Abstract
By labeling the β′ subunit of RNA polymerase (RNAP), we used fluorescence microscopy to study the spatial distribution and diffusive motion of RNAP in live Escherichia coli cells for the first time. With a 40-ms time resolution, the spatial distribution exhibits two or three narrow peaks of 300- to 600-nm full width at half-maximum that maintain their positions within 60 nm over 1 s. The intensity in these features is 20 to 30% of the total. Fluorescence recovery after photobleaching (FRAP) measures the diffusive motion of RNAP on the 1-μm length scale. Averaged over many cells, 53% ± 19% of the RNAP molecules were mobile on the 3-s timescale, with a mean apparent diffusion constant 〈DRNAP〉 of 0.22 ± 0.16 μm2-s−1. The remaining 47% were immobile even on the 30-s timescale. We interpret the immobile fraction as arising from RNAP specifically bound to DNA, either actively transcribing or not. The diffusive motion of the mobile fraction (fmobile) probably involves both one-dimensional sliding during nonspecific binding to DNA and three-dimensional hopping between DNA strands. There is significant cell-to-cell heterogeneity in both DRNAP and fmobile.
INTRODUCTION
In prokaryotes such as Escherichia coli, several thousand copies of a single RNA polymerase (RNAP) are responsible for all transcription. Previous work has used fluorescence microscopy to study the spatial distribution of green fluorescent protein (GFP)-labeled RNAP in both E. coli (4) and Bacillus subtilis (15) cells that were chemically fixed following growth in rich medium. In both species, narrow regions of high local GFP-RNAP concentration are observed atop a broader background concentration. Such “RNAP foci” are thought to be transcription foci, i.e., localized regions of intensive transcription, including rRNA. Accordingly, the narrow features were not observed for growth in minimal medium or following treatments designed to mimic nutrient starvation (4). The underlying cause of such spatial organization in prokaryotes is not clear (17). Quantifying the number, intensity, and spatial extent of RNAP foci in live cells is difficult due to the small size of the E. coli nucleoid, the presence of a background RNAP distribution, the dependence of the features on growth conditions, and the dynamic nature of the spatial distributions.
Here we present a detailed quantitative study of the spatial distribution and movement of RNAP in live bacterial cells, specifically a K-12 strain of E. coli. The spatially narrow, immobile RNAP features that we observe appear qualitatively similar to the foci in fixed cells reported earlier. To capture the underlying dynamics, we imaged RNAP with a 40-ms time resolution for 0.8-s intervals. Each cell exhibits two or three narrow RNAP foci with a full width at half-maximum intensity (FWHM) of about 500 nm, significantly broader than the diffraction limit. These narrow features account for 20 to 30% of the total intensity and maintain their positions to within 60 nm over 1 s, during which time the broader background RNAP distribution fluctuates substantially. These features are not observed in cells where transcription has been halted by the addition of rifampin, nor are they present in the distribution of the globular, soluble protein Kaede.
In addition, long-range RNAP motion on the 1-μm length scale was monitored by fluorescence recovery after photobleaching (FRAP) (6, 12, 13, 32). On average, roughly half the population of RNAP molecules is mobile over the 2- to 3-μm length of the cell on a timescale of 5 to 10 s, with an effective diffusion constant DRNAP of approximately 0.2 μm2-s−1. Remarkably, the DRNAP is comparable to diffusion constants measured in vitro for RNAP sliding along the contour of bare, double-stranded DNA (dsDNA) while nonspecifically bound (10, 26, 30). The other half of the population is immobile on a timescale of at least 30 to 60 s. We attribute the immobile fraction to RNAP copies that are specifically bound to DNA; at least some of these copies are presumably actively transcribing DNA.
MATERIALS AND METHODS
Strain construction.
pRM222, a derivative of pRM103 (21), was engineered by making an in-frame fusion of GFP (from pEGFP-N3; Clontech) to rpoC to make a plasmid carrying rpoC::GFP. To construct the β′::GFP fusion strain, pRM222 was transformed into RL324 (21) and recombination was selected by growth on LB-kanamycin (Kan) plates and repeated growth in LB-Kan liquid culture (25). Plasmid loss from single-colony isolates was confirmed by loss of ampicillin resistance. Replacement of rpoC with the rpoC::GFP fusion was confirmed by PCR amplification of genomic DNA and by fluorescence imaging of GFP. A P1vir lysate grown on this strain, RL1201, was used to transduce RL301 (a W3110 derivative [21]) to give strain RL1314. The growth of RL1314 was compared to that of RL301; no significant differences were found. At 30°C, the doubling time of RL1314 was 60 min in Lennox L broth (LB) and 90 min in a defined complete rich media (EZRDM).
Media and growth conditions for microscopy.
Rich, defined growth medium (EZRDM) contained the following per 100 ml of volume: 20 ml 10× supplement EZ, 10 ml 5× ACGU, 10 ml 10× MOPS (morpholinepropanesulfonic acid)-modified rich buffer without potassium or NaCl, 1 ml 0.132 M K2HPO4, 1 ml 20% (wt/vol) glucose, and 1.52 ml 5 M NaCl (23). The medium was supplemented with kanamycin to a final concentration of 50 μg/ml just prior to use. Three-milliliter cultures were grown overnight at 30°C with shaking at 200 rpm. Medium components (supplement EZ, ACGU, and MOPS-modified buffer) were purchased from Teknova (Hollister, CA), and the remaining chemicals were purchased from Sigma-Aldrich (St. Louis, MO).
Sample chamber preparation.
Coverslips (15 mm diameter, no. 1 thickness; BellCo Glass, Vineland, NJ) were sonicated for 30 min in acetone, rinsed with ultrapure water, sonicated for 30 min in 1 M KOH, and rinsed a second time. To improve cell adherence, the coverslips were soaked in 0.01% (wt/vol) 150,000- to 300,000-MW poly-l-lysine in water for 20 min and then rinsed with ultrapure water to remove excess polylysine solution. Coverslips were prepared not more than 48 h before each experiment. The clean, polylysine-coated coverslips were used to construct a 70-μl, temperature-controlled sample chamber (RC-20H; Warner Instruments, Hamden, CT). The whole assembly was brought into contact with the microscope objective and warmed to 30°C.
A 15- to 30-μl volume of an overnight culture was added to 3 ml prewarmed medium and grown with shaking (200 rpm) to mid-log phase (optical density at 600 nm [OD600] of ∼0.4). From these cultures, 20 μl was added to the sample chamber at 30°C. The chamber was sealed with a second coverslip, and warm, aerated growth medium was allowed to flow through the chamber (0.2 to 0.5 ml/min). The sample chamber and medium were maintained at 30°C by a dual-channel temperature controller (TC-344B; Warner Instruments). Cells adhered to the coverslip grew and divided with a doubling time of about 100 min, compared with a 90-min doubling time in liquid culture. As a control for the effects of polylysine, some cells were studied by sandwiching them between a thin 1% agar pad and a clean coverslip without polylysine.
For the fixation study, 900 μl of cells at an OD600 of 0.4 were added to 100 μl of 37% formaldehyde and left to fix for 1 h at room temperature. Cells were spun down and resuspended in 1× phosphate-buffered saline (PBS) (pH 7.4) before plating in the flow cell.
To test for possible phototoxic effects, a field of view containing roughly 10 cells was exposed to the probe beam for 25 min. This bleached >75% of the enhanced GFP (EGFP) in the cells. During imaging for an additional 50 min, the integrated intensity in the cells rose continuously (indicating production and folding of new EGFP), and the cells continued to elongate and divide normally.
Optical microscopy.
The optical setup for microscopy was similar to that previously reported (12). Samples were imaged on an inverted microscope (Eclipse TE300; Nikon) with a 100×, 1.3-numerical-aperture (NA) phase-contrast objective (Plan Fluor; Nikon) equipped with a dichroic mirror (Chroma 51019bs) and emission filter (Chroma HQ510LP). An argon ion laser (488 nm; Melles Griot 532-AP-A01) was split into two beams using standard optics. For the FRAP experiments, a focused beam (1.0-μm full width at half-maximum height [FWHM], 18-kW/cm2 peak intensity, 150 ms in duration) was used to selectively bleach one end of the cell at t = 0. A broader beam (23-μm FWHM, 350-W/cm2 peak intensity, 100 to 200 ms of exposure at 2 to 5 Hz) provided uniform illumination used to probe the recovering fluorescence distribution versus time. Movies of the same cell at different frame rates in time-lapse mode allowed access to multiple timescales while minimizing photobleaching effects of the probe laser. For the fast movies used for narrow-peak localization and fitting, a more intense probe beam (11-μm FWHM, 2.3-kW/cm2 peak intensity, 1.9 ms of exposure at 500 Hz) was used in order to freeze possible motion.
To compare DNA and RNAP spatial distributions in the same cell, two-color fluorescence was used to image RNAP in the green channel and the DNA-binding dye DRAQ5 (BioStatus Limited, Shepshed, Leicestershire, United Kingdom) in the red channel. Cells were grown to mid-log phase, and a 100-μl aliquot of cells was mixed with 100 μl of 4 μM DRAQ5 for a final concentration of 2 μM DRAQ5. The cells were allowed to stain for 10 min at room temperature and then adhered to a polylysine-coated coverslip for imaging. Both an argon ion laser (13-μm FWHM, 2.5-kW/cm2 peak intensity) and a helium-neon laser (633 nm; Melles Griot 1144P) (26-μm FWHM, 180-W/cm2 peak intensity) were coupled into the microscope using a dual-band-pass dichroic mirror (Chroma z488/633rpc). The fluorescence was collected with a 100× objective and passed through the same mirror. Each color of fluorescence was imaged separately by selecting one of two emission filters using a filter changer (FW102; ThorLabs, Newton, NJ): for GFP detection, HQ510/20 M, and for DRAQ5 detection, HQ700/75 M.
The timing of the laser pulses was controlled by a digital delay generator (DG535; Stanford Research Systems) and fast mechanical shutters (LS2; Uniblitz). An additional shutter (VS25; Uniblitz) prevented fluorescence intensity during the bleach pulse from reaching the camera. Images were captured on an electron-multiplying charge-coupled device (EMCDD), either DV-860 or DV-897 (Andor Technologies, Belfast, Northern Ireland), and stored as multiplane tif files. Additional image magnification after the microscope was realized with a home-built optical system. The total magnification of the imaging system was calibrated using a USAF 1951 resolution test target and fitting the intensity profiles to sine waves. The effective pixel size was typically 100 nm for the DV-860 and 65 nm for the DV-897. Postmicroscope magnification varied in the range from ×2.5 to ×2.7 from experiment to experiment due to realignment of optics.
Image analysis for FRAP experiments.
As shown in Fig. 1 and 2, the RNAP spatial distribution typically exhibits two distinct lobes of intensity which we label A and B, with A being the lobe that is photobleached at t = 0. We obtained the effective RNAP diffusion constant by measuring the intensities in lobe A (IA) and in lobe B (IB) as a function of time. The fractional intensity in lobe A (Fig. 1) is calculated as fA(t) = IA(t)/[IA(t) + IB(t)]. The ratio removes the effect of overall photobleaching by the probe laser. We obtained an effective RNAP diffusion constant by fitting the fractional intensity to the form
| (1) |
with adjustable constants c1, τ, and c2. Here c1 is a measure of the fraction of the population of RNAP that redistributes from lobe B to lobe A, τ is the time constant of the redistribution, and c2 = fA(∞) is a measure of the asymmetry of the distribution at long times.
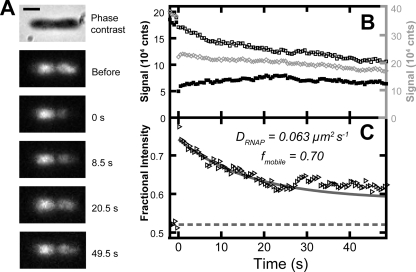
Fig. 1.
(A) Time course of fluorescence micrographs of RL1314 before and after photobleaching. Each 500-ms frame is normalized by rescaling to the maximum intensity to correct for photobleaching by the probe laser. The scale bar is 1 μm. (B) The integrated intensity in each region of the cell as a function of time. The left half [IA(t), black open squares] and right half [IB(t), black filled squares] of the cell use the ordinate scale to the left, and the total intensity in the cell (gray open diamonds) uses the ordinate scale to the right. (C) Fractional intensity fA(t) as a function of time after the photobleach. The best-fit exponential decay to the first 30 s is plotted as a solid gray line, and the fractional intensity value before the bleach is plotted as a dashed gray line.
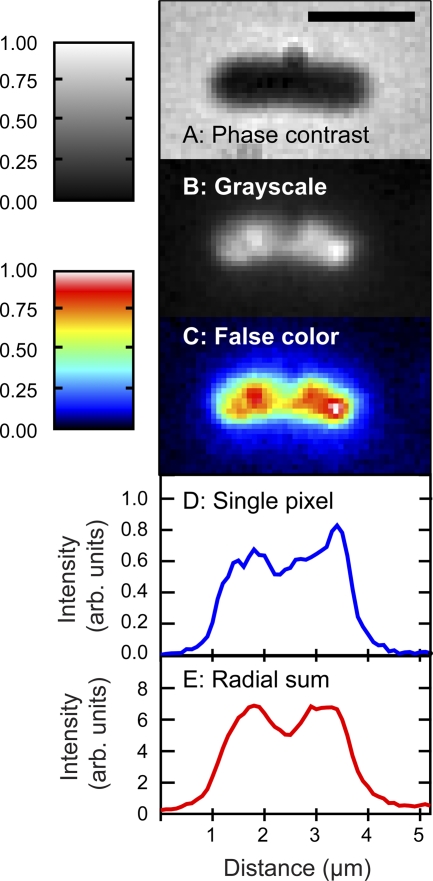
Fig. 2.
(A) Phase-contrast image of a characteristic cell. (B) Green fluorescence image of the same cell, showing the distribution of RNAP. The grayscale applies to panels A and B. (C) False-color representation for the image in panel B, with corresponding color scale. (D) One-pixel-wide axial intensity profile. (E) Axial intensity profile summed over the entire transverse dimension. Scale bar is 2 μm. The integration time was 200 ms.
We wrote custom functions to perform the image analysis operations in Matlab (R2010a; Mathworks, Natick, MA) and ImageJ (1.44d http://rsb.info.nih.gov/ij/). Multiplane tif files were read into Matlab and the metadata parsed to retrieve time stamp information. A region of interest was drawn around each cell, and the average background was subtracted from it. The cell was rotated so that its long axis was parallel to the x axis. The same region and orientation were used for the phase-contrast image of the cell. To locate the dividing line between lobes A and B, a one-dimensional longitudinal intensity profile I(x) was obtained by summing the intensities of all pixels in the (transverse) y direction at each axial position x (Fig. 2E). The pixel corresponding to the minimum of I(x) between the two lobes was used as the dividing line which separated the cell into bins A and B. The intensity in this central pixel was divided equally between bins A and B. The outside edges of the bins were the pixels corresponding to 10% of the maximum height of the axial profile I(x). The integrated intensities in bins A and B were used to calculate fA(t) and also to measure the time-dependent degree of photobleaching of the cell due to the probe laser. The delay time between the end of the bleach pulse and the start of the first frame was at most 20 ms, much shorter than the 200 to 500 ms between frames.
Obtaining an accurate estimate of the effective diffusion constant DRNAP from FRAP recovery data in the case of a nonuniform axial intensity distribution is not straightforward. In practice, we fit the first 20 to 30 s of the recovery of fA(t) to the model function in equation 1. The first frame after photobleaching is excluded from the analysis. The time constant τ was converted to an effective diffusion constant using D = Leff2/π2τ. The effective cell length Leff was approximated as the full width at half-maximum intensity (FWHM) of the longitudinal intensity profile I(x) (32). Each half-height was measured relative to the nearest peak using a cubic smoothing spline to interpolate between pixels.
We choose this method instead of the Fourier decomposition used previously (12, 13) primarily for its simplicity. For the more uniform axial intensity distribution typically observed for cytoplasmic GFP or Kaede (a GFP-sized fluorescent protein), the method using fA(t) gives diffusion constants in excellent agreement with those from the more rigorous Fourier analysis of the first cosine mode (12, 13). For FRAP data on nine cells expressing cytoplasmic Kaede, the two methods gave results that were statistically indistinguishable from each other (paired two-sided t test, P = 0.46). Evidently, for the cylindrical problem with end caps, fA(t) is an excellent proxy for the lowest cosine mode. Possible systematic errors in DRNAP are difficult to assess. As described below, the region of the nucleoid has roughly cylindrical symmetry. The dip in RNAP intensity between lobes A and B is due to a lower density of RNAP, not to a narrowing of the DNA spatial distribution. The low-density region at midcell may be a region of faster or slower RNAP diffusion than the body of the lobes.
In general, the best-fit asymptote c2 does not match the prebleach value fA(−∞). The deviation is usually in the direction of incomplete recovery, as if some fraction of RNAP is mobile on the 20- to 30-s time scale and another fraction is not. The mobile fraction fmobile is calculated as fmobile  [c2 − fA(0)]/[fA(−∞) − fA(0)], where fA(−∞) is the mean value before the bleach pulse, fA(0) is the value in the first frame after the bleach, and c2 is the fitting parameter defined above.
[c2 − fA(0)]/[fA(−∞) − fA(0)], where fA(−∞) is the mean value before the bleach pulse, fA(0) is the value in the first frame after the bleach, and c2 is the fitting parameter defined above.
Movies of fast RNAP motion.
In order to obtain fast snapshots of the RNAP distribution and to characterize RNAP motion with subsecond resolution, we omitted the bleach pulse and acquired 400-frame movies with a frame duration of only 2 ms. This required a 10-fold-higher illumination intensity. A rectangular region surrounding the cell was selected, and the average pixel value from a three-pixel-wide frame around the border of this region was subtracted from the image. The integrated intensity within each frame was scaled to make the total intensity constant, which corrects for photobleaching. To improve the signal/noise ratio, we averaged 20 successive frames to make a movie of 20 40-ms frames. Because of this averaging, we are unable to assess the dynamics on any shorter timescale.
These averaged images were then fit to the sum of multiple two-dimensional Gaussian functions. In practice, eight or more Gaussian functions were needed to capture all of the features in the images. These eight Gaussian functions are characterized by 49 parameters whose values were determined by a weighted least-squares fit. In brief, this computationally intensive fitting process yields information on the position, spatial extent, shape, and integrated intensity in each of the features. For a more detailed description of the fitting method, see “Quantifying features present in RNAP spatial distribution” in the supplemental material.
RESULTS
Coarse RNA polymerase distribution in live cells.
The distribution of RNA polymerase in live cells grown in defined rich medium seems qualitatively consistent with images of fixed cells grown in rich media (3, 4, 15). Fluorescence images of cells expressing RNA polymerase fused to EGFP typically exhibit a dual-lobed spatial distribution (Fig. 2). Under rich conditions, these two lobes occur for essentially all cells, whether or not the phase-contrast image exhibits pinching indicative of septation. This coarse distribution is qualitatively similar to that of DNA stained by DRAQ5, including a region of decreased intensity near the midplane of the cell (Fig. 3). However, within each RNAP lobe, we also observe narrow features not seen in the DNA distribution. These are observed as peaks and shoulders in one-pixel-wide axial intensity profiles (Fig. 2D). A grayscale image accentuates the intensity differences between the two lobes (Fig. 2B), while the narrow features can be seen more clearly on a false-color image (Fig. 2C).
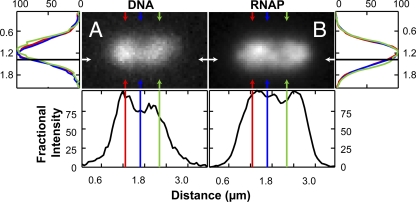
Fig. 3.
Two fluorescence images of the same cell stained with the DNA-specific dye DRAQ5 (A) and expressing β′-GFP (B). The axial intensity profile is plotted at the location of the white arrowheads; three transverse profiles are plotted for the left (red), center (blue), and right (green) portions of the cell. The profiles are averages over 300-nm-wide swaths (5 pixels) and are normalized to the maximum intensity. The image integration time is 450 ms.
We briefly studied cells grown at a higher growth rate in LB (doubling time of 60 min, compared with 90 min in EZRDM), which is similar to the growth conditions in the earlier fixed-cell studies (4). In LB, the narrow features are even more prominent and perhaps more numerous (see Fig. S1 in the supplemental material). However, the large amount of background fluorescence from the LB medium itself presented severe imaging difficulties. We chose to limit the dynamical imaging experiments to a single medium that could be used for both growth and imaging.
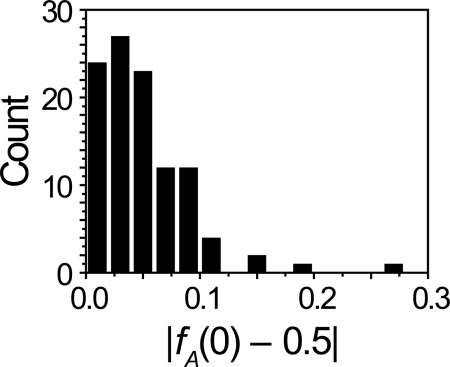
Figure 3 shows red (DRAQ5-stained DNA) and green (β′-GFP) fluorescence images of a representative cell and axial and transverse intensity distributions through each image. Both axial traces show a local minimum in intensity near the midcell plane. When the three transverse distributions are all scaled to have a maximum intensity of 100, they are nearly superimposable. That is, the shapes and widths of the transverse DNA and RNAP distributions are very similar at three axial positions: within lobe A, within lobe B, and at the axial intensity minimum. The full width at half-maximum intensity (FWHM) of the transverse intensity distribution is essentially constant within the cylindrical part of the cell, while the amplitude varies with axial position x. Thus, the central dip in the RNAP spatial distribution arises not from a narrowing of the distribution but rather from an overall decrease in RNAP concentration near the midplane (Fig. 3B). This phenomenon is also observed in the DNA distribution (Fig. 3A), indicating that the local concentration of DNA is smaller by a factor of about 1.25 near the midplane. The prebleach fraction of RNAP intensity within one lobe need not be 0.5, and, indeed, cells show a variable degree of left-right intensity asymmetry. A histogram of |fA − 0.5| for unbleached cells, which measures deviations of fA from 0.5, is shown for 106 cells in Fig. 4. In 25% of the cells, fA is more than 0.068 away from 0.5. In 10% of cells, the deviation is more than 0.096. We estimate the accuracy of fA to be ±0.01 from the average standard deviation in fA over a sequence of 40-ms frames taken for the same cell.
Fig. 4.
Histogram of observed fractional intensity for cells without photobleaching. The absolute value of the difference between the measured fractional intensity and the benchmark of equal intensity in both halves (fA = 0.5) is plotted.
Diffusion of RNAP on the micrometer length scale and 30-s timescale.
For FRAP experiments, we bleached most of the population of RNAP in one half of the cell (i.e., most of one intensity lobe) and measured fA versus time. For one cell whose behavior highlights the complexities inherent in imaging RNAP in live, growing cells, Fig. 1 shows a sequence of fluorescence images and the corresponding plot of fA(t). The recovery is clearly incomplete on the 50-s timescale studied. In addition, at t = 26 s the fraction fA takes a sudden, significant jump upward over a 5-s interval before continuing its smooth decay. Such jumps are unusual on a 30-s timescale but are not rare (3 of 39 cells). They are evidently not due to movement of the entire cell relative to the camera. When the dividing line between the two lobes is measured as the minimum in the cubic spline fit to I(x), its position remains fixed to an accuracy of 0.3 pixel = 30 nm. Rather, we suspect that the jumps are due to directed transport of DNA and the associated RNAP across the dividing line, causing a sudden shift in fA.
For the particular cell pictured in Fig. 1, we fit the decay from 2 to 30 s to a single exponential function with offset, as described in Materials and Methods. The best-fit decay time is 16.7 s, and the FWHM of I(x) gives an effective cell length of 3.22 μm. The resulting effective diffusion constant is DRNAP = 0.063 μm2-s−1. The mobile fraction is 0.70. A movie of the fluorescent images after photobleaching and the recovery curve is available as Movie S8 in the supplemental material. A similar analysis of another live cell is shown in Fig. S2 in the supplemental material.
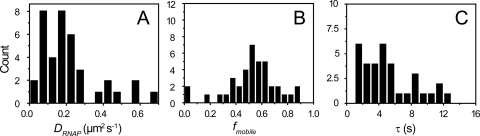
For 39 cells adhered to polylysine with growth medium flowing over them, Fig. 5 shows histograms of τ, DRNAP, and fmobile. The mean time constant for FRAP recovery, the mean diffusion constant, and the mean mobile and immobile fractions are 〈τ〉 = 5.8 ± 4.4 s, 〈DRNAP〉 = 0.22 ± 0.16 μm2-s−1, and 〈fmobile〉 = 0.53 ± 0.19. Each value is the mean ± one standard deviation of the distribution of measured values across cells. The values of DRNAP and fmobile are uncorrelated (see Fig. S3 in the supplemental material). In 4 out of the 63 cells that were examined by FRAP, there was almost no recovery (mobile fraction near zero), prohibiting accurate estimation of the apparent diffusion constant. These cells were not included in the diffusion constant distribution, but they were included in the distributions of fmobile in Fig. 5C and in Fig. S4 in the supplemental material.
Fig. 5.
(A) Histogram of effective diffusion constants for RNAP::GFP measured by FRAP. DRNAP was measured for cells plated on polylysine coverslips with 30°C aerated growth medium flowing over them. (B) Histogram of observed fractions of RNAP molecules able to redistribute to the other half of the cell on the 30-s timescale. (C) Histogram of time constants from fits of fractional intensity to equation 1.
Similarly, the results for 24 cells sandwiched between a clean coverslip and a pad made from 1% agar are 〈τ〉 = 7.1 ± 4.5 s, 〈DRNAP〉 = 0.21 ± 0.17 μm2-s−1, and 〈fmobile〉 = 0.44 ± 0.13. Figure S4 in the supplemental material shows the corresponding distributions. Evidently the polylysine coating has no effect on RNAP diffusion.
In spite of the uncertainties inherent in the analysis, we believe that the wide variations in DRNAP and fmobile are real effects dominated by cell-to-cell heterogeneity. From the raw data, it is clear that cells closely matched in Leff exhibit different partial recovery times. Such cell-to-cell heterogeneity is commonplace in protein diffusion measurements in live cells (12, 13, 19, 22, 34).
In some cells, the estimate of the mobile fraction is affected by abrupt or gradual shifts in the baseline (Fig. 1C). To test for the natural variability of fA(t), nine cells were exposed to the probe laser as before but did not receive a bleach pulse. The standard deviation of fA over a 60-s period was only 0.01, suggesting that for most cells time-dependent shifts in the underlying distribution of RNAP do not greatly affect the accuracy of fmobile. We therefore believe that the broad distribution of fmobile across cells is also a real effect.
We presume that those RNAP molecules that are immobile in FRAP measurements on the 30-s timescale would eventually redistribute themselves across the entire cell. As a control, we made time-lapse movies of cells over a period of 0 to 50 min. On this timescale, the cells grow substantially and eventually begin to pinch and divide. The original midplane of the cell moves relative to the camera, and the baseline value of fA drifts. These effects make it difficult to assess the several-minute timescale over which complete redistribution of RNAP may occur.
Cells that had been fixed for 1 h in 3.7% formaldehyde generally showed no clear evidence of recovery of fA(t) due to diffusion (see Fig. S5 in the supplemental material). The total intensity in these cells increased slightly over the first few seconds following the bleach. We attribute this to a small amount of reversible photobleaching. This phenomenon results in a small apparent recovery in fA(t) (estimated fmobile, <0.15) as the lobe that was bleached recovers. In unfixed cells, we measured no increase in total fluorescence following the bleach pulse. This reversible photobleaching effect, if it exists at all in live cells, is much smaller in magnitude and does not impact our observed recovery of fA(t).
RNAP movement on the submicrometer length scale and subsecond timescale.
The dynamics of RNAP on a subsecond timescale were examined by taking images at 500 Hz (1.96 ms/frame) for 800 ms using higher laser power (see Materials and Methods). Averaging 20 consecutive frames together yielded high signal-to-noise RNAP distributions at a total exposure time of 40 ms/frame, which was fast enough to track the changing distribution smoothly over time. The grayscale (Fig. 2B) and false-color scale (Fig. 2C) snapshots show the existence of several narrow, intense features atop the two broad lobes of intensity at an integration time of 200 ms. To our eyes, the intensity distribution appears to be dynamic on the subsecond timescale, but characterizing the nature of the motion is nontrivial.
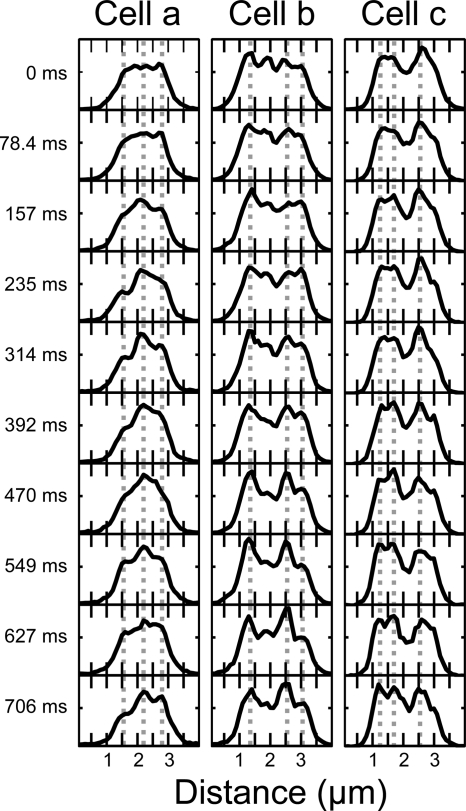
In Fig. 6, we show one-pixel-wide axial intensity profiles as a function of time for three cells. We use this method to display the dynamics of the spatial distribution. The vertical lines are drawn to guide the eye. These traces reveal a time-varying pattern of peaks and valleys in the RNAP distribution. Any two successive 40-ms frames look quite similar, but gradual changes do occur on a 100- to 400-ms timescale. Close scrutiny suggests that peaks in the axial intensity profiles maintain their positions over the entire 800 ms, while a fluctuating background alters the local peak/valley ratio on a timescale of approximately 100 ms. This is roughly consistent with a model that includes small, immobile transcription foci within a broad, fluctuating “background” of mobile (hopping and sliding) RNAPs with an effective diffusion constant of about 0.2 μm2-s−1, the mean bulk diffusion constant obtained from the FRAP studies. The root mean square displacement of mobile molecules in a two-dimensional image over 100 ms would then be 280 nm. This is comparable to the diffraction limit and thus is sufficiently large to explain fluctuations in the background intensity surrounding narrow peaks.
Fig. 6.
Three-hundred-nanometer-wide axial fluorescence intensity distributions as a function of time for three different cells imaged at 78.4 ms/frame. Peaks seem to remain in the same position while the background fluctuates. Vertical dotted lines are drawn to guide the eye.
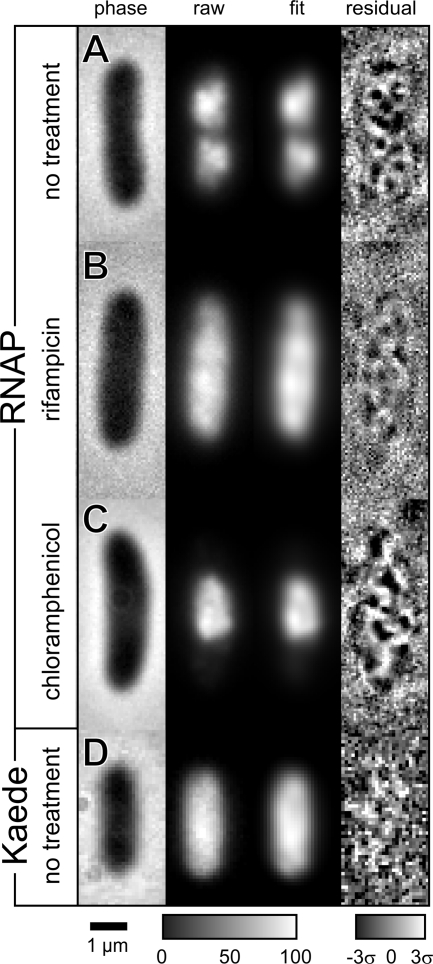
We also attempted to track possible motion of the narrowest peaks by fitting the entire dual-lobed, two-dimensional intensity distribution to a sum of eight Gaussian functions for each 40-ms time interval. Qualitatively good fits require at least eight Gaussian functions, which vary widely in width, aspect ratio, orientation, and amplitude. With fewer Gaussian functions, the narrowest peaks are not well recovered. Figure 7A shows an example of a fit to eight Gaussian functions. The raw image is shown in the second column and the best fit in the third. A weighted residual map showing the difference between the image and the fit is shown in the fourth. An additional example is shown in Fig. S6 in the supplemental material.
Fig. 7.
Snapshots (250 ms long) of green fluorescence from E. coli cells under different treatments. (A) RNAP distribution without drug treatment. (B) RNAP distribution following 90 min of treatment with 200 μg/ml rifampin. (C) RNAP distribution following 90 min treatment with 200 μg/ml of chloramphenicol. (D) Kaede, a uniformly distributed cytoplasmic protein. In each row is shown the phase-contrast image, the fluorescence image, the least-squares fit to a sum of eight Gaussian functions, and the weighted residuals plot.
The best fits include a broad distribution of Gaussian widths. Usually 1 or 2 Gaussian functions per cell are very broad (FWHM of greater than 1.5 μm); these account for 35% of the total RNAP intensity on average. Typically two or three peaks are narrow and round. We define narrow as having a geometric mean of the FWHM along the two principal axes of the Gaussian function of less than 600 nm. This is significantly larger than the diffraction limit of 250-nm FWHM. We define round as having an aspect ratio (ratio of FWHM values along the two principal axes) of less than 1.4. These peaks tend to remain in place and maintain their fractional intensity over the entire 800-ms movie. The standard deviation in the position of these peaks within a cell during a 20-frame movie (800 ms) is only 60 nm. The average intensity in these narrow peaks is 9% per peak, so that the combined intensity of the two to three narrowest peaks is some 20 to 30% of the total RNAP intensity.
While the fits are qualitatively good (Fig. 7), each entire set of fitting parameters is not unique. The χ2 surface is quite flat with many shallow local minima, meaning that some aspects of the fitting results depend sensitively on the initial guess. In addition, after an exhaustive search of fitting parameter space for the first frame of each movie, we biased the fits in later frames by starting the minimization with the best parameter set from the previous frame. Nevertheless, when we sample more and more of the parameter space near the minimum of χ2, the exact parameters of each fit vary but the overall picture of the narrow features remains essentially the same. In each cell, the two or three narrowest features maintain their intensities and positions over the entire 800 ms of the movie.
As controls, we applied the same routine of fitting eight Gaussian functions to cells that had been treated with rifampin (Fig. 7B) or chloramphenicol (Fig. 7C) or to a cell line expressing the cytoplasmic fluorescent protein Kaede instead of β′-GFP (Fig. 7D). Following treatment with rifampin, the RNAP distribution expands to fill the entire cell volume (3). Of the four cells analyzed with the fitting routine, only one showed a narrow and round feature as defined above, and it had only 6% of the total intensity. Following treatment with chloramphenicol, the RNAP distribution becomes much more compact and centrally located (3). Of the three cells analyzed, only one had narrow, round features. It contained two such features each with 6% of the total intensity. In the three analyzed cells expressing the uniformly distributed fluorescent protein Kaede, the number of narrow, round features was 0, 1, or 2. Each feature contributed less than 3% of the total intensity in the cell.
DISCUSSION
Interpretation of narrow RNAP features as transcription foci.
For both E. coli (4) and B. subtilis (15) cells grown in rich medium and then chemically fixed, narrow features have been observed within the distribution of GFP-labeled RNAP. These features have been interpreted as transcription foci, sites in which a high local concentration of RNAP transcribes stable RNA. The spatially narrow, immobile RNAP features that we have observed in live cells appear qualitatively similar to the foci reported earlier but are somewhat less pronounced and numerous. This may be due to the lower growth rate in our study. For comparison, in Fig. S1 in the supplemental material we show an image of a cell grown in LB in our labs, fixed, and imaged by our methods.
To further test the transcription focus hypothesis, we have attempted to quantify the size, shape, and fractional intensity of the narrow RNAP features in live cells for the first time. This is important not only because fixation is known to induce artifacts in the structure of the nucleoid, and thus in the RNAP spatial distribution (27), but also because measurements on live cells afford the possibility of observing movement of the narrow foci in real time.
Each cell exhibits two or three narrow features comprising roughly 20 to 30% of the total RNAP population. Previous biochemical measurements estimate that depending on the growth rate, 10 to 25% of total RNAP is involved in active transcription of stable RNA (1). These RNAP molecules constitute a large fraction of the actively transcribing RNAP molecules, which itself is estimated as 20 to 30% of the total population (1). In addition, the narrow features do not move appreciably over 800 ms, consistent with expectations for transcription foci. Our results seem consistent with the hypothesis of transcription foci in which many RNAP copies are actively transcribing a small number of operons. These narrow and round features are not present following the halting of transcription by rifampin. The fast, short-range fluctuations in the background RNAP spatial distribution observed in the 800-ms-long movies (Fig. 6) are sensibly consistent with the movement of the mobile RNAP fraction on the micrometer length scale and 5-s timescale observed by FRAP. It remains to be seen whether or not the RNAP foci observed here colocalize with rrn operons.
Interpretation of FRAP data.
The FRAP study shows that on average roughly half of the fluorescent, GFP-labeled β′ subunits (the mobile fraction) interchange between one lobe of intensity and the other lobe on a timescale of 5 s. The other half (the immobile fraction) do not interchange even on a 30- to 60-s timescale. In addition, both the effective diffusion constant of the mobile molecules and the fraction of mobile molecules vary substantially from cell to cell, but they are not correlated with each other. This cell-to-cell heterogeneity occurs both for cells plated on polylysine-coated coverslips and for cells sandwiched between agar and the coverslip.
The first question is the extent to which the fluorescent, GFP-labeled β′ subunits in the cytoplasm are incorporated into RNAP, the multisubunit (α2bb′ω) complex that binds DNA either specifically or nonspecifically. Both the mobile fraction and the immobile fraction observed by FRAP almost surely arise from β′-EGFP that is properly incorporated into the core polymerase. The complex is assembled stepwise, with the β′ subunit added to preassembled α2b. Because we have substituted rpoC-egfp for the normal β′ gene rpoC, we expect all copies of β′ to bear the label. For the subset of molecules that we can observe, those having β′ fused to fluorescent, mature EGFP, we expect the incorporation percentage to be even higher than the 90% estimated for all β′ (11). The reason is that the EGFP maturation time of 60 min (31) far exceeds the 90-s timescale of incorporation of new β′ subunits into nucleoid-associated core enzyme, as measured by pulse-chase experiments with radioactive labels (28). For these reasons, we speak of RNAP diffusion and the RNAP spatial distribution even though we are actually measuring the fluorescence from β′-EGFP. In addition, because we are tagging the large RNAP by a single copy of the small EGFP, we expect the dynamics measured to be indicative of the true RNAP dynamics (24).
For growth in rich media, a substantial fraction of RNA polymerases are involved in active transcription (1). We expect these molecules to be immobile in the 30- to 60-s-long FRAP experiments. Processive transcription could not account for the observed 5-s recovery time over a 2-μm length scale. Transcription occurs at less than 100 nucleotides (nt)/s = 34 nm/s along the DNA contour (2, 16). Over 5 s, transcribing RNAP moves at most 170 nm along the DNA contour, far too short a distance to contribute to the FRAP recovery. In addition, based on the measured motional properties of specifically labeled DNA loci in live E. coli (7), RNAP molecules which “ride” on DNA as it is segregated can make only a minor contribution to DRNAP. Even the most mobile of these loci underwent confined diffusive motion with a diffusion coefficient DDNA at least two orders of magnitude smaller than the DRNAP values of ∼0.2 μm2-s−1 observed here.
The immobile fraction fimmobile does not recover for at least 30 s and accounts for 47% of the RNAP on average. In our scheme, these are RNA polymerase molecules specifically bound to DNA. On average, the 〈 fimmobile〉 of 0.47 from the FRAP experiments is roughly 0.20 larger than the fraction of intensity contained in the narrow, immobile peaks in fast movies. However, these two fractions could not be measured for the same cell because both imaging methods photobleach a substantial fraction of the total molecules. It is roughly 0.20 larger than the estimated total fraction of 0.2 to 0.3 of RNAPs that are actively transcribing (1). We interpret this “extra” population of immobile RNAPs as molecules bound at specific DNA sites but not actively transcribing. This fraction may include some of the RNAPs that are actively transcribing nonstable RNA; 0.2 may somewhat overestimate the fraction of RNAP bound to specific sites. These specific binding locations could be initiation sites, lengthy pause sites within transcribed genes, or promoter-like sequences. If so, typical dwell times are roughly 10 s or longer. Some of these RNA polymerase molecules may be waiting at initiation sites for a signal associated with a change in nutrient status, temperature, or pH. Such a stalled population may be related to the fraction of RNAP which cross-links to locations from which no mRNA is detected in chromatin immunoprecipitation on chip (ChIP/chip) experiments (8, 9). If our interpretation is essentially correct, then the data indicate significant cell-to-cell variations in the combined populations of RNAP involved in active transcription and stalled while specifically bound to DNA (Fig. 5B).
We interpret the mobile population of RNAP as arising from fully assembled RNAP enzymes that are nonspecifically bound to DNA or that are not bound to DNA at all. The mobile fraction averages 0.53 and varies from 0.3 to 0.9 across cells (Fig. 5B). Background fluctuations in fast movies suggest that the motion of these RNAPs is diffusive on the timescale of 0.2 s. The FRAP experiments indicate that the motion is also diffusive on the timescale of 5 to 10 s. The same effective diffusion coefficient can explain both observations.
In a single-molecule study of the diffusion of Lac repressor in E. coli, Elf and coworkers (5) were able to separate contributions from sliding and free diffusion rather cleanly by combining measurements of DLacI in the cell with independent measurements of the one-dimensional sliding diffusion constant D1D in vitro and of the three-dimensional diffusion constant D3D in vivo. They inferred that Lac repressor moves long distances across the cell primarily by diffusing freely in the three-dimensional cytoplasm, even though it spends on average only 13% of its time doing so.
The movement of the mobile RNAPs in the complex cytoplasm presumably involves intervals of one-dimensional diffusive sliding on the DNA contour interspersed with intervals of unbinding, hopping from DNA strand to DNA strand by three-dimensional diffusion, and rebinding in a new location, followed by additional sliding. This composite motion would look like free diffusion on the length and timescales studied here. Accordingly, we follow Elf et al. and write DRNAP as a time-weighted average:
| (2) |
Here θns is the average fraction of total RNAP that is nonspecifically bound to DNA and sliding in one dimension (1D), θfree is the average fraction of total RNAP that is unbound and diffusing in 3D, α is the mean projection of the DNA contour onto the measurement axis, D1D is the one-dimensional sliding diffusion coefficient along the DNA contour, and D3D is the three-dimensional RNAP diffusion coefficient in cytoplasm (5). For a more detailed discussion, see “One-dimensional diffusion along DNA contour” in the supplemental material.
We lack sufficient quantitative information to permit a clean separation of DRNAP into 1D and 3D diffusive components. Based on the available evidence, the measured DRNAP for the mobile fraction seems remarkably large. For T7 RNAP diffusing on stretched λ DNA in vitro, Kim and Larson found D1D = 0.12 μm2-s−1 (10). For E. coli RNAP, early biochemical studies estimated D1D = 0.13 to 0.15 μm2-s−1 (26, 30). These results are about two times smaller than typical values of DRNAP that we have measured. Estimates of θfree, the fraction of the total RNAP population that is freely diffusing in 3D, are in the range of 0.01 to 0.15 (11, 18, 29). Assuming the large estimate θfree = 0.15, the mobile population would spend about 70% of its time sliding in 1D and about 30% diffusing in 3D. Assuming a smaller θfree value of 0.02, the mobile population would spend more than 95% of its time sliding in 1D. We also lack a good estimate of D3D for RNAP in cytoplasm. A recent study measured the diffusion constants of a variety of globular proteins in the E. coli cytoplasm and found a fairly smooth correlation between D3D and molar mass (14). The largest protein studied, cyan fluorescent protein (CFP)-CheA-yellow fluorescent protein (YFP), has a mass of 250 kDa and D3D = 0.45 μm2-s−1. The mass of core RNAP is 390 kDa, suggesting that for free diffusion of unbound RNAP in the cytoplasm D3D may be ∼0.3 μm2-s−1. As one illustrative example, insertion into equation 2 of the combination of parameters α = 1, θfree = 0.15, θns = 0.30, fmobile = 0.45, D1D = 0.15 μm2-s−1, and D3D = 0.3 μm2-s−1 yields the value DRNAP = 0.2 μm2-s−1, consistent with the mean observed value.
This type of thinking suggests that in the cytoplasm, the effective sliding/hopping diffusion coefficient for RNAP near the DNA is at least as large as D1D in vitro for RNAP sliding on bare DNA. That is a surprising result in view of cytoplasmic crowding and the presence of protein obstacles on the DNA. Perhaps the physical idea embodied in equation 2 is too coarse grained in that it partitions the motion into periods of 1D sliding on DNA and distinct periods of 3D diffusion in an “averaged” cytoplasm. Due to its strong propensity to bind DNA and the supercoiled nature of the DNA itself, RNAP may remain essentially always in a special subvolume of the cytoplasm around the DNA. In this DNA-rich subvolume, free, globular protein crowders may be relatively sparse due to excluded volume effects (20, 33). If so, the impediment to sliding due to immobile protein obstacles bound to DNA may be compensated for by facile diffusion in a relatively uncrowded “tube” surrounding the DNA. In the future, tracking studies of single RNAP copies with high localization accuracy will shed additional light on the nature of RNAP transport in the cytoplasm.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Rick Gourse, Tamas Gaal, and Robert Landick (University of Wisconsin—Madison, Departments of Bacteriology and Biochemistry) for technical discussions and critical readings of the manuscript.
B.P.B. and J.C.W. were supported by the National Institute of General Medical Sciences under grant R01-GM086468 (an American Recovery and Reinvestment Act grant) and by the National Science Foundation under grant CHE-0452375. B.P.B. was supported in part by a National Institutes of Health training grant (T32-GM08293). R.A.M. was supported by NIH grant GM38660 to Robert Landick.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 22 July 2011.
REFERENCES
- 1. Bremer H., Dennis P. P. 1996. Modulation of chemical composition and other parameters of the cell by growth rate, p. 1553–1569 In Neidhardt F. C., et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed ASM Press, Washington, DC [Google Scholar]
- 2. Bremer H., Yuan D. 1968. RNA chain growth-rate in Escherichia coli. J. Mol. Biol. 38:163–180 [DOI] [PubMed] [Google Scholar]
- 3. Cabrera J. E., Cagliero C., Quan S., Squires C. L., Jin D. J. 2009. Active transcription of rRNA operons condenses the nucleoid in Escherichia coli: examining the effect of transcription on nucleoid structure in the absence of transertion. J. Bacteriol. 191:4180–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cabrera J. E., Jin D. J. 2003. The distribution of RNA polymerase in Escherichia coli is dynamic and sensitive to environmental cues. Mol. Microbiol. 50:1493–1505 [DOI] [PubMed] [Google Scholar]
- 5. Elf J., Li G.-W., Xie X. S. 2007. Probing transcription factor dynamics at the single-molecule level in a living cell. Science 316:1191–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Elowitz M. B., Surette M. G., Wolf P. E., Stock J. B., Leibler S. 1999. Protein mobility in the cytoplasm of Escherichia coli. J. Bacteriol. 181:197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Espeli O., Mercier R., Boccard F. 2008. DNA dynamics vary according to macrodomain topography in the E. coli chromosome. Mol. Microbiol. 68:1418–1427 [DOI] [PubMed] [Google Scholar]
- 8. Grainger D. C., Hurd D., Harrison M., Holdstock J., Busby S. J. W. 2005. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc. Natl. Acad. Sci. U. S. A. 102:17693–17698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Herring C. D., et al. 2005. Immobilization of Escherichia coli RNA polymerase and location of binding sites by use of chromatin immunoprecipitation and microarrays. J. Bacteriol. 187:6166–6174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim J. H., Larson R. G. 2007. Single-molecule analysis of 1D diffusion and transcription elongation of T7 RNA polymerase along individual stretched DNA molecules. Nucleic Acids Res. 35:3848–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Klumpp S., Hwa T. 2008. Growth-rate-dependent partitioning of RNA polymerases in bacteria. Proc. Natl. Acad. Sci. U. S. A. 105:20245–20250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Konopka M. C., Shkel I. A., Cayley S., Record M. T., Weisshaar J. C. 2006. Crowding and confinement effects on protein diffusion in vivo. J. Bacteriol. 188:6115–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Konopka M. C., et al. 2009. Cytoplasmic protein mobility in osmotically stressed Escherichia coli. J. Bacteriol. 191:231–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kumar M., Mommer M. S., Sourjik V. 2010. Mobility of cytoplasmic, membrane, and DNA-binding proteins in Escherichia coli. Biophys. J. 98:552–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lewis P. J., Thaker S. D., Errington J. 2000. Compartmentalization of transcription and translation in Bacillus subtilis. EMBO J. 19:710–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Manor H., Goodman D., Stent G. S. 1969. RNA chain growth rates in Escherichia coli. J. Mol. Biol. 39:1–27 [DOI] [PubMed] [Google Scholar]
- 17. Marenduzzo D., Micheletti C., Cook P. R. 2006. Entropy-driven genome organization. Biophys. J. 90:3712–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McClure W. R. 1985. Mechanism and control of transcription initiation in prokaryotes. Annu. Rev. Biochem. 54:171–204 [DOI] [PubMed] [Google Scholar]
- 19. Mika J. T., Bogaart G. v. d., Veenhoff L., Krasnikov V., Poolman B. 2010. Molecular sieving properties of the cytoplasm of Escherichia coli and consequences of osmotic stress. Mol. Microbiol. 77:200–207 [DOI] [PubMed] [Google Scholar]
- 20. Mondal J., Bratton B. P., Li Y., Yethiraj A., Weisshaar J. C. 2011. Entropy-based mechanism of ribosome-nucleoid segregation in E. coli cells. Biophys. J. 100:2605–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mooney R. A., Landick R. 2003. Tethering σ70 to RNA polymerase reveals high in vivo activity of σ factors and σ70-dependent pausing at promoter-distal locations. Genes Dev. 17:2839–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mullineaux C. W., Nenninger A., Ray N., Robinson C. 2006. Diffusion of green fluorescent protein in three cell environments in Escherichia coli. J. Bacteriol. 188:3442–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Neidhardt F. C., Bloch P. L., Smith D. F. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nenninger A., Mastroianni G., Mullineaux C. W. 2010. Size dependence of protein diffusion in the cytoplasm of Escherichia coli. J. Bacteriol. 192:4535–4540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Oden K. L., DeVeaux L. C., Vibat C. R. T., Croman J. E. 1990. Genomic replacement in Escherichia coli K-12 using covalently closed circular plasmid DNA. Gene 96:29–36 [DOI] [PubMed] [Google Scholar]
- 26. Ricchetti M., Metzger W., Heumann H. 1988. One-dimensional diffusion of Escherichia coli DNA-dependent RNA polymerase: a mechanism to facilitate promoter location. Proc. Natl. Acad. Sci. U. S. A. 85:4610–4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Robinow C., Kellenberger E. 1994. The bacterial nucleoid revisited. Microbiol. Mol. Biol. Rev. 58:211–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saitoh T., Ishihama A. 1977. Biosynthesis of RNA polymerase in Escherichia coli. VI. Distribution of RNA polymerase subunits between nucleoid and cytoplasm. J. Mol. Biol. 115:403–416 [DOI] [PubMed] [Google Scholar]
- 29. Shepherd N., Dennis P., Bremer H. 2001. Cytoplasmic RNA polymerase in Escherichia coli. J. Bacteriol. 183:2527–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Singer P., Wu C. W. 1987. Promoter search by Escherichia coli RNA polymerase on a circular DNA template. J. Biol. Chem. 262:14178–14189 [PubMed] [Google Scholar]
- 31. Sniegowski J. A., Lappe J. W., Patel H. N., Huffman H. A., Wachter R. M. 2005. Base catalysis of chromophore formation in Arg96 and Glu222 variants of green fluorescent protein. J. Biol. Chem. 280:26248–26255 [DOI] [PubMed] [Google Scholar]
- 32. Sochacki K. A., Shkel I. A., Record M. T., Weisshaar J. C. 2011. Protein diffusion in the periplasm of E. coli under osmotic stress. Biophys. J. 100:22–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Valkenburg J. A., Woldringh C. L. 1984. Phase separation between nucleoid and cytoplasm in Escherichia coli as defined by immersive refractometry. J. Bacteriol. 160:1151–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van den Bogaart G., Hermans N., Krasnikov V., Poolman B. 2007. Protein mobility and diffusive barriers in Escherichia coli: consequences of osmotic stress. Mol. Microbiol. 64:858–871 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.