Abstract
Many bacteria, in particular Gram-positive bacteria, contain high proportions of non-N-acetylated amino sugars, i.e., glucosamine (GlcN) and/or muramic acid, in the peptidoglycan of their cell wall, thereby acquiring resistance to lysozyme. However, muramidases with specificity for non-N-acetylated peptidoglycan have been characterized as part of autolytic systems such as of Clostridium acetobutylicum. We aim to elucidate the recovery pathway for non-N-acetylated peptidoglycan fragments and present here the identification and characterization of an acetyltransferase of novel specificity from C. acetobutylicum, named GlmA (for glucosamine/glucosaminide N-acetyltransferase). The enzyme catalyzes the specific transfer of an acetyl group from acetyl coenzyme A to the primary amino group of GlcN, thereby generating N-acetylglucosamine. GlmA is also able to N-acetylate GlcN residues at the nonreducing end of glycosides such as (partially) non-N-acetylated peptidoglycan fragments and β-1,4-glycosidically linked chitosan oligomers. Km values of 114, 64, and 39 μM were determined for GlcN, (GlcN)2, and (GlcN)3, respectively, and a 3- to 4-fold higher catalytic efficiency was determined for the di- and trisaccharides. GlmA is the first cloned and biochemically characterized glucosamine/glucosaminide N-acetyltransferase and a member of the large GCN5-related N-acetyltransferases (GNAT) superfamily of acetyltransferases. We suggest that GlmA is required for the recovery of non-N-acetylated muropeptides during cell wall rescue in C. acetobutylicum.
INTRODUCTION
The glycan chains of the peptidoglycan of the bacterial cell wall are composed of alternating, β-1,4-glycosidically linked amino sugars N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) (30). The two glycosidic bonds of these glycans are targeted by muramidases, such as lysozymes that hydrolyze the MurNAc-GlcNAc linkages, and by (endo-)N-acetylglucosaminidases that cleave the GlcNAc-MurNAc bonds. Many bacteria, in particular Gram-positive pathogens, have been reported to acquire resistance against cell lysis due to the action of lysozyme and N-acetylglucosaminidases by N-deacetylation of a great portion of the amino sugars of the peptidoglycan of their cell wall (29). Araki et al. showed that the majority of glucosamine residues of the peptidoglycans of Bacillus cereus, B. subtilis, and B. megaterium have free amino groups and that non-N-acetylated glucosamine residues accounted for the resistance of these strains to lysozyme (1, 2, 12). The peptidoglycan of Streptococcus pneumoniae, which contains 40 to 80% glucosamine (GlcN) and up to 10% muramic acid (19), is N-deacetylated at the GlcNAc residues in the peptidoglycan by the GlcNAc deacetylase PgdA, the first characterized peptidoglycan deacetylase (31). Clostridium acetobutylicum, which is an endospore-forming, anaerobic firmicute that is closely related to bacilli, presumably also contains N-deacetylated peptidoglycan since orthologs of pgdA are present on its chromosome. Moreover, an autolytic muramidase has been identified in this organism that is active only on non-N-acetylated peptidoglycan, generating peptidoglycan fragments (muropeptides) that contain a GlcN residue at the nonreducing terminus (10).
We have been studying the peptidoglycan recycling and recognized a muropeptide rescue pathway in B. subtilis that involves the sequential hydrolysis of muropeptides (GlcNAc-MurNAc peptides) by N-acetylglucosaminidase and N-acetylmuramic acid-l-Ala amidase in the wall compartment (16). We recognized that a muropeptide rescue pathway is likely also present in C. acetobutylicum, which, however, appears to display some distinct features. This organism contains an ortholog of the cytoplasmic MurNAc–6-phosphate etherase MurQ (13) that allows the conversion of MurNAc-6-phosphate to GlcNAc-6-phosphate, which subsequently can enter the nag pathway for further catabolism (32, 33). However, C. acetobutylicum apparently lacks a specific phosphotransferase system for MurNAc that is present in B. subtilis. In an accompanying paper (22), we showed that C. acetobutylicum instead possesses a cytoplasmic kinase that phosphorylates both GlcNAc and MurNAc. The cell wall sugars presumably are released from cell wall fragments in the cytoplasm by orthologs of the N-acetylglucosaminidase NagZ and the N-acetylmuramyl-l-Ala amidase AmiE of B. subtilis (16), which apparently are not a secreted in C. acetobutylicum but cytoplasmic. We recently have investigated the structure-function relationship of the B. subtilis NagZ and revealed that this enzyme is highly specific for the N-acetyl group of the substrate (17). Hence, a proposed recovery of non-N-acetylated sugars would require the N-acetylation of GlcN residues at the nonreducing end of peptidoglycan fragments (GlcN-MurNAc peptides) prior to NagZ cleavage. We describe here the cloning and biochemical characterization of an enzyme from C. acetobutylicum that has a unique glucosamine/glucosaminide N-acetyltransferase activity.
MATERIALS AND METHODS
Bacterial strains, plasmids, chemicals, and growth conditions.
The Escherichia coli and B. subtilis strains used in the present study were grown aerobically at 37°C in LB medium (5) that was supplemented with an appropriate antibiotic for plasmid maintenance. Kanamycin and ampicillin were used at final concentrations of 50 and 100 μg/ml, respectively. All chemicals were obtained from Sigma-Aldrich, except as otherwise noted. MurNAc was purchased from Bachem (Weil am Rhein, Germany), chitosan from Medac (Wedel, Germany), and [acetyl-1-14C]coenzyme A ([acetyl-1-14C]CoA) from Hartmann Analytic (Wedel, Germany).
Plasmid construction.
ORF0184 of C. acetobutylicum ATCC 824 was amplified by PCR from chromosomal DNA kindly provided by G. Bennett (Rice University, Houston, TX) and Hubert Bahl (University of Rostock, Rostock, Germany) using the primers 5′-GTGGGATCCCATGGAAATTAAAGAGACATATGATTTTAGTAGC-3′ and 5′-ATCCTGCAGATCTACCCTTGCCATCTTTTACCTCTTTG-3′ (the recognition sites for the restriction endonucleases NcoI and BglII are underlined). The resulting 963-bp DNA fragment was cloned into the expression vector pCS19 (ampicillin resistance cassette [Ampr]) (26), yielding plasmid pGlmA, which allows the cytoplasmic overexpression of glmA as a C-terminal His6 fusion protein under the control of the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible T5 promoter. Cloning was performed using E. coli DH5α (6).
Overexpression and purification of GlmA.
E. coli strain M15(pREP4) (kanamycin resistance cassette [Kanr]; Qiagen) harboring pGlmA was cultivated under vigorous shaking in LB medium supplemented with kanamycin and ampicillin. At an optical density at 600 nm (OD600) of 0.6, the glmA expression was induced by the addition of IPTG at a final concentration of 0.2 mM. After a further 3 h of incubation, the cells were harvested by centrifugation (5,000 × g, 4°C), resuspended in binding buffer (50 mM NaH2PO4 · H2O, 300 mM NaCl, 10% [vol/vol] glycerol, 20 mM imidazole [pH 7.5, 4°C]), and finally disrupted using a French press cell. Cell debris and unbroken cells were removed from the cell extract by ultracentrifugation (150,000 × g for 1 h, 4°C). The His-tagged GlmA was purified from the soluble cell extract by Ni2+ affinity chromatography on a 5-ml His-Trap column (GE Healthcare), preequilibrated with 50 ml of binding buffer. The elution of GlmA from the column was achieved by applying a linear gradient to 500 mM imidazole using binding buffer and elution buffer (50 mM NaH2PO4 · H2O, 300 mM NaCl, 10% [vol/vol] glycerol, 500 mM imidazole [pH 7.5, 4°C]) for 200 ml with a flow rate of 2 ml/min. GlmA eluted from the column with 200 mM imidazole. The elution fractions were analyzed for enzyme purity by SDS-PAGE. Pure GlmA containing fractions were pooled, and the pool was then dialyzed against 5 liters of 50 mM NaH2PO4 · H2O–300 mM NaCl–10% (vol/vol) glycerol at 4°C. The protein concentration was determined by UV absorption at 280 nm and according to the method of Bradford (8). The protein solution was stored at −80°C.
Acetylation assays.
The activity of GlmA with different amino sugars (GlcN, muramic acid [MurN], N-acetylgalactosamine [GalN], and N-acetylmannosamine [ManN]), as well chitosan oligomers (degrees of polymerization of 2, 3 and 5), was determined by incubation of a 5 mM concentration of the sugar substrate with 1.5 μg of GlmA (39 pmol; 1.95 μM in the assay) in 20-μl reaction volumes containing 50 mM Tris-HCl (pH 7.5) and 45 μM [acetyl-1-14C]CoA (1,800 Bq). The reaction mixtures were incubated at 30°C, and at different times 2-μl portions of the reaction mixtures were spotted onto a thin-layer chromatography (TLC) plate (silica gel 60 F254; Merck, Darmstadt, Germany). The sugars in the reaction mixture were subsequently separated by TLC with the eluant butanol-methanol-ammonia-water (5:4:2:1 [vol/vol/vol/vol]). Acetylated, radioactive samples were detected and quantified by autoradiography using an FLA-9000 phosphorimager (Fuji Film) and the software Multi Gauge. Reaction mixtures without a sugar substrate were used as controls. To determine the position of acetylation of GlmA, the reaction product was incubated with GlcNAc/MurNAc kinase MurK (from C. acetobutylicum, prepared as described previously [22]) and N-acetylglucosaminidase NagZBs (from B. subtilis, prepared as described previously [16]). Three-microgram portions of each enzyme preparation were added to the 20-μl reaction mixtures and analyzed as described above.
GlmA activity was also studied by a nonradioactive acetylation assay. Portions (70 μg [1.8 nmol; 91.0 μM in the assay]) of GlmA were incubated with 50 mM sugar substrate and 100 mM acetyl-CoA in a 20-μl reaction volume buffered with 200 mM Tris-HCl (pH 7.5) at 37°C. At different times, 5-μl samples of the mixture were spotted onto a TLC plate and separated as described above. Visualization of the sugars on the TLC plate was accomplished by dipping the plate in methanol containing 2% (vol/vol) concentrated H2SO4, followed by drying and charring for 5 min at 180°C.
Stability assay and pH dependency.
We first controlled the stability of GlmA at different pH values. Concentrated GlmA was at first diluted in buffers with a pH ranging from 4.5 to 10.5. After incubation for 30 min at 25°C, the reaction mixture was diluted in phosphate-citrate buffer at pH 7.5, and the GlmA activity was determined after further incubation for 30 min at 25°C as mentioned below. The pH optimum of GlmA was determined by the radioactive acetylation assay in 0.2 M phosphate–0.1 M citrate buffer at pH 4.5 to 8.0, in 0.2 M Tris-HCl buffer at pH 7.5 to 9.0, and in 0.2 M glycine-NaOH buffer at pH 9.0 to 10.5. The 20-μl reaction mixtures contained 100 μM GlcN and 120 μM [acetyl-1-14C]CoA (5,550 Bq) and were incubated with 1 μg (26 pmol) of GlmA at 37°C. After 25 min, the reactions were stopped by spotting 2-μl samples onto a TLC plate. Finally, radioactively labeled reaction products were analyzed and quantified as described above for the acetylation assays.
Kinetic parameters.
The activity of GlmA was determined with a continuous assay that relies on the quantification of CoA, generated from acetyl-CoA in the acetyl transfer reaction. CoA reacts stoichiometrically with Ellman reagent [5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB)] (24). The product of this reaction, 2-nitro-5-thiobenzoate, was monitored by an increase in absorbance at 412 nm using microcuvettes and a spectrophotometer (Ultrospec 3000; Pharmacia Biotech). For a 100-μl assay, sugar substrates [e.g., GlcN, (GlcN)2 or (GlcN)3] at various concentrations and 4 mM acetyl-CoA were diluted in 100 mM sodium phosphate (pH 7.27). The assay further contained 1.25 mM DTNB, and the reactions were initiated by addition of 0.3 μg (7.8 pmol) of GlmA. When the kinetic parameters for acetyl-CoA were measured, 4 mM GlcN was used with various concentrations of acetyl-CoA. The absorbance at 412 nm was monitored for 3 min upon incubation of the reaction mixture at 30°C. To confirm that DTNB did not inhibit GlmA activity, we also used discontinuous assays as a control. In these assays the reaction was run in the absence of DTNB. After 30, 60, and 90 s, the assay was stopped by the addition of 6.4 M GuHCl in 100 mM sodium phosphate (pH 7.32). DTNB was then added, and the absorbance was measured at 412 nm in a SpectraMax M2 microplate reader (Molecular Devices). The kinetic parameters were determined by nonlinear regression using the program Prism4 (GraphPad). The molar extinction coefficient for DTNB in phosphate buffer at pH 7.27 was 14.15 × 103 M−1 cm−1, and in high salt concentrations (GuHCl) at pH 7.32 it was 13.7 × 103 M−1 cm−1.
B. subtilis and E. coli cell wall preparation.
Cell wall was prepared from B. subtilis 168 (Bacillus Genetic Stock Center) and E. coli MG 1655 (7) cells. Exponential-phase cells were harvested from 50-ml cultures at an OD600 of 0.9 by centrifugation (5,000 × g, 45 min, 4°C). The cell pellets were suspended in 1 ml of ice-cold distilled water, followed by immediate boiling in a water bath for 10 min. The insoluble material was collected (13,000 × g, 10 min), washed three times with water, and then resuspended in 1 ml of 50 mM Tris-HCl (pH 7.0). A mixture of purified peptidoglycan hydrolases (10 μg of each) was added and allowed to degrade the peptidoglycan content of the insoluble material at 30°C for 18 h. The peptidoglycan hydrolases used were mutanolysin (from Streptomyces globisporus [Sigma-Aldrich], 4,000 U/mg), N-acetylmuramic acid-l-Ala amidase AmiD (from E. coli, prepared as described previously [27]), and/or N-acetylglucosaminidase NagZBs (from B. subtilis, prepared as described previously [16]). After incubation, the enzymes were heat inactivated for 10 min at 95°C. The supernatants of the digestions were collected by centrifugation (13,000 × g, 10 min) and evaporated in a Speed-Vac. Finally, the dried pellets were resuspended in 100 μl of water. From this, 5-μl samples were used for analysis of GlmA activity as described below.
RESULTS
Cloning and purification of GlmA of C. acetobutylicum.
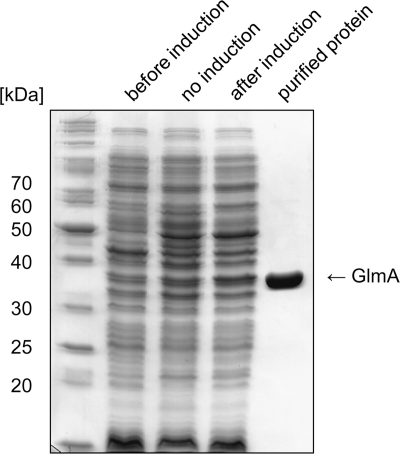
We recently characterized the GlcNAc/MurNAc kinase MurK of C. acetobutylicum (22). The encoding gene is located in an operon that presumably is involved in the rescue of peptidoglycan fragments (muropeptides). Since C. acetobutylicum processes non-N-acetylated murein sugars, in particular GlcN, we further proposed that N-acetylation of the cell wall amino sugars should occur in the reutilization process. The open reading frame downstream of murK on the genome of C. acetobutylicum is predicted to encode an N-acetyltransferase. This putative acetyltransferase was cloned, overexpressed as a cytoplasmic His6 fusion construct in E. coli and was purified by Ni2+ affinity chromatography. By SDS-PAGE analysis (Fig. 1) the protein, GlmA, appeared as one band of high purity and with an apparent molecular mass of 35 to 40 kDa, which is in agreement with the calculated mass (Mr) of 38,447 of GlmA-His6 fusion protein.
Fig. 1.
Purity of recombinant GlmA analyzed by SDS-PAGE and Coomassie brilliant blue staining. The protein was overproduced in E. coli M15 [pREP4] carrying pGlmA. Lane 1, protein size standard, lane 2, E. coli cell extract before induction; lane 3, E. coli cell extract, no induction; lane 4, E. coli cell extract after induction of GlmA; lane 5, 4 μg of purified GlmA.
GlmA is an N-acetyltransferase with specificity for GlcN.
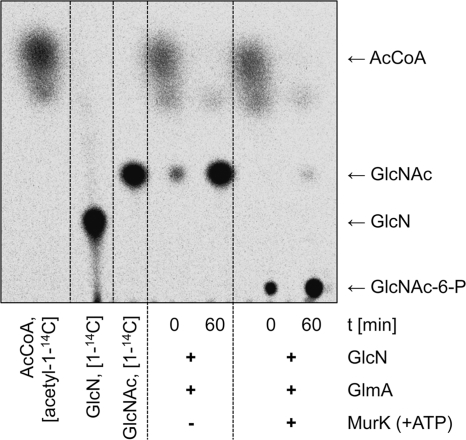
In a first attempt to examine the role of GlmA in the N-acetylation of non-N-acetylated peptidoglycan fragments, we tested the amino sugar GlcN as a possible substrate. Purified GlmA generated a radioactively labeled product with GlcN using [acetyl-1-14C]CoA as the cosubstrate (Fig. 2, lanes 4 and 5). With MurK, the GlcNAc/MurNAc kinase of C. acetobutylicum (22), and ATP, the product of the acetylation reaction was converted to a sugar phosphate only when GlcN had been previously acetylated by GlmA (Fig. 2, lanes 6 and 7). This indicates that GlcNAc is generated from GlcN by the action of GlmA, which can be further processed to GlcNAc-6-phosphate by MurK. Thus, GlmA is a GlcN N-acetyltransferase.
Fig. 2.
Acetylation of GlcN by GlmA with [acetyl-1-14C]CoA. Lanes 1 to 3, radioactive standards as indicated; lanes 5 and 6, time-dependent acetylation of GlcN; lanes 6 and 7, the acetylated reaction product was identified as GlcNAc by phosphorylation upon treatment with the GlcNAc/MurNAc kinase MurK and ATP, yielding GlcNAc-6-P. AcCoA, acetyl-CoA.
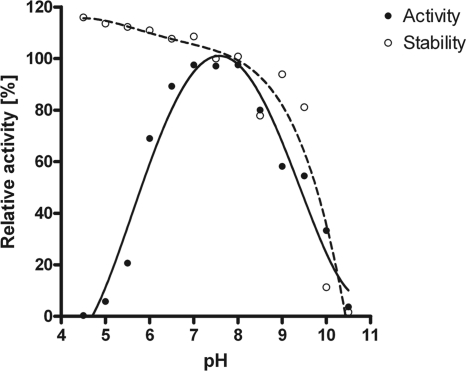
The pH dependency of GlmA was determined by quantification of the reaction product GlcNAc, as described in Materials and Methods. The relative activity was plotted against pH, which resulted in a bell-shaped curve (Fig. 3; see also Fig. S1 in the supplemental material). GlmA was fully active between pH 7 and pH 8, retained half-maximal activity at about pH 6, and was inactive below pH 5. GlmA was stable in the pH range 4.5 to 8. At greater than pH 8, the enzyme rapidly lost activity. To investigate whether GlmA requires divalent metal ions for its activity, the effect of EDTA was studied. GlmA activity was not inhibited by treatment with EDTA (data not shown) in either the presence or the absence of Mg2+. Thus, GlmA activity apparently does not require divalent metal ions.
Fig. 3.
pH profile of GlmA activity and stability. GlmA was incubated in various buffers at different pH and activity was determined with GlcN and [acetyl-1-14C]CoA by applied TLC as described in Materials and Methods. The buffers used were phosphate-citrate at pH 4.5 to 8.0, Tris-HCl at pH 7.5 to 9.0, and glycine-NaOH at pH 9.0 to 10.5. The data are means of four independent experiments; errors were <5%.
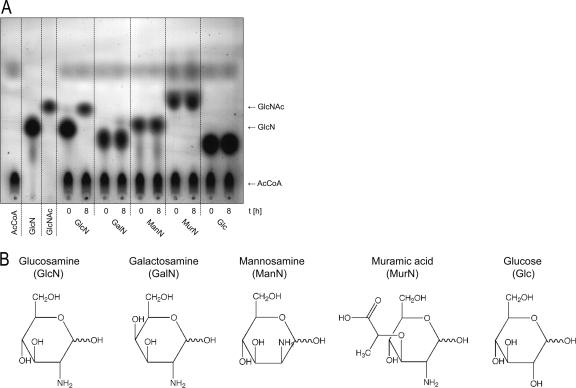
We determined the substrate specificity of GlmA and tested whether MurN and other amino sugars are recognized as substrates, thereby applying a nonradioactive acetylation assay as described in Materials and Methods. GlmA only acetylated GlcN, yielding GlcNAc of the tested amino sugar substrates (Fig. 4A). Both the 4-enantiomer GalN and the 2-enantiomer ManN, as well as MurN, the 3-lactic acid derivative of glucosamine, were not acetylated by GlmA (Fig. 4B). These results were also confirmed by the applied more sensitive radioactive acetylation assay (see Fig. S2 in the supplemental material), showing that GlmA of C. acetobutylicum is a GlcN-specific N-acetyltransferase.
Fig. 4.
GlmA is an N-acetyltransferase specific for GlcN. (A) Only GlcN (not GalN, ManN, or MurN amino sugars and not glucose [Glc]) was acetylated by GlmA using nonradioactive acetyl-CoA (AcCoA) as an acetyl donor. Lanes 1 to 3, standards as indicated. (B) Structures of the tested sugars.
Acetylation of chitosan oligomers and determination of the acetylation site.
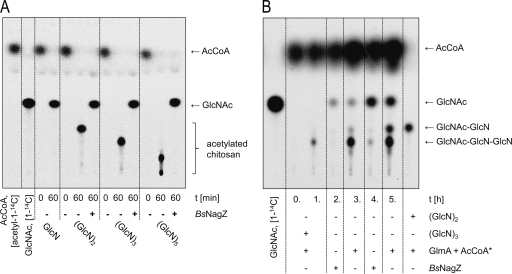
If GlmA is required for muropeptide rescue in C. acetobutylicum, the enzyme should be able to acetylate not only GlcN but also terminal GlcN residues of glycosides, which could then be processed by NagZ. To investigate whether GlmA also converted di- or oligosaccharides, we tested commercially available dimeric to pentameric fragments of chitosan, a linear homoglycan of β-1,4-glycosidically linked GlcN residues, as substrates. We observed that GlmA could acetylate all tested chitosan oligomers. Subsequent treatment with N-acetylglucosaminidase NagZBs released GlcNAc (Fig. 5A). To further examine the specific acetylation site in the chitosan glycan strand, we acetylated chitosan trimer with GlmA as before but inactivated GlmA prior to treatment with NagZBs. With this procedure, the remaining dimeric product (GlcN)2 could be detected by radioactive acetylation only if GlmA was freshly added (Fig. 5B). This indicates that GlmA acetylated only the terminal nonreducing glucosamine residue of chitosan.
Fig. 5.
(A) Acetylation of GlcN and dimeric to pentameric β-1,4-glycosidically linked GlcN residues (chitosan oligosaccharides) by GlmA. The acetylated reaction products, using [acetyl-1-14C]CoA, were separated by TLC as described in Materials and Methods. Treatment with N-acetylglucosaminidase NagZBs released radioactively labeled GlcNAc. Lanes 1 to 3, standards as indicated. (B) The terminal nonreducing GlcN residue of chitosan was acetylated by GlmA. Lane 1, standard as indicated. After acetylation of chitosan trimer, using [acetyl-1-14C]CoA, with GlmA but prior to treatment with NagZBs, GlmA was inactivated. Therefore, the terminal, radioactively labeled GlcNAc residue was released from the trimer. The remaining dimeric chitosan product could be detected by repeated radioactive acetylation with newly added GlmA. AcCoA, acetyl-CoA.
GlmA acetylates non-N-acetylated muropeptides.
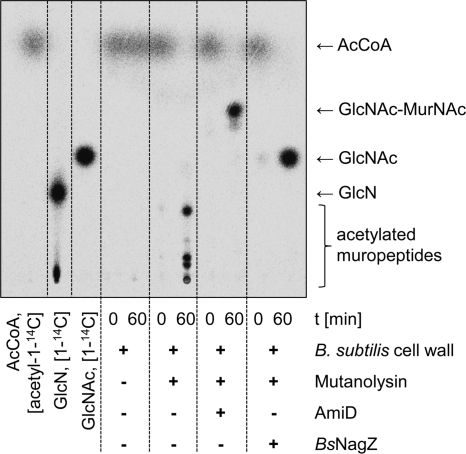
We used the radioactive acetylation assay to determine whether GlmA can acetylate non-N-acetylated glucosamine residues in muropeptides derived from Gram-positive peptidoglycan. Cell wall preparations from B. subtilis were first degraded with the purified lytic enzyme mutanolysin (an N-acetylmuramidase) and then further processed by AmiD (an N-acetylmuramic acid-l-Ala amidase) and/or NagZBs (an N-acetylglucosaminidase). Thereby released muropeptides, disaccharides and monosaccharides were subjected to acetylation with GlmA (Fig. 6). We observed that the terminal nonreducing GlcN residue of mutanolysin formed muropeptides had to be acetylated by GlmA prior to the release of terminal GlcNAc by N-acetylglucosaminidase NagZBs (see Fig. S3 in the supplemental material). Compared to B. subtilis, peptidoglycan of Gram-negative E. coli does not contain N-deacetylated modifications. These cell wall preparations could not be acetylated by GlmA (see Fig. S4 in the supplemental material).
Fig. 6.
GlmA acetylates terminal GlcN of non-N-acetylated muropeptides prepared from B. subtilis cell wall. Muropeptides were generated by mutanolysin digestion of the cell wall peptidoglycan that could be acetylated, using [acetyl-1-14C]CoA, by GlmA at GlcN residues of N-deacetylated peptidoglycan. After treatment with mutanolysin and AmiD, an acetylated disaccharide product can be obtained upon GlmA acetylation. The radioactively labeled GlcNAc of this product could be released after treatment with NagZBs, which was added subsequent to the acetylation reaction. Lanes 1 to 3, standards as indicated. AcCoA, acetyl-CoA.
Kinetic parameters of GlmA.
The kinetic parameters of the glucosamine N-acetyltransferase GlmA were determined with a nonradioactive, continuous spectrophotometric assay. Thereby, GlmA activity was measured by using Ellman's reagent to quantitate the release of free CoA. The results were evaluated by nonlinear regression and followed the Michaelis-Menten equation (see Fig. S5 in the supplemental material). Kinetic data in Table 1 display that GlmA revealed a slightly lower Vmax value for GlcN compared to the chitosan oligosaccharides (GlcN)2 and (GlcN)3. The Km value of GlcN was about 2- to 3-fold higher than for chitosans composed of more than one GlcN residue. The higher affinity and reaction rate, as well as a 3- to 4-fold higher catalytic efficiency, for chitosan oligomers indicated the preference for glycoside substrates for the acetyltransferase GlmA.
Table 1.
Kinetic parameters of GlmAa
| Substrate | Vmax (μmol/min/mg) | Km (μM) | kcat (1/s) | kcat/Km (1/s/mM) |
|---|---|---|---|---|
| GlcN | 2.11 | 114.3 | 1.35 | 11.8 |
| (GlcN)2 | 2.69 | 64.5 | 1.72 | 26.7 |
| (GlcN)3 | 2.60 | 39.2 | 1.66 | 42.4 |
| AcCoA | 2.10 | 46.4 | 1.34 | 28.9 |
Kinetic parameters were determined in 100 mM sodium phosphate (pH 7.27) at 30°C as described in Materials and Methods. Data represent the means of two independent experiments. Standard errors were <5%. AcCoA, acetyl-CoA.
DISCUSSION
Cell wall turnover is a catabolic pathway of bacteria by which up to 50% of the cell wall is turned over in one generation (20). Whether cell wall fragments released in this way are also recovered in Gram-positive bacteria is currently unclear (cf. our recent review [23]). The Gram-positive firmicute C. acetobutylicum is an important solvent producer strain and widely used for metabolic engineering. The initiation of the solvent production in this organism is coupled with endospore formation and associated with the thinning of the peptidoglycan layer, which apparently results in the increased resistance against the solvents (15). A better knowledge of cell wall modifications, turnover, and recycling is mandatory for optimization of solvent production in C. acetobutylicum by rational approaches. In our effort to contribute to the understanding of cell wall catabolic processes in clostridia and Gram-positive bacteria in general, we cloned and characterized an N-acetyltransferase of C. acetobutylicum, GlmA, that is capable of acetylating GlcN and glucosaminides to GlcNAc and N-acetylglucosaminides, respectively. GlmA belongs to the GNAT (GCN5-related N-acetyltransferases) superfamily of functionally diverse acetyltransferases that catalyze the transfer of an acetyl group from acetyl-CoA to the primary amine of a wide range of acceptor substrates (11). Members of this enzyme superfamily include aminoglycoside N-acetyltransferases, serotonin N-acetyltransferase, the glucosamine-6-phosphate N-acetyltransferase of yeast and mammals, the histone acetyltransferases, mycothiol synthase, and the Fem family of amino acyl transferases, among others (28). GlmA shows only very weak amino acid sequence homology with characterized enzymes of the GNAT family but exhibits an overall identity of up to 36% with predicted N-acetyltransferases of various species of the Clostridium and Thermoanaerobacter genera, including the two extremely thermophilic ethanol- and acetate-producing organisms T. mathranii strain A3 and T. tengcongensis strain MB4 (14, 34). GlcN N-acetyltransferase activity has been identified in crude pigeon liver extracts and membrane fractions isolated from oat coleoptile segments, which are so far the only descriptions of such an enzymatic activity (9, 21). This is thus the first report of a cloned and characterized GlcN N-acetyltransferase (EC 2.3.1.3). Our results indicate that the substrates of GlmA other than GlcN are GlcN-containing muropeptides, as well as GlcN glycosides such as β-1,4-linked chitosan oligomers. These substrates were acetylated by the enzyme at the terminal nonreducing GlcN. Kinetic analysis of GlmA showed that the catalytic efficiency of the enzyme is about 3- to 4-fold higher for chitosan oligomers (GlcN)2-3 compared to the monomeric GlcN. This indicates that GlmA is primarily involved in the acetylation of di- or oligosaccharides that are transported into the cytoplasm. Similar observations have previously indicated that heparin/heparan sulfate-α-glucosamine N-acetyltransferases from purified lysosomal membrane fractions of human placenta and rat liver N-acetylate with high efficiency terminal α-linked GlcN residues of sulfated di- and tetraglycosaminoglycan chains derived from heparin and heparan sulfate (4, 18). De-N-acetylated GlcNAc residues are found primarily in the peptidoglycan of Gram-positive bacterial species (29), such as S. pneumoniae, B. cereus, B. subtilis, and L. monocytogenes, as well as in the Gram-negative bacterium Rhodopseudomonas viridis (25). Our data confirmed that GlcN residues are not present in the cell wall peptidoglycan of the Gram-negative bacterium E. coli.
GlmA from C. acetobutylicum apparently did not act on intact peptidoglycan isolated from B. subtilis, presumably because terminal nonreducing GlcN residues are not present. These, however, can be generated by the action of mutanolysin, which is a muramidase with broad specificity that acts on non-N-acetylated peptidoglycan similar to the lyc gene encoding autolytic lysozyme of C. acetobutylicum. The autolytic extracellular muramidase Lyc of C. acetobutylicum is active on nonacetylated cell wall peptidoglycan and generates GlcN-containing muropeptides such as GlcN-MurNAc-peptide compounds (10). NagZ-like N-acetylglucosaminidases involved in muropeptide processing are highly specific for N-acetylated substrates (17), which is presumably also true for the NagZ ortholog of C. acetobutylicum. Therefore, N-acetylation of terminal GlcN residues is required for processing of muropeptides by NagZ-like glycosidases. In an accompanying study, we show that both amino sugars GlcNAc and MurNAc, presumably released by NagZ of C. acetobutylicum, are phosphorylated by the MurK kinase, yielding GlcNAc- and MurNAc-6-phosphate in the cytoplasm of C. acetobutylicum (22). We conclude therefore that GlmA is required for the recovery of de-N-acetylated muropeptides during putative cell wall recycling in C. acetobutylicum. Interestingly, a glucosamine/glucosaminide N-acetyltransferase should also be present in B. subtilis. The Bacillus enzyme is expected to be not cytoplasmic like GlmA but secreted, since muropeptide catabolism occurs in the wall compartment in this organism (16). However, no apparent ortholog of glmA was found on the chromosome of B. subtilis. Furthermore, both GlcN and MurN residues are present in the peptidoglycan of B. subtilis (3), suggesting that a MurN N-acetyltransferase may also be present in this organism. GlmA of C. acetobutylicum had no specificity for MurN. Further studies are required to clarify the role of GlmA in cell wall rescue in C. acetobutylicum.
Supplementary Material
ACKNOWLEDGMENTS
We thank G. Bennett (Rice University) and Hubert Bahl (University of Rostock) for kindly providing C. acetobutylicium genomic DNA.
This study was supported by grant MA2346/5 and Heisenberg stipend MA2346/3 to C.M. from the German Research Foundation (DFG).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 22 July 2011.
REFERENCES
- 1. Araki Y., Fukuoka S., Oba S., Ito E. 1971. Enzymatic deacetylation of N-acetylglucosamine residues in peptidoglycan from Bacillus cereus cell walls. Biochem. Biophys. Res. Commun. 45:751–758 [DOI] [PubMed] [Google Scholar]
- 2. Araki Y., Nakatani T., Hayashi H., Ito E. 1971. Occurrence of non-N-substituted glucosamine residues in lysozyme-resistant peptidoglycan from Bacillus cereus cell walls. Biochem. Biophys. Res. Commun. 42:691–697 [DOI] [PubMed] [Google Scholar]
- 3. Atrih A., Bacher G., Allmaier G., Williamson M. P., Foster S. J. 1999. Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP 5 in peptidoglycan maturation. J. Bacteriol. 181:3956–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bame K. J., Rome L. H. 1985. Acetyl coenzyme A:α-glucosaminide N-acetyltransferase: evidence for a transmembrane acetylation mechanism. J. Biol. Chem. 260:11293–11299 [PubMed] [Google Scholar]
- 5. Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bethesda Research Laboratories 1986. BRL pUC host: Escherichia coli DH5α competent cells. Focus 8:9 [Google Scholar]
- 7. Blattner F. R., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462 [DOI] [PubMed] [Google Scholar]
- 8. Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 9. Chou T. C., Soodak M. 1952. The acetylation of d-glucosamine by pigeon liver extracts. J. Biol. Chem. 196:105–109 [PubMed] [Google Scholar]
- 10. Croux C., Canard B., Goma G., Soucaille P. 1992. Purification and characterization of an extracellular muramidase of Clostridium acetobutylicum ATCC 824 that acts on non-N-acetylated peptidoglycan. Appl. Environ. Microbiol. 58:1075–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dyda F., Klein D. C., Hickman A. B. 2000. GCN5-related N-acetyltransferases: a structural overview. Annu. Rev. Biophys. Biomol. Struct. 29:81–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hayashi H., Araki Y., Ito E. 1973. Occurrence of glucosamine residues with free amino groups in cell wall peptidoglycan from bacilli as a factor responsible for resistance to lysozyme. J. Bacteriol. 113:592–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jaeger T., Arsic M., Mayer C. 2005. Scission of the lactyl ether bond of N-acetylmuramic acid by Escherichia coli “etherase.” J. Biol. Chem. 280:30100–30106 [DOI] [PubMed] [Google Scholar]
- 14. Larsen L., Nielsen P., Ahring B. K. 1997. Thermoanaerobacter mathranii sp. nov., an ethanol-producing, extremely thermophilic anaerobic bacterium from a hot spring in Iceland. Arch. Microbiol. 168:114–119 [DOI] [PubMed] [Google Scholar]
- 15. Linhová M., et al. 2010. Development of flow cytometry technique for detection of thinning of peptidoglycan layer as a result of solvent production by Clostridium pasteurianum. Folia Microbiol. 55:340–344 [DOI] [PubMed] [Google Scholar]
- 16. Litzinger S., et al. 2010. Muropeptide rescue in Bacillus subtilis involves sequential hydrolysis by β-N-acetylglucosaminidase and N-acetylmuramyl-l-alanine amidase. J. Bacteriol. 192:3132–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Litzinger S., et al. 2010. Structural and kinetic analysis of Bacillus subtilis N-acetylglucosaminidase reveals a unique Asp-His dyad mechanism. J. Biol. Chem. 285:35675–35684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meikle P. J., Whittle A. M., Hopwood J. J. 1995. Human acetyl-coenzyme A:α-glucosaminide N-acetyltransferase: kinetic characterization and mechanistic interpretation. Biochem. J. 308:327–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ohno N., Yadomae T., Miyazaki T. 1982. Identification of 2-amino-2-deoxyglucose residues in the peptidoglucan of Streptococcus pneumoniae. Carbohydr. Res. 107:152–155 [DOI] [PubMed] [Google Scholar]
- 20. Park J. T., Uehara T. 2008. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol. Mol. Biol. Rev. 72:211–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Piro G., Buffo M., Dalessandro G. 1994. Membrane-bound glucosamine acetyltransferase in coleoptile segments of Avena sativa. Physiol. Plant 90:181–186 [Google Scholar]
- 22. Reith J., Berking A., Mayer C. 2011. Characterization of an N-acetylmuramic acid/N-acetylglucosamine kinase of Clostridium acetobutylicum J. Bacteriol. 193:5386–5392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reith J., Mayer C. 2011. Peptidoglycan turnover and recycling in Gram-positive bacteria. Appl. Microbiol. Biotechnol. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 24. Riddles P. W., Blakeley R. L., Zerner B. 1979. Ellman's reagent: 5,5′-dithiobis(2-nitrobenzoic acid): a reexamination. Anal. Biochem. 94:75–81 [DOI] [PubMed] [Google Scholar]
- 25. Schmelzer E., Weckesser J., Warth R., Mayer H. 1982. Peptidoglycan of Rhodopseudomonas viridis: partial lack of N-acetyl substitution of glucosamine. J. Bacteriol. 149:151–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Spiess C., Beil A., Ehrmann M. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97:339–347 [DOI] [PubMed] [Google Scholar]
- 27. Uehara T., Park J. T. 2007. An anhydro-N-acetylmuramyl-l-alanine amidase with broad specificity tethered to the outer membrane of Escherichia coli. J. Bacteriol. 189:5634–5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vetting M. W., et al. 2005. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 433:212–226 [DOI] [PubMed] [Google Scholar]
- 29. Vollmer W. 2008. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol. Rev. 32:287–306 [DOI] [PubMed] [Google Scholar]
- 30. Vollmer W., Blanot D., De Pedro M. A. 2008. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32:149–167 [DOI] [PubMed] [Google Scholar]
- 31. Vollmer W., Tomasz A. 2000. The pgdA gene encodes for a peptidoglycan N-acetylglucosamine deacetylase in Streptococcus pneumoniae. J. Biol. Chem. 275:20496–20501 [DOI] [PubMed] [Google Scholar]
- 32. White R. J. 1968. Control of amino sugar metabolism in Escherichia coli and isolation of mutants unable to degrade amino sugars. Biochem. J. 106:847–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. White R. J., Pasternak C. A. 1967. The purification and properties of N-acetylglucosamine 6-phosphate deacetylase from Escherichia coli. Biochem. J. 105:121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xue Y., Xu Y., Liu Y., Ma Y., Zhou P. 2001. Thermoanaerobacter tengcongensis sp. nov., a novel anaerobic, saccharolytic, thermophilic bacterium isolated from a hot spring in Tengcong, China. Int. J. Syst. Evol. Microbiol. 51:1335–1341 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.