by 5′-RACE. RNA was derived from cultures of wild-type, ΔdosR mutant (ΔdevR mutant), or ΔmprAB mutant strains of M. tuberculosis that were either grown with shaking or were grown statically to induce expression of dosR (devR) (64). For each reaction, two gene-specific reverse primers generated to the 5′ end of Rv0081 were used in combination with a forward primer generated to the 5′-RACE adaptor. Two TSPs were identified for wild-type M. tuberculosis grown with shaking (Table 2). The TSP for ∼60% of the clones examined occurred at position −46 (cytosine) relative to the Rv0081 translational start site. The TSP for the remaining ∼40% of clones was positioned at −15 (cytosine) relative to the Rv0081 translational start site. The cytosine at −46 (designated P1) lays within IR subunit 2 and is positioned downstream of the DosR/DevR binding site but within the region bound by Rv0081 and MprA. In contrast, the cytosine at −15 (designated P2) lays downstream of the location where Rv0081, MprA, and DosR/DevR bind (Fig. 4). When strains were deleted of dosR (devR), 100% of the clones examined initiated transcription at −15 relative to the Rv0081 translational start site (Table 2). In contrast, 100% of the clones examined initiated transcription at −46 relative to the Rv0081 translational start site when strains were deleted of mprAB (Table 2). The same two TSPs were also identified in the Rv0081 upstream region following growth of M. tuberculosis strains under static conditions. However, 100% of clones from wild-type M. tuberculosis possessed a TSP at −46 relative to the Rv0081 translational start site under these conditions (Table 2), likely reflecting the influence of DosR/DevR at this promoter element. Similar to that observed previously, 100% of clones examined from the ΔdosR mutant (ΔdevR mutant) initiated transcription at −15 relative to the Rv0081 translational start site (Table 2). In contrast, nearly 100% of clones possessed a TSP at −46 in the ΔmprAB mutant (Table 2). When taken together, these results indicate that P1 is the promoter positively regulated by DosR/DevR but is negatively regulated following binding by Rv0081 or MprA, while P2 is the promoter positively regulated by MprA. Furthermore, P1 is the predominant promoter mediating expression of Rv0081 under static growth conditions.
Table 2.
Frequency of TSP utilization in the Rv0081 promoter region
| Strain | Condition | Total no. of clones screened | No. of clones with TSP at position: |
|
|---|---|---|---|---|
| −46 | −15 | |||
| Wild type | Shaking | 7 | 4 | 3 |
| ΔdosR mutant | Shaking | 4 | 0 | 4 |
| ΔmprAB mutant | Shaking | 20 | 20 | 0 |
| Wild type | Static | 13 | 13 | 0 |
| ΔdosR mutant | Static | 4 | 0 | 4 |
| ΔmprAB mutant | Static | 14 | 13 | 1 |
Fig. 4.
Location of TSP and binding sites for Rv0081, DosR, and MprA in the Rv0081 promoter region. Arrows indicate TSP (P1) at −46 (black arrow) or the TSP (P2) at −15 (gray arrow). Predicted binding sites for Rv0081 are shown in bold. Predicted binding sites for DosR (underlined in black) and MprA (underlined in gray) are also shown. Binding sites for Rv0081 and MprA, but not DosR, overlap the TSP at −46.
Rv0081 is negatively regulated by Rv0081 in M. tuberculosis and is cotranscribed with downstream genes.
Rv0081-lacZ reporter studies conducted in M. smegmatis indicated that Rv0081 negatively regulated its own expression. To determine whether a similar pattern of regulation was observed in M. tuberculosis, lacZ reporter construct pTZ1111 was introduced into a ΔRv0081 mutant of M. tuberculosis in which Rv0081 was (TB229) or was not (TB228) expressed from hsp60 carried on an integrated vector. β-Galactosidase levels in TB229 were ∼2-fold lower than levels observed in TB228 (Fig. 5A). Therefore, Rv0081 also represses expression from its own promoter in M. tuberculosis. The Rv0081-Rv0088 locus contains intergenic regions that are small or overlapping in M. tuberculosis and other closely related species of Mycobacterium (Fig. 1A). To determine whether Rv0081-Rv0088 may be expressed as a single transcriptional unit, RT-PCRs were carried out on RNA from wild-type M. tuberculosis using primer sets that amplified intergenic regions. PCR products of the expected sizes were observed with each primer set in reaction mixtures containing reverse transcriptase (Fig. 5B). In contrast, PCR products were not observed in reaction mixtures lacking reverse transcriptase or template (Fig. 5B). Thus, Rv0081 can be expressed on a single transcriptional unit along with other genes comprising the Rv0081-Rv0088 locus.
Fig. 5.
Rv0081 is negatively regulated by Rv0081 in M. tuberculosis and is cotranscribed with downstream genes. (A) Rv0081-lacZ reporter construct pTZ1111 was introduced into a ΔRv0081 mutant of M. tuberculosis in which Rv0081 was (TB229) or was not (TB228) expressed from hsp60 carried on an integrated vector. Cultures were grown in Sauton's medium, and β-galactosidase assays were performed. The asterisk denotes statistical significance (P < 0.05). (B) RT-PCR was used to assess cotranscription of genes downstream of Rv0081. Primer pairs were designed to span intergenic regions and to amplify 300- to 500-bp fragments. Template for PCRs included total RNA incubated in the presence (+RT) or absence (-RT) of reverse transcriptase. Control reactions included genomic DNA as template (+) or water only (−).
Rv0081 and downstream genes are upregulated by NO in M. tuberculosis but repressed following Rv0081 overproduction.
Previous studies had indicated that Rv0081 was upregulated in M. tuberculosis following exposure to NO (66). To confirm this observation and determine whether genes downstream of Rv0081 were also responsive to this stimulus, qRT-PCR was conducted on wild-type M. tuberculosis that was either left untreated or was exposed to 100 μM DETA/NO for 40 min (66). As expected, exposure to NO significantly upregulated expression of dosR (devR) in M. tuberculosis relative to the untreated culture control (Fig. 6A). Importantly, NO exposure also upregulated expression of Rv0081 as well as representative genes from this locus, including Rv0082, Rv0083, and hycD (Fig. 6A). hycP was also upregulated following NO exposure; however, the extent of upregulation failed to reach statistical significance compared to the untreated control (Fig. 6A). Thus, several members of the Rv0081-Rv0088 locus are induced by NO. To determine whether Rv0081 could repress expression of Rv0081 following induction by NO, qRT-PCR expression analysis was carried out on wild-type M. tuberculosis or an isogenic ΔRv0081 mutant constitutively expressing Rv0081 from the hsp60 promoter (TB222). The primer set used to assess Rv0081 expression amplified a region downstream of the Rv0081 P1 promoter but upstream of the generated deletion. Similar to that previously observed (Fig. 6A), exposure of H37Rv to NO induced expression from the Rv0081 promoter region relative to the untreated control (Fig. 6B). Rv0081 expression was also upregulated in TB222 following NO exposure (Fig. 6B). However, relative expression from the Rv0081 promoter was lower in TB222 than in wild-type H37Rv following NO treatment (Fig. 6B). Thus, Rv0081 represses its own expression, even under conditions that induce the upregulation of this locus.
Fig. 6.
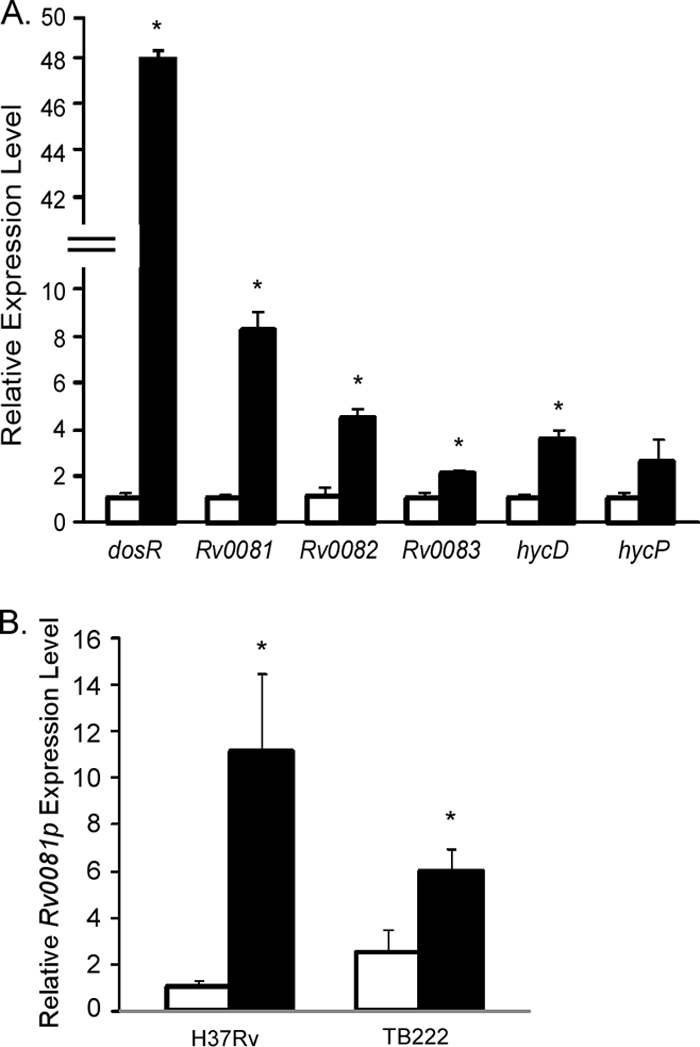
Rv0081 is upregulated by NO in M. tuberculosis but is repressed following Rv0081 overproduction. (A) qRT-PCR was performed on RNA derived from wild-type M. tuberculosis H37Rv grown in the absence (white bars) or presence (black bars) of NO. Expression levels for various genes, including dosR and representatives of the Rv0081-Rv0088 locus, were normalized to rrs. Expression levels in NO-treated cultures are presented as the fold induction relative to those from untreated cultures, which were set to 1.0. Asterisks denote statistical significance between NO-treated versus untreated cultures (P < 0.05). (B) Relative expression levels of Rv0081 in M. tuberculosis H37Rv or an isogenic ΔRv0081 mutant expressing hsp60-Rv0081 from an integrated vector (TB222). Cultures were grown in the absence (white bars) or presence (black bars) of NO. Expression levels were normalized to rrs. Expression was presented as the fold induction relative to that observed in the wild-type strain in the absence of treatment, which was set to 1.0. Asterisks denote statistical significance between NO-treated versus untreated cultures (P < 0.05).
DISCUSSION
It has become increasingly clear that M. tuberculosis utilizes complex and multifaceted regulatory networks to mediate adaptation processes in response to potentially detrimental stimuli within the host. Two such regulatory systems, MprAB and DosRS-DosT/DevRS-Rv2027c, are responsive to conditions likely to be present within the granuloma and have been shown to play a role in the virulence of M. tuberculosis in vitro and in vivo. Interestingly, microarray studies of M. tuberculosis mprA and dosR (devR) mutants have indicated that there may be a subset of genes that are coregulated by these response regulators (28, 43, 45). This observation suggests that either there is some overlap between these regulons, or that MprAB directly regulates dosRS (devRS) or vice versa. Here, we demonstrated that Rv0081 and determinants immediately downstream of Rv0081 are directly regulated by Rv0081, as well as DosR/DevR and MprA.
Bioinformatic analyses indicate that Rv0081 is a member of the ArsR/SmtB family of metal-dependent transcriptional repressors (11). Members of this family sense and are responsive to divalent and multivalent heavy metal ions and control the expression of genes involved in metal uptake, efflux, sequestration, or detoxification (10, 38, 63). ArsR/SmtB family members are typically homodimers and contain a winged helix-turn-helix DNA binding motif (10). In the absence of metal, these proteins bind operator/promoter regions and block access by RNA polymerase, leading to transcriptional repression of target genes. Association with metal allosterically inhibits DNA binding (10), allowing derepression of genes when a minimum metal concentration is achieved. Importantly, ArsR/SmtB family members recognize one or two imperfect 12-2-12 inverted repeat elements, often within their own promoter/operator region (10).
At least 12 metal-sensing repressors belonging to this family have been predicted in the M. tuberculosis genome (11). The large number of metal-sensing repressors from this family suggests that divalent and/or heavy metal adaptation may play an important role in the physiology and/or pathogenesis of the tubercle bacillus. However, in spite of their potential importance, only 3 of the 12 encoded repressors have been characterized to date. KmtR is responsive to nickel and cobalt and represses transcription of Rv0826 and Rv2025, encoding a hypothetical protein and a deduced transmembrane cation diffusion facilitator family metal ion transporter, respectively (11). NmtR also senses nickel and cobalt but regulates expression of nmtA, encoding a predicted P1-type ATPase (13, 67). Finally, CmtR is responsive to cadmium (and possibly lead) and regulates expression of its own operon, which includes downstream genes Rv1993c and cmtA, encoding a predicted P1-type ATPase (12, 14).
In vitro studies with purified Rv0081 indicate that this protein is monomeric in solution and that it binds cooperatively to a 12-2-12 inverted repeat element in its own upstream region. Rv0081 preferentially binds the IR2 subunit and once bound cooperatively recruits another monomer of Rv0081 to IR1. The physical location of IR2 overlaps the TSP (P1), which is positioned 46 nucleotides upstream of the Rv0081 translational start site. Consistent with this mapping, lacZ and qRT-PCR expression analyses indicated that Rv0081 functions as a transcriptional repressor of its own expression. While it is currently unclear whether Rv0081 is responsive to metal(s), sequence clustering analyses indicate that Rv0081 falls into an SmtB/ArsR subfamily that lacks a defined metal binding site (11). Interestingly, key residues within domains important for metal binding in other SmtB/ArsR family members (α3 and α5) are absent from the corresponding regions in Rv0081 (data not shown), indicating that if this protein binds metal the mechanism may be novel. Studies to delineate the ability of Rv0081 to bind metal and regulate gene expression in response to metals are under way.
In addition to Rv0081, response regulators DosR/DevR and MprA also directly bind the Rv0081 promoter region and positively regulate its expression. DosR/DevR recognizes a region that partially overlaps that bound by Rv0081. Based on this location, it is likely that DosR positively regulates transcription from the P1 promoter. Consistent with this idea, transcripts initiating from promoter P1 are not observed by 5′-RACE when using RNA extracted from ΔdosR mutants of M. tuberculosis. Importantly, expression of Rv0081 and downstream genes is upregulated in M. tuberculosis following exposure to NO, a stimulus known to activate signaling through the DosRS-DosT/DevRS-Rv2027c TCSS (66). While the sequence recognized by DosR/DevR in the Rv0081 upstream region contains several highly conserved residues that are present in the first half of the palindromic sequence from other defined DosR/DevR binding sites (15–17, 26, 45), homology to sequences in the second half of the palindromic motif is more limited. Thus, DosR/DevR may tolerate greater variation in its recognition site than previously appreciated. The Rv0081 upstream region is also subject to direct regulation by MprA through recognition of a well-conserved MprA binding motif. Interestingly, this motif is contained completely within the two IR subunits recognized by Rv0081, indicating that Rv0081 and MprA likely compete for binding sites at this locus and that binding by MprA also limits accessibility to P1. Thus, MprA may act to limit expression from P1 even under conditions when Rv0081 may not be functional to bind DNA. However, given that expression from the Rv0081 promoter region is reduced in M. tuberculosis ΔmprAB relative to the isogenic wild-type parent, it is likely that MprA also positively regulates expression from the P2 promoter. Consistent with this observation, transcripts initiating nearly exclusively from P1 are observed in 5′-RACE when RNA is derived from the ΔmprAB mutant strain of M. tuberculosis. Thus, expression of Rv0081 is likely influenced by multiple factors, including the relative concentrations of Rv0081, DosR/DevR, and MprA, the relative affinity of each transcription factor for its recognition sequence, and the environmental conditions in which the bacterium finds itself.
Rv0081 is cotranscribed with seven additional determinants in M. tuberculosis and other closely related mycobacteria that are predicted to encode components of an enzyme complex involved in formate metabolism. In E. coli, three enzyme complexes are able to metabolize formate to CO2, depending on growth conditions (7, 46, 49). During aerobic growth, formate is metabolized by formate hydrogenase O (FDH-O) by using oxygen as a terminal electron acceptor (46). When growing anaerobically and when nitrate is available as a terminal electron acceptor, E. coli metabolizes formate via formate hydrogenase N (FDH-N) (32, 46). Finally, in the absence of both oxygen and nitrate, formate hydrogenase H (FDH-H) and components of the Hyd-3 or Hyd-4 enzyme complex associate to form the FHL enzyme complex, which converts formate to CO2 and couples this to the production of H2 (7, 37, 49). Genes required for FHL synthesis in E. coli are regulated by two transcription factors (FhlA and HycA) and are induced in response to high concentrations of formate (31, 37, 53–55, 57). FhlA is a positive regulator of these genes and is a member of the NtrC group of transcriptional regulators. This protein binds formate directly, and upon binding, associates with upstream activating sequences located within the promoter regions for these genes to activate transcription in a σ54-dependent manner (reviewed in reference 37). In contrast, HycA negatively regulates expression of these genes, likely through negative interactions with FhlA (53).
Currently, nothing is known about the function of Rv0081 or downstream genes in the physiology and/or pathogenesis of M. tuberculosis. While most sequenced Mycobacterium genomes except for Mycobacterium leprae encode Rv0081 orthologs, Blast searches indicate that the downstream genes encoded at this locus are restricted to certain mycobacterial species, particularly those associated with respiratory disease and/or granuloma formation in their respective host(s). Thus, the ability of these organisms to metabolize formate may be a characteristic that evolved to enhance survival within specific growth environments. M. tuberculosis is predicted to encode FDH-H (Rv2900c-Rv2899c) but lacks determinants for the other two formate dehydrogenases. The bacterium also lacks apparent orthologs of FhlA and HycA. While Rv0082-Rv0088 is predicted to encode a Hyd enyzme complex, this appears to be the only Hyd complex produced in M. tuberculosis. Additionally, its components exhibit homology to members of both Hyd-3 and Hyd-4 from E. coli. Interestingly, a subset of genes comprising the Rv0081-Rv0088 locus are also predicted to be essential for growth on laboratory medium (52), suggesting that this system may be necessary for M. tuberculosis growth even in the presence of oxygen. Thus, formate metabolism in M. tuberculosis appears to differ significantly from the paradigm seen in enteric bacteria, including the enzymatic components involved, the regulatory determinants utilized, and the environmental conditions under which this system is expressed.
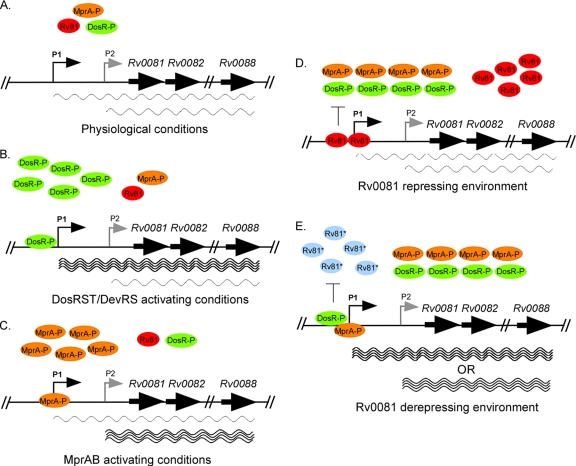
Based on evidence presented in this study and the work published in E. coli, we present a model (Fig. 7) in which M. tuberculosis encodes components of a predicted FHL enzyme complex that is positively regulated by the DosRS-DosT/DevRS-Rv2027c and MprAB TCSS and negatively regulated by Rv0081. During growth of M. tuberculosis under normal physiological conditions, levels of DosR-P/DevR-P and MprA-P are expected to be low, resulting in basal-level expression of Rv0081-Rv0088 from promoters P1 and P2 (Fig. 7A). However, upon exposure of M. tuberculosis to environmental conditions that activate DosRS-DosT/DevRS-Rv2027c, levels of DosR-P/DevR-P are expected to increase, resulting in upregulation of Rv0081-Rv0088 from the P1 promoter (Fig. 7B). Similarly, upon exposure of M. tuberculosis to environmental conditions that activate MprAB, levels of MprA-P are expected to increase, leading to the upregulation of Rv0081-Rv0088 from the P2 promoter (Fig. 7C). As Rv0081 is a negative regulator, we postulate that the role of this protein may be to serve as a rheostat to allow fine-tuning of expression of Rv0081-Rv0088 within the cell (Fig. 7D and E). Elevated levels of Rv0081 produced following the upregulation of Rv0081-Rv0088 are likely to repress expression of this locus by (i) Rv0081 binding to its recognition sequence in the Rv0081 upstream region and inhibiting transcription from the P1 promoter element (by physically blocking DosR-P/DevR-P binding and/or preventing access by RNA polymerase) and (ii) Rv0081 binding to its recognition sequence in the Rv0081 upstream region and dampening transcription from the P2 promoter element (by physically blocking MprA-P binding). However, binding by Rv0081 to its consensus sequence in the Rv0081 upstream region may be abrogated following the recognition of an as-yet-unidentified stimulus, possibly a metal(s). Deregulation by Rv0081 could lead to increased expression of Rv0081-Rv0088 expression from either P1 or P2. Many of the FHL enzyme components in E. coli and other organisms contain metal cofactors, including iron, nickel, and molybdenum (48, 55, 65). Therefore, it is possible that recognition of metal by Rv0081 may help ensure that sufficient levels of these cofactors are available for incorporation into FHL enzyme components prior to their synthesis. Studies to address this possibility are under way, as are experiments to investigate the role for Rv0081-Rv0088 in the physiology and virulence of M. tuberculosis.
Fig. 7.
Model for the regulation of Rv0081-Rv0088 in M. tuberculosis in response to growth under different environmental conditions. The Rv0081-Rv0088 locus is transcribed by two promoters upstream of Rv0081; P1 is activated by DosR but repressed by Rv0081 and MprA, and P2 is activated by MprA. During growth under physiological conditions (i.e., no environmental stimuli) (A), expression from P1 and P2 is low due to the general lack of DosR-P and MprA-P within the bacterium. Upon conditions that activate the DosRS-DosT/DevRS-Rv2027c TCSS, levels of DosR-P/DevR-P increase, leading to increased expression of Rv0081-Rv0088 from P1 (B). Similarly, growth of M. tuberculosis under conditions that activate the MprAB TCSS lead to increased levels of MprA-P and increased expression of Rv0081-Rv0088 from P2 (C). Over time, high levels of Rv0081 accumulate. In the absence of as-yet-undefined stimuli, Rv0081 binds its recognition sequence in the Rv0081 upstream region, physically blocking access by both DosR-P or MprA-P and repressing transcription from P1. However, in response to specific stimuli (i.e., high concentrations of metal, possibly), Rv0081 dissociates from the DNA, leading to derepression of Rv0081-Rv0088 expression from P1 or P2. For all panels, thick arrows indicate genes while thin arrows indicate sites of transcription. Wavy lines below genes indicate transcripts initiating from the designated promoter elements. DosR-P, green; MprA-P, orange; Rv0081 unmodified, red; Rv0081 modified, blue.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant RO1 AI51669 and a Burroughs Wellcome Fund Investigator in Pathogenesis Award to T.C.Z.
We are grateful to Shelley Haydel for providing pJEM15 and to Bill Jacobs for providing pYUB854 and pYUB870.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 5 August 2011.
REFERENCES
- 1. Allen B. W. 1998. Mycobacteria: general culture methodology and safety considerations, p. 15–30 In Parish T., Stoker N. G. (ed.), Mycobacteria protocols, vol. 101 Humana Press Inc., Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 2. Anonymous 2010. Global tuberculosis control 2010. World Health Organization, Geneva, Switzerland [Google Scholar]
- 3. Anonymous 2010. Treatment of tuberculosis guidelines. World Health Organization, Geneva, Switzerland: [PubMed] [Google Scholar]
- 4. Bardarov S., et al. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007–3017 [DOI] [PubMed] [Google Scholar]
- 5. Barik S., Sureka K., Mukherjee P., Basu J., Kundu M. 2010. RseA, the SigE specific anti-sigma factor of Mycobacterium tuberculosis, is inactivated by phosphorylation-dependent ClpC1P2 proteolysis. Mol. Microbiol. 75:592–606 [DOI] [PubMed] [Google Scholar]
- 6. Betts J. C., Lukey P. T., Robb L. C., McAdam R. A., Duncan K. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717–731 [DOI] [PubMed] [Google Scholar]
- 7. Birkmann A., Zinoni F., Sawers G., Bock A. 1987. Factors affecting transcriptional regulation of the formate-hydrogen-lyase pathway of Escherichia coli. Arch. Microbiol. 148:44–51 [DOI] [PubMed] [Google Scholar]
- 8. Black G. F., et al. 2009. Immunogenicity of novel DosR regulon-encoded candidate antigens of Mycobacterium tuberculosis in three high-burden populations in Africa. Clin. Vaccine Immunol. 16:1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boon C., Dick T. 2002. Mycobacterium bovis BCG response regulator essential for hypoxic dormancy. J. Bacteriol. 184:6760–6767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Busenlehner L. S., Pennella M. A., Giedroc D. P. 2003. The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 27:131–143 [DOI] [PubMed] [Google Scholar]
- 11. Campbell D. R., et al. 2007. Mycobacterial cells have dual nickel-cobalt sensors: sequence relationships and metal sites of metal-responsive repressors are not congruent. J. Biol. Chem. 282:32298–32310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cavet J. S., Graham A. I., Meng W., Robinson N. J. 2003. A cadmium-lead-sensing ArsR-SmtB repressor with novel sensory sites. Complementary metal discrimination by NmtR AND CmtR in a common cytosol. J. Biol. Chem. 278:44560–44566 [DOI] [PubMed] [Google Scholar]
- 13. Cavet J. S., et al. 2002. A nickel-cobalt-sensing ArsR-SmtB family repressor. Contributions of cytosol and effector binding sites to metal selectivity. J. Biol. Chem. 277:38441–38448 [DOI] [PubMed] [Google Scholar]
- 14. Chauhan S., Kumar A., Singhal A., Tyagi J. S., Krishna Prasad H. 2009. CmtR, a cadmium-sensing ArsR-SmtB repressor, cooperatively interacts with multiple operator sites to autorepress its transcription in Mycobacterium tuberculosis. FEBS J. 276:3428–3439 [DOI] [PubMed] [Google Scholar]
- 15. Chauhan S., Tyagi J. S. 2008. Cooperative binding of phosphorylated DevR to upstream sites is necessary and sufficient for activation of the Rv3134c-devRS operon in Mycobacterium tuberculosis: implication in the induction of DevR target genes. J. Bacteriol. 190:4301–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chauhan S., Tyagi J. S. 2008. Interaction of DevR with multiple binding sites synergistically activates divergent transcription of narK2-Rv1738 genes in Mycobacterium tuberculosis. J. Bacteriol. 190:5394–5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chauhan S., Tyagi J. S. 2009. Powerful induction of divergent tgs1-Rv3131 genes in Mycobacterium tuberculosis is mediated by DevR interaction with a high-affinity site and an adjacent cryptic low-affinity site. J. Bacteriol. 191:6075–6081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cole S. T., et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544 [DOI] [PubMed] [Google Scholar]
- 19. Converse P. J., et al. 2009. Role of the dosR-dosS two-component regulatory system in Mycobacterium tuberculosis virulence in three animal models. Infect. Immun. 77:1230–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dasgupta N., et al. 2000. Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuber. Lung Dis. 80:141–159 [DOI] [PubMed] [Google Scholar]
- 21. Domenech P., Kolly G. S., Leon-Solis L., Fallow A., Reed M. B. 2010. Massive gene duplication event among clinical isolates of the Mycobacterium tuberculosis W/Beijing family. J. Bacteriol. 192:4562–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dona V., et al. 2008. Evidence of complex transcriptional, translational, and posttranslational regulation of the extracytoplasmic function sigma factor σE in Mycobacterium tuberculosis. J. Bacteriol. 190:5963–5971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Estorninho M., et al. 2010. ClgR regulation of chaperone and protease systems is essential for Mycobacterium tuberculosis parasitism of the macrophage. Microbiology 156:3445–3455 [DOI] [PubMed] [Google Scholar]
- 24. Fallow A., Domenech P., Reed M. B. 2010. Strains of the East Asian (W/Beijing) lineage of Mycobacterium tuberculosis are DosS/DosT-DosR two-component regulatory system natural mutants. J. Bacteriol. 192:2228–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fisher M. A., Plikaytis B. B., Shinnick T. M. 2002. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J. Bacteriol. 184:4025–4032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Florczyk M. A., et al. 2003. A family of acr-coregulated Mycobacterium tuberculosis genes shares a common DNA motif and requires Rv3133c (dosR or devR) for expression. Infect. Immun. 71:5332–5343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haydel S. E., Clark-Curtiss J. E. 2004. Global expression analysis of two-component system regulator genes during Mycobacterium tuberculosis growth in human macrophages. FEMS Microbiol. Lett. 236:341–347 [DOI] [PubMed] [Google Scholar]
- 28. He H., Hovey R., Kane J., Singh V., Zahrt T. C. 2006. MprAB is a stress-responsive two-component system that directly regulates expression of sigma factors SigB and SigE in Mycobacterium tuberculosis. J. Bacteriol. 188:2134–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. He H., Zahrt T. C. 2005. Identification and characterization of a regulatory sequence recognized by Mycobacterium tuberculosis persistence regulator MprA. J. Bacteriol. 187:202–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Honaker R. W., Dhiman R. K., Narayanasamy P., Crick D. C., Voskuil M. I. 2010. DosS responds to a reduced electron transport system to induce the Mycobacterium tuberculosis DosR regulon. J. Bacteriol. 192:6447–6455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hopper S., et al. 1994. Regulated expression in vitro of genes coding for formate hydrogenlyase components of Escherichia coli. J. Biol. Chem. 269:19597–19604 [PubMed] [Google Scholar]
- 32. Hopper S., Bock A. 1995. Effector-mediated stimulation of ATPase activity by the sigma 54-dependent transcriptional activator FHLA from Escherichia coli. J. Bacteriol. 177:2798–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jacobs W. R., Jr., et al. 1991. Genetic systems for mycobacteria. Methods Enzymol. 204:19. [DOI] [PubMed] [Google Scholar]
- 34. Karakousis P. C., et al. 2004. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J. Exp. Med. 200:647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kumar A., et al. 2008. Heme oxygenase-1-derived carbon monoxide induces the Mycobacterium tuberculosis dormancy regulon. J. Biol. Chem. 283:18032–18039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Leistikow R. L., et al. 2010. The Mycobacterium tuberculosis DosR regulon assists in metabolic homeostasis and enables rapid recovery from nonrespiring dormancy. J. Bacteriol. 192:1662–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Leonhartsberger S., Korsa I., Bock A. 2002. The molecular biology of formate metabolism in enterobacteria. J. Mol. Microbiol. Biotechnol. 4:269–276 [PubMed] [Google Scholar]
- 38. Lucarelli D., et al. 2007. Crystal structure and function of the zinc uptake regulator FurB from Mycobacterium tuberculosis. J. Biol. Chem. 282:9914–9922 [DOI] [PubMed] [Google Scholar]
- 39. Malhotra V., et al. 2004. Disruption of response regulator gene, devR, leads to attenuation in virulence of Mycobacterium tuberculosis. FEMS Microbiol. Lett. 231:237–245 [DOI] [PubMed] [Google Scholar]
- 40. Manganelli R., Voskuil M. I., Schoolnik G. K., Smith I. 2001. The Mycobacterium tuberculosis ECF sigma factor σE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41:423–437 [DOI] [PubMed] [Google Scholar]
- 41. Mehra S., Dutta N. K., Mollenkopf H. J., Kaushal D. 2010. Mycobacterium tuberculosis MT2816 encodes a key stress-response regulator. J. Infect. Dis. 202:943–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Morby A. P., Turner J. S., Huckle J. W., Robinson N. J. 1993. SmtB is a metal-dependent repressor of the cyanobacterial metallothionein gene smtA: identification of a Zn inhibited DNA-protein complex. Nucleic Acids Res. 21:921–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pang X., et al. 2007. Evidence for complex interactions of stress-associated regulons in an mprAB deletion mutant of Mycobacterium tuberculosis. Microbiology 153:1229–1242 [DOI] [PubMed] [Google Scholar]
- 44. Parish T., et al. 2003. Deletion of two-component regulatory systems increases the virulence of Mycobacterium tuberculosis. Infect. Immun. 71:1134–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Park H. D., et al. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48:833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pecher A., et al. 1983. On the redox control of synthesis of anaerobically induced enzymes in enterobacteriaceae. Arch. Microbiol. 136:131–136 [DOI] [PubMed] [Google Scholar]
- 47. Riley L. 1996. Phagocytosis of M. tuberculosis, p. 281–289 In Rom W., Garay S. (ed.), Tuberculosis. Little Brown, Boston, MA [Google Scholar]
- 48. Rosentel J. K., Healy F., Maupin-Furlow J. A., Lee J. H., Shanmugam K. T. 1995. Molybdate and regulation of mod (molybdate transport), fdhF, and hyc (formate hydrogenlyase) operons in Escherichia coli. J. Bacteriol. 177:4857–4864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rossmann R., Sawers G., Bock A. 1991. Mechanism of regulation of the formate-hydrogenlyase pathway by oxygen, nitrate, and pH: definition of the formate regulon. Mol. Microbiol. 5:2807–2814 [DOI] [PubMed] [Google Scholar]
- 50. Rustad T. R., Harrell M. I., Liao R., Sherman D. R. 2008. The enduring hypoxic response of Mycobacterium tuberculosis. PLoS One 3:e1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 52. Sassetti C. M., Boyd D. H., Rubin E. J. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77–84 [DOI] [PubMed] [Google Scholar]
- 53. Sauter M., Bohm R., Bock A. 1992. Mutational analysis of the operon (hyc) determining hydrogenase 3 formation in Escherichia coli. Mol. Microbiol. 6:1523–1532 [DOI] [PubMed] [Google Scholar]
- 54. Sawers R. G. 2005. Formate and its role in hydrogen production in Escherichia coli. Biochem. Soc. Trans. 33:42–46 [DOI] [PubMed] [Google Scholar]
- 55. Schlensog V., Lutz S., Bock A. 1994. Purification and DNA-binding properties of FHLA, the transcriptional activator of the formate hydrogenlyase system from Escherichia coli. J. Biol. Chem. 269:19590–19596 [PubMed] [Google Scholar]
- 56. Schnappinger D., et al. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Skibinski D. A., et al. 2002. Regulation of the hydrogenase-4 operon of Escherichia coli by the σ54-dependent transcriptional activators FhlA and HyfR. J. Bacteriol. 184:6642–6653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stover C. K., et al. 1991. New use of BCG for recombinant vaccines. Nature 351:456–460 [DOI] [PubMed] [Google Scholar]
- 59. Sureka K., et al. 2007. Polyphosphate kinase is involved in stress-induced mprAB-sigE-rel signalling in mycobacteria. Mol. Microbiol. 65:261–276 [DOI] [PubMed] [Google Scholar]
- 60. Talaat A. M., Lyons R., Howard S. T., Johnston S. A. 2004. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc. Natl. Acad. Sci. U. S. A. 101:4602–4607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Taneja N. K., Dhingra S., Mittal A., Naresh M., Tyagi J. S. 2010. Mycobacterium tuberculosis transcriptional adaptation, growth arrest and dormancy phenotype development is triggered by vitamin C. PLoS One 5:e10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Timm J., Lim E. M., Gicquel B. 1994. Escherichia coli-mycobacteria shuttle vectors for operon and gene fusions to lacZ: the pJEM series. J. Bacteriol. 176:6749–6753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tottey S., Harvie D. R., Robinson N. J. 2005. Understanding how cells allocate metals using metal sensors and metallochaperones. ACC Chem. Res. 38:775–783 [DOI] [PubMed] [Google Scholar]
- 64. Vasudeva-Rao H. M., McDonough K. A. 2008. Expression of the Mycobacterium tuberculosis acr-coregulated genes from the DevR (DosR) regulon is controlled by multiple levels of regulation. Infect. Immun. 76:2478–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vignais P. M., Colbeau A. 2004. Molecular biology of microbial hydrogenases. Curr. Issues Mol. Biol. 6:159–188 [PubMed] [Google Scholar]
- 66. Voskuil M. I., et al. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang Y., Hemmingsen L., Giedroc D. P. 2005. Structural and functional characterization of Mycobacterium tuberculosis CmtR, a PbII/CdII-sensing SmtB/ArsR metalloregulatory repressor. Biochemistry 44:8976–8988 [DOI] [PubMed] [Google Scholar]
- 68. White M. J., He H., Penoske R. M., Twining S. S., Zahrt T. C. 2010. PepD participates in the mycobacterial stress response mediated through MprAB and SigE. J. Bacteriol. 192:1498–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zahrt T. C., Deretic V. 2001. Mycobacterium tuberculosis signal transduction system required for persistent infections. Proc. Natl. Acad. Sci. U. S. A. 98:12706–12711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zahrt T. C., Wozniak C., Jones D., Trevett A. 2003. Functional analysis of the Mycobacterium tuberculosis MprAB two-component signal transduction system. Infect. Immun. 71:6962–6970 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.