Abstract
YdiV acts as an anti-FlhD4C2 factor, which negatively regulates the class 2 flagellar operons in poor medium in Salmonella enterica serovar Typhimurium. On the other hand, one of the class 2 flagellar genes, fliZ, encodes a positive regulator of the class 2 operons. In this study, we found that the FliZ-dependent activation of class 2 operon expression was more profound in poor medium than in rich medium and not observed in the ydiV mutant background. Transcription of the ydiV gene was shown to increase in the fliZ mutant. Purified FliZ protein was shown in vitro to bind to the promoter region of the nlpC gene, which is located just upstream of the ydiV gene, and to repress its transcription. These results indicate that FliZ is a repressor of the nlpC-ydiV operon and activates the class 2 operons by repressing ydiV expression. Therefore, the fliZ and ydiV genes form a regulatory loop.
INTRODUCTION
Salmonella enterica serovar Typhimurium cells swim by means of rotating flagella through their environment. The individual flagellum consists of three structural parts, a basal body, a hook, and a filament. More than 50 genes are specifically required for flagellar formation and function (1, 2, 4, 26). These genes are organized into at least 15 operons, and their transcriptional expression forms a highly organized three-tiered cascade called a flagellar regulon (22, 27). The flhDC operon is the sole one belonging to class 1 and encodes FlhD and FlhC, which assemble into an FlhD4C2 heterohexamer acting as an activator of class 2 operons (31, 42). Class 2 contains operons encoding component proteins for the hook-basal body structure and the flagellum-specific type III export apparatus as well as a gene encoding the flagellum-specific sigma factor σ28 (FliA), essential for class 3 expression (33). Class 3 operons encode proteins involved in filament assembly and flagellar function. σ28 activity is negatively controlled by an anti-σ28 factor, FlgM (15, 24, 34), which is excreted from the cell through the flagellum-specific type III export apparatus upon completion of hook-basal body assembly (11, 19).
Class 2 operons are transcribed by σ70 RNA polymerase in the presence of the FlhD4C2 complex, which binds to the DNA region upstream of the class 2 promoter (13, 30, 31). Two genes within the flagellar regulon, fliT and fliZ, have been shown to be involved in fine control of the class 2 operons (23). The fliT and fliZ genes belong to the fliDST and fliAZ operons, respectively, both of which are transcribed from both class 2 and class 3 promoters (12, 21, 46). FliT acts as an anti-FlhD4C2 factor, which binds to the FlhD4C2 complex through interaction with the FlhC subunit and inhibits its binding to the class 2 promoter, resulting in decreased expression of the class 2 operons (45). On the other hand, FliZ was shown to be a positive regulator of class 2 operons (23) and to participate in a positive-feedback loop that induces a kinetic switch in class 2 operon expression (38). Although class 2 operon expression is inhibited in vivo by the fliZ mutation (23), transcription from the class 2 promoter occurs efficiently in vitro in the absence of FliZ (13, 31). This suggests that FliZ affects indirectly the activation process of the class 2 operons. On the basis of the observation that the fliZ mutation decreased the amount of FlhC without having a significant effect on the transcription and translation of the flhC gene, Saini et al. (37) proposed that FliZ is a posttranslational activator of FlhD4C2. However, the molecular mechanism underlying FliZ control of the class 2 operons has remained obscure.
The fliZ mutation has also been shown to impact several cellular processes unrelated to flagellar biogenesis in Salmonella. For example, the fliZ mutation decreases the transcriptional expression of Salmonella pathogenicity island 1 (SPI1) genes (14) through affecting the activity of their positive regulator, HilD (5, 17). On the other hand, the fliZ mutation increases type 1 fimbrial gene expression through affecting posttranscriptionally the activity of the positive regulator FimZ (6, 39). Furthermore, in Escherichia coli, FliZ was shown to affect curli expression by interfering with RpoS, the stationary-phase-specific σ factor (35). However, the precise molecular mechanisms underlying these FliZ controls are also still unclear.
Recently, we found that a nonflagellar gene, ydiV, encodes another anti-FlhD4C2 factor (41). YdiV binds to FlhD4C2 by interacting with the FlhD subunit, resulting in inhibition of class 2 operon expression. The intracellular level of YdiV is higher in poor medium than in rich medium, which allows YdiV to act as a mediator of the nutritional level of the environment in the expression of the flagellar regulon. Nutritional control of YdiV expression was shown to be executed mainly at the translational level (41). On the other hand, Wozniak et al. (43) reported that ydiV transcription is under the negative regulation of the flhDC genes, suggesting that some gene(s) within the flagellar regulon may act as a negative regulator of the ydiV gene.
In this study, we showed that FliZ-dependent activation of class 2 operon expression was more profound in poor medium than in rich medium and not observed in the ydiV mutant background. This suggested the possibility that FliZ control of class 2 operon expression is exerted via the regulatory pathway of ydiV expression. Gel mobility shift assay and in vivo and in vitro transcription analyses revealed that FliZ binds to the promoter region of the nlpC-ydiV operon and acts as a repressor of its transcription. Therefore, FliZ-dependent activation of class 2 operon expression is achieved by repression of the ydiV gene by FliZ.
MATERIALS AND METHODS
Bacterial strains, plasmids, chemicals, and culture media.
The Salmonella strains and plasmids used in this study are listed in Tables 1 and 2, respectively. All the Salmonella strains used were derivatives of a standard laboratory strain, LT2. Their construction procedures are described below. P22-mediated transduction was performed as described previously (25, 28). Unless otherwise specified, all the chemicals and DNA enzymes used were purchased from Nacalai Tesque (Kyoto, Japan) and Toyobo (Osaka, Japan), respectively. Oligonucleotide primers used were purchased from Life Technologies (Tokyo, Japan) and are summarized in Table 3. The rich and poor media used were Luria broth (LB) and M9 minimal medium (32) supplemented with 0.2% glycerol and 0.2% Casamino Acids (MGC), respectively (41). Hard-agar and motility agar plates were prepared by adding 1.2% and 0.25% agar (Shoei, Tokyo, Japan), respectively, to LB or MGC. Ampicillin, kanamycin, and tetracycline were used at final concentrations of 50, 50, and 20 μg/ml, respectively. Where required, arabinose or isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the culture media at final concentrations of 0.2% or 0.1 mM, respectively.
Table 1.
Salmonella enterica strains used in this study
| Strain | Relevant characteristic(s) | Source or reference |
|---|---|---|
| KK1004 | LT2 Δ(hin-fljBA) ΔFels-2 | 25, 28 |
| KK1004hDC | KK1004 flhD2140::Tn10 | 28 |
| KK1004iA | KK1004 ΔfliA::FRT (in-frame) | This study |
| KK1004iZ | KK1004 ΔfliZ::FRT (in-frame) | This study |
| KK1004iAZ | KK1004 ΔfliAZ::FRT (in-frame) | This study |
| KK1004V | KK1004 ΔydiV::FRT (in-frame) | 41 |
| KK1004ViA | KK1004V ΔfliA::FRT (in-frame) | This study |
| KK1004ViZ | KK1004V ΔfliZ::FRT (in-frame) | This study |
| KK1004ViAZ | KK1004V ΔfliAZ::FRT (in-frame) | This study |
| KK1004 flhC-3F | KK1004 flhC-3×FLAG | 41 |
| KK1004 ydiV-lac | KK1004 ΔydiV::FRT::pKG137 | 41 |
Table 2.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pKD13 | FRT-kan-FRT cassette template, amp | 7 |
| pKD46 | Red expression plasmid, amp | 7 |
| pCP20 | Flp expression plasmid, amp cat | 7 |
| pQE80L | Expression vector, His6 tag, amp | Qiagen |
| pQE80L-flhCD | pQE80L flhC flhD (His6-FlhC FlhD) | This study |
| pQE80L-fliZ | pQE80L fliZ (His6-FliZ) | This study |
| pQE80L-ydiV | pQE80L ydiV (His6-YdiV) | 41 |
| pQE80L-ΔPT5-PnlpC | pQE80L ΔPT5 PnlpC-T0 | This study |
| pQE80K | pQE80L Δamp::kan | Laboratory stock |
| pQE80K-fliZ | pQE80K fliZ (His6-FliZ) | This study |
| pFZY1 | Promoter probe vector, amp | 18 |
| pFZY1-PfliA-lacZ | pFZY1 PfliA-lacZ | This study |
| pFZY1-PnlpC-lacZ | pFZY1 PnlpC-lacZ | This study |
| pFZY1-ΔPnlpC-lacZ | pFZY1-PnlpC-lacZ ΔPnlpC | This study |
Table 3.
Oligonucleotide primers used in this study
| Use and primer name | Nucleotide sequence (5′ to 3′)a |
|---|---|
| Construction of chromosomal deletion mutants | |
| fliAH1P1 | TAGAAACGGATAATCATGCCGATAACTCATTTAAC |
| GCAGGGCTGTTTATCGTGTAGGCTGGAGCTGCTTC | |
| fliAH2P4 | GGTAGTCTATACGTTGTGCGGCACTTTTCGGGTGC |
| GATCATGCGCGACCTATTCCGGGGATCCGTCGACC | |
| fliZH1P1 | TCGCACCCGAAAAGTGCCGCACAACGTATAGACT |
| ACCAGGAGTTCTCATGGTGTAGGCTGGAGCTGCTTC | |
| fliZH2P4 | AGGTTTGCCACGTTTCACCAACACGACTCTGCTA |
| CATCTTATGCTTTTTTATTCCGGGGATCCGTCGACC | |
| Construction of plasmids | |
| IAEIB | GGGGGATCCTCTGACGATACTCCGC |
| KpnIfliA0 | GGGGGTACCGCTACAGGTTACATAA |
| IZP1 | GGGGATCCACGGTGCAGCAACCTAAAAG |
| SalIfliZR | GGGGTCGACTTAATATATATCAGAA |
| HCf1B | GGGGATCCAGTGAAAAAAGCATTGTTCAGG |
| HCr1S | GGGGTCGACTTAAACAGCCTGTTCGATCTG |
| HDf1S | GGGGTCGACACATCACGGGGTGCGGCTA |
| HDr1P | GGGCTGCAGTCATGCCCTTTTCTTACGCGC |
| PnlpCf1E | GGGGAATTCATTTATGTCAGCCAGGAATTG |
| PnlpCf2E | GGGGAATTCCTACGGTTTGCGCTTTCGACG |
| NLPCF | ACCAGAATTCAAACAGAGGATTGTTGC |
| PnlpCr1B | GGGGGATCCAAAAACGCATGCCGCAACAAT |
| PnlpCr2B | GGGGGATCCCTGCAACTGATCGTTCAGACC |
Nucleotides corresponding to the created restriction enzyme cleavage sites are shown in italics.
Construction of chromosomal deletion mutants.
The λ Red recombination system (7) was used for construction of chromosomal in-frame deletion mutants. Double deletion mutants were constructed by P22-mediated transduction.
An in-frame deletion mutant of the fliA gene (KK1004iA) was constructed as follows. The kanamycin resistance Flp recombination target (FRT) cassette was PCR amplified from pKD13 with primers fliAH1P1 and fliAH2P4, which possess sequences homologous to the 5′- and 3′-end-flanking regions of the fliA gene, respectively. The amplified product was introduced into KK1004 harboring pKD46 by electroporation, and kanamycin-resistant colonies were selected on LB agar plates containing kanamycin and arabinose. After the correct replacement of the fliA gene with the kanamycin resistance gene cassette was confirmed, this mutation was moved to fresh KK1004 via P22-mediated transduction. The kan gene was removed through site-specific recombination between the flanking FRT sequences by transient exposure of the mutant cells to pCP20.
Similarly, an in-frame deletion of the fliZ gene (resulting in strain KK1004iZ) was carried out using primers fliZH1P1 and fliZH2P4, which possess sequences homologous to the 5′- and 3′-end-flanking regions of the fliZ gene, respectively. Through the same procedure, an in-frame deletion of the fliAZ operon (resulting in strain KK1004iAZ) was carried out using primers fliAH1P1 and fliZH2P4.
Plasmid construction.
The fliZ gene was PCR amplified with primers IZP1 and SalIfliZR using genomic DNA from KK1004 as a template. The amplified product was digested with BamHI and SalI and inserted into the corresponding site of pQE80L or pQE80K to obtain pQE80L-fliZ or pQE80K-fliZ, respectively. They encode an N-terminally hexahistidine-tagged FliZ protein (His6-FliZ). Introduction of these plasmids restored full motility to the cells of KK1004iZ (ΔfliZ) (data not shown), indicating that the His6-FliZ protein is functional.
The flhC and flhD genes were separately PCR amplified with primers HCf1B and HCr1S and HDf1S and HDr1P, respectively, using genomic DNA from KK1004 as a template. The amplified flhC DNA was digested with BamHI and SalI, while the amplified flhD DNA was digested with SalI and PstI. These two DNA fragments were inserted together into the BamHI/PstI site of pQE80L to obtain pQE80L-flhCD. This plasmid encodes an N-terminally hexahistidine-tagged FlhC protein (His6-FlhC) and a nontagged FlhD protein. Introduction of this plasmid restored full motility to the cells of KK1004hDC (flhD::Tn10) (data not shown), indicating that both the His6-FlhC and FlhD proteins encoded by this plasmid are functional.
The promoter region of the fliAZ operon was PCR amplified with primers KpnIfliA0 and IAEIB using genomic DNA from KK1004 as a template. The amplified product was digested with KpnI and BamHI and inserted into the corresponding site of pFZY1 to obtain pFZY1-PfliA-lacZ. In this plasmid, the lacZ gene is transcribed from the class 2 and class 3 promoters of the fliAZ operon (PfliA).
The promoter region of the nlpC gene was PCR amplified with primers PnlpCf1E and PnlpCr1B using the genomic DNA from KK1004 as a template. The amplified product was digested with EcoRI and BamHI and inserted into the corresponding site of pQE80L. In order to delete the T5 promoter on the vector sequence, the resulting plasmid was digested with XhoI and EcoRI, blunt ended with the Klenow fragment, and then self-ligated. In the resulting plasmid, pQE80L-ΔPT5-PnlpC, transcription initiates at the nlpC promoter (PnlpC) and terminates at the T0 terminator on the vector sequence, yielding an RNA product of 184 or 183 bases.
Similarly, the same region was PCR amplified with primers PnlpCf2E and PnlpCr2B or NLPCF and PnlpCr2B, digested with EcoRI and BamHI, and inserted into the corresponding site of pFZY1 to obtain pFZY1-PnlpC-lacZ or pFZY1-ΔPnlpC-lacZ, respectively. In pFZY1-PnlpC-lacZ, the lacZ gene is transcriptionally fused to PnlpC. pFZY1-ΔPnlpC-lacZ has the same structure as pFZY1-PnlpC-lacZ except that it lacks the PnlpC region.
β-Galactosidase enzyme assay.
β-Galactosidase activity was assayed as described previously (20) using cells grown aerobically to exponential phase at 37°C in LB or MGC containing appropriate antibiotics. Each sample was assayed in triplicate.
Protein analysis.
Cells were grown to exponential phase at 37°C in LB or MGC, and the cultures were directly subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). SDS-PAGE and Western blotting of proteins were performed according to the method described previously (24). FliC (flagellin) was detected with an anti-FliC polyclonal antibody and an anti-rabbit horseradish peroxidase (HRP)-conjugated antibody (Santa Cruz Biotechnology, CA) using the ECL Plus Western blotting detection system (GE Healthcare, NJ). FLAG-tagged proteins were detected with an anti-FLAG M2 monoclonal antibody (Sigma, MO) and an anti-mouse HRP-conjugated antibody (Santa Cruz Biotechnology).
Affinity purification of His6-FliZ.
His6-FliZ protein synthesized in E. coli strain BL21 ΔslyD (laboratory stock) harboring pQE80L-fliZ was affinity purified according to the method described previously (44). The purified protein exhibited a single band at a position corresponding to approximately 21 kDa in an SDS-PAGE gel (data not shown).
In vitro transcription.
In vitro RNA synthesis was performed using E. coli RNA polymerase holoenzyme (Epicenter, WI) according to the method described previously (41, 44). The reaction mixture contained 40 mM Tris-HCl, pH 7.5, 150 mM KCl, 1 mM dithiothreitol (DTT), 10 mM MgCl2, 50 U of RNase inhibitor, 100 μM (each) ATP, GTP, and CTP, 50 μM UTP with 4 × 105 Bq [α-32P]UTP (Institute of Isotopes, Budapest, Hungary), 1 μg of template DNA, 1 U of RNA polymerase, and various concentrations (0 to 1,250 nM) of His6-FliZ. Mixtures without substrates were prepared in a 45-μl volume and incubated at 37°C for 10 min. The reaction was initiated by addition of 5 μl prewarmed substrate mixture. After incubation at 37°C for 10 min, the reaction was terminated by adding 50 μl of a stop solution (0.6 M sodium acetate [pH 5.5], 20 mM EDTA, 200 μg tRNA/ml). The transcripts were extracted with phenol and then precipitated with ethanol. The precipitated materials were electrophoretically separated in an 8% polyacrylamide gel containing 6 M urea. The labeled transcripts were visualized by autoradiography.
Gel mobility shift assay.
A 287-bp DNA fragment containing PnlpC was PCR amplified from the KK1004 genomic DNA using primers PnlpCf2E and PnlpCr2B. The amplified product was gel purified and then labeled at the 5′ end with [γ-32P]ATP (Institute of Isotopes) by T4 polynucleotide kinase and used as a probe. The DNA-binding activity of His6-FliZ was assayed according to a method described previously (41). The binding reaction mixture (20 μl) contained a 0.2 nM concentration of the labeled probe DNA, 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 0.1 mM EDTA, 1 mM dithiothreitol, 1 mM MgCl2, 0.1 mg/ml bovine serum albumin, 5% glycerol, and various concentrations (0 to 250 nM) of His6-FliZ. The reaction mixture was incubated at 37°C for 30 min and subjected to electrophoresis on a native 5% polyacrylamide gel in 0.5× TBE buffer (25 mM Tris, 25 mM boric acid, 1 mM EDTA). Labeled DNAs were detected by autoradiography.
RESULTS
FliZ is required for efficient flagellar production in poor medium.
We showed previously that a disruption of the fliZ gene reduced class 2 operon expression more than 5-fold (23), whereas Saini et al. (37) reported that the fliZ mutation had only a moderate effect on class 2 operon expression. We noted that the culture media used were different between these two studies; that is, Kutsukake et al. used MGC, whereas Saini et al. used LB. This suggested the possibility that the effect of the fliZ mutation on flagellar production is influenced by the nutritional condition. In order to test this possibility, we examined first more carefully the effect of the fliZ mutation on class 2 transcription with the cells grown in LB and MGC. For this purpose, we used the PfliA-lacZ transcriptional fusion gene on the single-copy plasmid pFZY1-PfliA-lacZ. The PfliA DNA contains both class 2 and class 3 promoters of the fliAZ operon. Therefore, in order to avoid transcription from the class 3 promoter, all the host strains that we used for this study possessed an in-frame deletion of the fliA gene. As shown in Table 4, the effect of the fliZ mutation on β-galactosidase activity was more pronounced in MGC (more than 5-fold) than in LB (less than 2-fold).
Table 4.
Effects of growth media and ydiV mutation on FliZ control of class 2 transcriptiona
| Medium | Genotypeb |
Plasmid | β-Galactosidase activity (Miller units) | ||
|---|---|---|---|---|---|
| ydiV | fliA | fliZ | |||
| LB | + | − | + | None | 236 ± 8 |
| + | − | − | None | 123 ± 21 | |
| − | − | + | None | 226 ± 60 | |
| − | − | − | None | 220 ± 26 | |
| MGC | + | − | + | None | 31 ± 6 |
| + | − | − | None | 6 ± 1 | |
| − | − | + | None | 248 ± 33 | |
| − | − | − | None | 251 ± 48 | |
| + | − | − | pQE80K | 76 ± 23 | |
| + | − | − | pQE80K-fliZ | 386 ± 60 | |
| − | − | − | pQE80K | 502 ± 59 | |
| − | − | − | pQE80K-fliZ | 519 ± 56 | |
β-Galactosidase activity was assayed with strains harboring pFZY1-PfliA-lacZ.
Strains used were ydiV+ fliA fliZ+ strain KK1004iA, ydiV+ fliA fliZ strain KK1004iAZ, ydiV fliA fliZ+ strain KK1004ViA, and ydiV fliA fliZ mutant KK1004ViAZ. −, the gene is deleted.
Next, we examined motility and flagellin (FliC) production in LB and MGC. As shown in Fig. 1, the effect of the fliZ mutation on motility and flagellin level was also more pronounced in MGC than in LB. These results indicate that Salmonella cells require FliZ for efficient flagellar synthesis more strictly in poor medium than in rich medium.
Fig. 1.
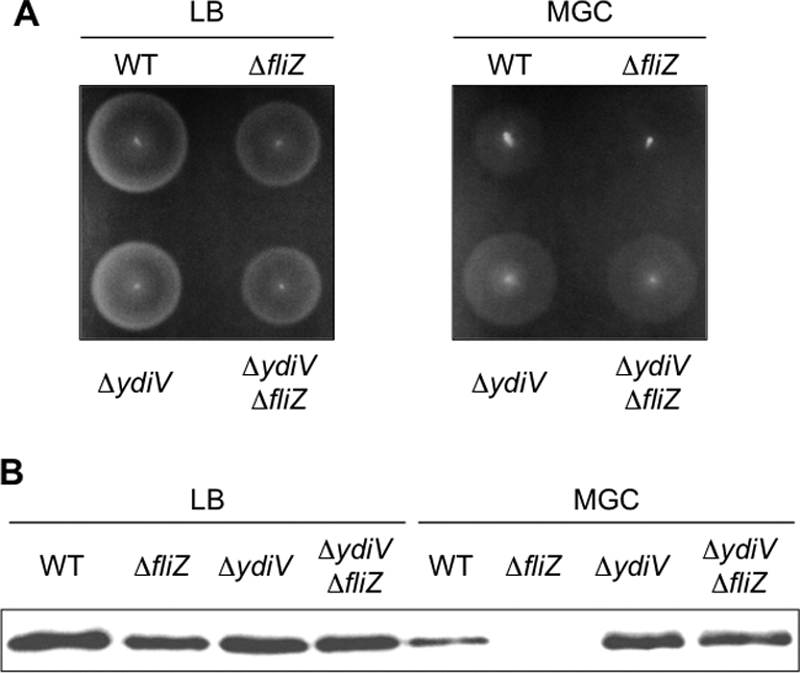
Effect of the fliZ mutation on motility (A) and the steady-state level of flagellin (B). (A) Single colonies were stabbed onto an LB motility agar plate (left panel) or an MGC motility agar plate (right panel) and incubated for 4 h at 30°C. Strains used include KK1004 (wild type [WT]), KK1004iZ (ΔfliZ), KK1004V (ΔydiV), and KK1004ViZ (ΔydiV ΔfliZ). (B) When the bacterial growth reached logarithmic phase, the cells were sampled and separated by SDS-PAGE. Flagellin was detected by Western blotting with an anti-FliC polyclonal antibody. Strains used were the same as those used for panel A.
FliZ acts in a regulatory pathway upstream of YdiV.
Saini et al. (37) reported that the fliZ deletion caused an approximately 50% decrease in the FlhC-3×FLAG protein level in rich medium. Therefore, there is a possibility that inhibition of class 2 operon expression by the fliZ mutation in poor medium might be due to the decreased level of the FlhC protein. However, we could not observe any significant effect of the fliZ mutation on the FlhC-3×FLAG protein level in MGC (data not shown). Therefore, FliZ-dependent activation of class 2 operon expression is unlikely to be mediated by modulation of the intracellular FlhD4C2 level, at least in the strains used here.
Since YdiV was shown to be a mediator of the nutritional cue to flagellar synthesis (41), the above-mentioned results raised the possibility that YdiV is involved in the fliZ-mediated regulation of the flagellar regulon. In order to test this, the effect of the fliZ mutation on flagellar gene expression was examined in the absence of YdiV. As shown in Fig. 1 and Table 4, the negative effects of the fliZ mutation on motility, flagellin production, and class 2 gene transcription in MGC were all relieved in the ydiV mutant background. These results suggest the hypothesis that FliZ acts upstream of YdiV on the common regulatory pathway.
In order to further confirm this, the effect of FliZ overexpression from pQE80K-fliZ on class 2 operon transcription was examined with the strains harboring pFZY1-PfliA-lacZ (Table 4). Introduction of the empty vector pQE80K enhanced PfliA-lacZ expression. This is consistent with a previous report showing that the lacI gene on the plasmid upregulates at least some flagellar genes (8). In the ydiV+ background, FliZ overexpression enhanced PfliA-lacZ expression 5-fold. In contrast, FliZ overexpression had no significant effect on PfliA-lacZ expression in the absence of YdiV. These results conform to the hypothesis mentioned above. Taken together, we conclude that the FliZ protein is a direct or indirect antagonist of the anti-FlhD4C2 factor YdiV in transcriptional control of the class 2 flagellar operons. Interestingly, overexpression of FliZ in the ydiV+ strain did not attain a level of expression of PfliA-lacZ equivalent to its expression level in the ydiV mutant. This is discussed below.
FliZ negatively regulates transcription of the ydiV gene.
As described in the introduction, Wozniak et al. (43) reported that ydiV transcription is under the negative regulation of the flhDC genes. Since their experiment was carried out in rich medium, we examined the effect of the flhD mutation on ydiV transcription in poor medium. As shown in Table 5, when the cells of the strain carrying the ydiV-lacZ transcriptional fusion gene on the chromosome were grown in MGC, the flhD::Tn10 mutation increased β-galactosidase activity 2.4-fold.
Table 5.
Effects of the flhD or fliZ mutation on the ydiV-lac expressiona
| Medium | Plasmid | β-Galactosidase activity (Miller units) with mutationb |
||
|---|---|---|---|---|
| None | flhD::Tn10 | fliZ | ||
| LB | None | 173 ± 61 | 404 ± 15 | 376 ± 34 |
| MGC | None | 156 ± 13 | 381 ± 10 | 290 ± 37 |
| MGC | pQE80L | 212 ± 19 | 336 ± 9 | 341 ± 9 |
| MGC | pQE80L-flhCD | 198 ± 17 | 191 ± 44 | 323 ± 20 |
| MGC | pQE80L-fliZ | 159 ± 10 | 188 ± 13 | 180 ± 22 |
| MGC | pQE80L-ydiV | 380 ± 6 | 385 ± 20 | 379 ± 35 |
All strains used carried the ydiV-lacZ fusion gene on the chromosome, which was transduced from KK1004 ydiV-lac.
Strains used were wild-type KK1004, flhD::Tn10 strain KK1004hDC, and fliZ strain KK1004iZ.
Since the fliZ gene is under the positive control of FlhD4C2 (12, 23), FliZ is one of the most plausible candidates of the regulator involved in FlhD4C2 control of the ydiV gene. In order to test this, the effect of the fliZ mutation on ydiV-lacZ expression was examined (Table 5). As expected, the fliZ mutation increased β-galactosidase activity about 2-fold. Ectopic expression of FliZ from pQE80L-fliZ in the flhD::Tn10 mutant resulted in decreased β-galactosidase activity, whereas that of FlhC and FlhD from pQE80L-flhCD in the fliZ mutant had no significant effect on enzyme activity (Table 5). These results indicate that FlhD4C2 affects ydiV expression through its positive control of fliZ expression. It should be noted that the chromosomal ydiV-lacZ fusion gene was not completely turned off by FliZ expression from pQE80L-fliZ (Table 5). This is consistent with the result shown in Table 4. These results suggest that both FliZ-sensitive and FliZ-insensitive transcriptions are responsible for ydiV expression. This is discussed further below.
Because YdiV inhibits fliZ expression by acting as an anti-FlhD4C2 factor (41), the above-described result suggests that the ydiV gene should be autogenously activated. As expected, YdiV overexpression from pQE80L-ydiV enhanced ydiV-lacZ expression, and the flhD or fliZ mutation did not enhance further its expression (Table 5).
FliZ represses ydiV transcription from the nlpC promoter.
The ydiV gene is located just downstream of the nlpC gene (Fig. 2). Jonas et al. (16) reported results suggesting that two promoters located within the nlpC-ydiV intergenic region may be responsible for ydiV transcription. Recently, however, we showed that these promoters are inactive in our strains and that the ydiV gene is transcribed mainly from a promoter, PnlpC, located upstream of the nlpC gene (41). Therefore, in this study, we examined the structure and function of PnlpC further. A bioinformatics study using the software Neural Network Promoter Prediction (http://www.fruitfly.org/seq_tools/promoter.html) (36) suggested one σ70-type promoter sequence, TTGAAA-N18-TAAATT, whose predicted transcriptional start site is nucleotide C or A at bp position 30 or 29 upstream of the start codon of the nlpC gene (Fig. 2A). Unfortunately, our attempts to identify the transcriptional start site of the nlpC gene by primer extension analysis of mRNA or by the RNA ligation-mediated reverse transcription-PCR method were unsuccessful for unknown reasons. So, we analyzed the activity of PnlpC using a single-copy promoter-probe vector, pFZY1.
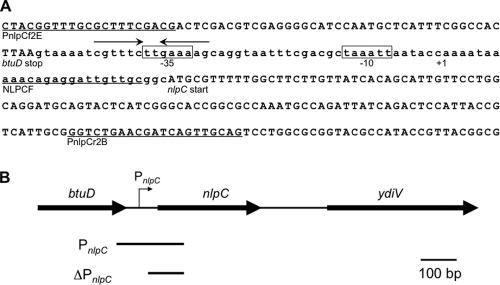
Fig. 2.
Sequence of the nlpC promoter region (A) and the structure of the nlpC-ydiV operon (B). (A) The coding sequences of the btuD and nlpC genes are written in uppercase, whereas the sequence of their intergenic region is written in lowercase letters. The putative −35 and −10 sequences of PnlpC are boxed. The predicted transcriptional start point of the nlpC promoter is labeled +1. The convergent arrows indicate the inverted repetitious sequences, which may act as a FliZ-binding site. Nucleotide sequences corresponding to the primers used for PCR amplification of the nlpC promoter region are underlined. (B) The btuD, nlpC, and ydiV genes are indicated by horizontal arrows. The chromosomal regions cloned on the recombinant reporter plasmids are shown by horizontal lines. The putative promoter sequence shown in panel A is present in the PnlpC fragment but absent from the ΔPnlpC fragment.
DNA fragments containing the upstream region of the nlpC gene with and without this promoter-like sequence were inserted into pFZY1 to obtain pFZY1-PnlpC-lacZ and pFZY1-ΔPnlpC-lacZ, respectively (Fig. 2B). In the wild-type cells grown in MGC, β-galactosidase activity was more than 20-fold higher from pFZY1-PnlpC-lacZ (310 ± 37 Miller units) than from pFZY1-ΔPnlpC-lacZ (15 ± 5 Miller units), suggesting that this promoter-like sequence is functional in Salmonella.
Next, we analyzed the effect of the fliZ mutation on β-galactosidase activity from pFZY1-PnlpC-lacZ. In order to avoid any possible effect of YdiV on transcription from PnlpC through the ydiV-fliZ regulatory loop, β-galactosidase activity was assayed in the ydiV mutant background. As a result, the enzyme activity was higher in the ydiV fliZ deletion strain (272 ± 51 Miller units) than in the ydiV fliZ+ strain (38 ± 9 Miller units), indicating that this promoter is responsive to negative regulation by FliZ. Note that, in the presence of FliZ, the enzyme activity was much lower in the ydiV mutant than in the wild-type strain. This indicates that YdiV activates its own expression by inhibiting FliZ expression. Furthermore, in the ydiV fliZ+ background, the enzyme activity from the chromosomal ydiV-lacZ fusion gene (Table 5) was higher than that from pFZY1-PnlpC-lacZ, which suggests an additional promoter for ydiV expression on the chromosome. This promoter may be responsible for the FliZ-insensitive transcription of the ydiV gene described above.
FliZ is a repressor for the nlpC promoter.
PnlpC activity was analyzed further by an in vitro transcription experiment using plasmid pQE80L-ΔPT5-PnlpC as a template. If the above prediction about the transcriptional start site is correct, a 184- or 183-nucleotide transcript is expected to be produced by transcription from PnlpC in this plasmid. In the presence of σ70 RNA polymerase, an RNA product of about 190 nucleotides was produced in addition to a 107-nucleotide RNA I derived from the vector sequence (Fig. 3, lane 2). This result indicates the validity of the above-mentioned prediction for PnlpC.
Fig. 3.
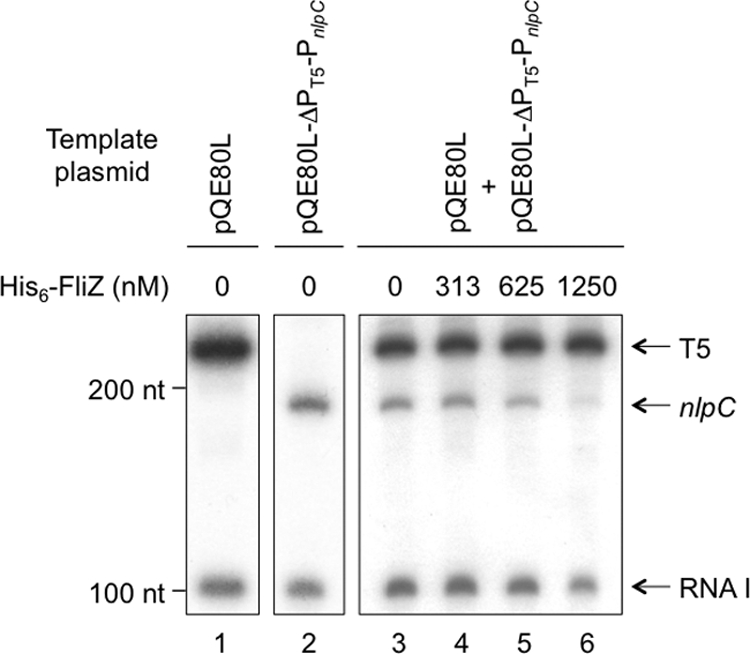
Effects of FliZ on nlpC transcription in vitro. A plasmid carrying the T5 or nlpC promoter was transcribed in vitro with E. coli RNA polymerase holoenzyme with or without His6-FliZ in the presence of [α-32P]UTP. His6-FliZ was added to the reaction mixture at the final concentrations indicated above the lanes. Synthesized RNAs were separated on an 8% polyacrylamide gel containing 6 M urea and detected by autoradiography. Positions of the T5, nlpC, and RNA I transcripts are indicated by arrows. nt, nucleotides.
In order to analyze the function of the FliZ protein in vitro, we purified the His6-FliZ protein from E. coli cells harboring pQE80L-fliZ and examined its effect on in vitro transcription from PnlpC. An empty vector, pQE80L, was also included in the reaction mixture as a control template, which produces a 230-nucleotide RNA from the T5 promoter. Addition of the purified His6-FliZ protein into the reaction mixture reduced the amount of the nlpC transcript in a concentration-dependent manner, whereas that of the RNA product from the T5 promoter was not affected (Fig. 3, lanes 3 to 6). This result indicates that FliZ specifically represses transcription from PnlpC. Interestingly, at the highest concentration of His6-FliZ used in this study, the amount of RNA I was also somewhat reduced (Fig. 3, lane 6). This is discussed below.
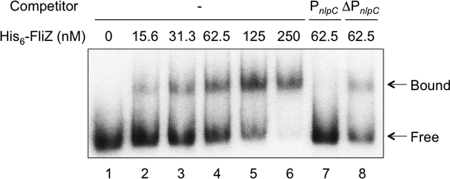
FliZ is known to contain a SAM (sterile alpha motif)-like phage integrase domain (37), suggesting the possibility that FliZ has a DNA-binding ability. In order to test this, gel mobility shift assay of the 32P-labeled DNA fragment containing PnlpC was performed in the presence of the purified His6-FliZ protein. A shifted band was observed in the presence of His6-FliZ, and its amount increased in a concentration-dependent manner (Fig. 4, lanes 2 to 5). Band shift was not observed with the 32P-labeled DNA fragment lacking PnlpC (data not shown), indicating that FliZ binds specifically to the DNA region containing PnlpC. Binding specificity was further examined by a competition experiment using unlabeled DNAs as competitors. An unlabeled DNA fragment containing PnlpC behaved as an effective competitor (Fig. 4, lane 7), whereas an unlabeled DNA fragment without PnlpC did not (Fig. 4, lane 8), indicating the high specificity of FliZ binding to the DNA region containing PnlpC.
Fig. 4.
Gel mobility shift assay of the nlpC promoter DNA by FliZ. A 287-bp PnlpC DNA fragment containing the nlpC promoter was 5′-end labeled with 32P and used as a probe. Each mixture contained 0.2 nM labeled DNA. After incubation with His6-FliZ at the final concentrations indicated above the lanes, the DNA-protein mixtures were separated on a native 5% polyacrylamide gel, and labeled DNA was detected by autoradiography. In the competition experiments (lanes 7 and 8), 2 nM concentrations of the unlabeled PnlpC and ΔPnlpC fragments shown in Fig. 2B were used. Positions of free and bound DNAs are indicated by arrows.
DISCUSSION
We showed that FliZ is required for efficient expression of the class 2 flagellar operons in poor medium in Salmonella (Table 4). Although Saini et al. (37) reported data suggesting that FliZ functions at the posttranslational level of FlhD4C2 by regulating its stability, we could not observe any significant effect of the fliZ mutation on the FlhC protein level (data not shown). Instead, we showed that FliZ binds to the promoter region of the nlpC-ydiV operon and represses its transcription (Fig. 3 and 4), which results in the decreased expression of the ydiV gene, encoding an anti-FlhD4C2 factor.
Our current model depicting a regulatory pathway involving FliZ and YdiV is summarized in Fig. 5A. According to this model, the fliZ and ydiV genes form a regulatory loop and are differentially expressed in response to nutritional cues. Since ydiV expression is enhanced in poor medium at the translational step (41), an increased level of the YdiV protein would activate its own transcription by inhibiting the FlhD4C2-dependent expression of the fliZ gene. Under this condition, the flagellar operons stay at a low expression level. On the other hand, in a rich nutritional environment where the amount of the YdiV protein is low, the FlhD4C2 complex is free from inhibition by YdiV, resulting in high expression of the FliZ protein, which in turn represses ydiV expression. This leads the flagellar operons to the actively expressed state. Thus, the FliZ-YdiV regulatory loop has a stable bipartite expression profile in switching between highly and weakly motile lifestyles in responding to nutrient availability (Fig. 5B).
Fig. 5.
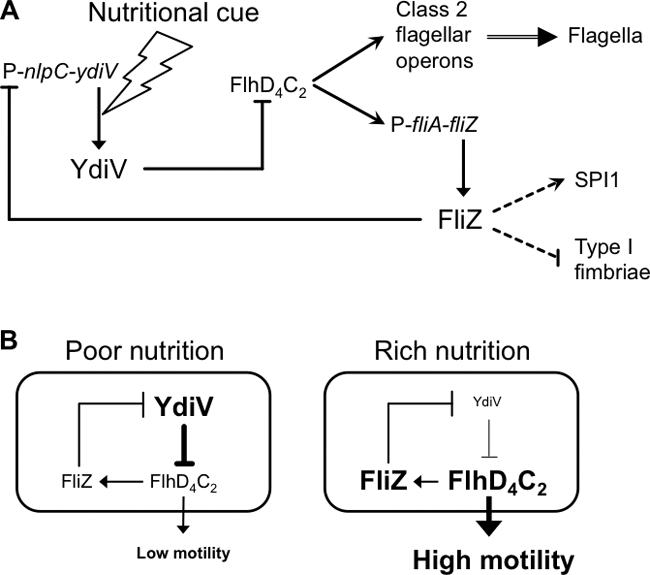
Models illustrating the FliZ-dependent activation of the flagellar class 2 operons via control of the expression of the ydiV gene (A) and the interplay between YdiV and FliZ in motility control in response to nutritional conditions (B). Details are provided in the text.
In this study, we analyzed ydiV expression by focusing on the PnlpC promoter, which is susceptible to FliZ repression. However, as described above, we suggest that an additional promoter insensitive to FliZ repression may also be involved in ydiV transcription. We anticipate that transcriptional readthrough from the btuCED operon upstream of the nlpC gene may be responsible for this transcription, because there is no apparent ρ-independent terminator-like sequence in the intergenic region between the btuD and nlpC genes (Fig. 2A).
FliZ is known to regulate the SPI1 genes positively (5, 14, 17) and the type 1 fimbrial genes negatively (6, 39). FliZ control of these genes was observed in rich medium, in which YdiV expression must be low. Therefore, it is reasonable to predict that this FliZ control may not be affected by the ydiV mutation. This prediction is consistent with the previous observation that the expression of one of the SPI1 genes, sipC, was not affected by the ydiV mutation (10). Therefore, we believe that FliZ control of these genes is not mediated by YdiV. FliZ may regulate these genes through a mechanism different from that of FliZ control of the flagellar genes.
FliZ from Xenorhabdus nematophila was shown to specifically bind to the promoter region of the flhDC operon and to function as an activator (29). Although X. nematophila FliZ shows 56% identity with Salmonella FliZ, they differ significantly from each other in their C-terminal sequences (data not shown). Since FliZ is not involved in the regulation of the flhDC operon in Salmonella (23), there is a possibility that the C-terminal portion of FliZ determines its DNA-binding specificity. This possibility is supported by our preliminary experiment showing that a C-terminally truncated FliZ protein does not retain DNA-binding ability (our unpublished result).
The C-terminal portion of FliZ contains a SAM-like phage integrase domain (37), which is also included in Cre, Flp, and XerD recombinases (9). These recombinases are known to recognize palindromic sequences (3, 40). Interestingly, a 14-bp palindromic sequence (CGTTTCTTGAAAAG, where underlining indicates complementary nucleotides) was found also at the nlpC promoter region (Fig. 2A). This sequence overlaps the putative −35 element of the nlpC promoter and thus may be a plausible target site of FliZ binding. A similar palindromic sequence (GAGTTCTTGAAGTG) exists around the −35 element of the RNA I promoter, whereas such a sequence is not observed at the T5 promoter region of pQE80L. This structural difference conforms to the results of Fig. 3 showing that the nlpC and RNA I promoters were susceptible to FliZ repression but that the T5 promoter was not. Identification of a recognition sequence of FliZ will help us to understand a molecular mechanism underlying FliZ control of the ydiV gene and reveal additional targets of FliZ on the bacterial genome.
ACKNOWLEDGMENTS
We thank Tatsuhiko Abo, Yuhei Chadani, and Sayumi Shintani for strains and invaluable discussion. Radioisotope facilities were provided by the Advanced Science Research Center Tsushima Division, Okayama University.
Footnotes
Published ahead of print on 29 July 2011.
REFERENCES
- 1. Aizawa S. I. 2001. Bacterial flagella and type III secretion systems. FEMS Microbiol. Lett. 202:157–164 [DOI] [PubMed] [Google Scholar]
- 2. Aldridge P., Hughes K. T. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5:160–165 [DOI] [PubMed] [Google Scholar]
- 3. Blakely G., Sherratt D. 1996. Determinants of selectivity in Xer site-specific recombination. Genes Dev. 10:762–773 [DOI] [PubMed] [Google Scholar]
- 4. Chilcott G. S., Hughes K. T. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chubiz J. E., Golubeva Y. A., Lin D., Miller L. D., Slauch J. M. 2010. FliZ regulates expression of the Salmonella pathogenicity island 1 invasion locus by controlling HilD protein activity in Salmonella enterica serovar Typhimurium. J. Bacteriol. 192:6261–6270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clegg S., Hughes K. T. 2002. FimZ is a molecular link between sticking and swimming in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:1209–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eswarappa S. M., Karnam G., Nagarajan A. G., Chakraborty S., Chakravortty D. 2009. lac repressor is an antivirulence factor of Salmonella enterica: its role in the evolution of virulence in Salmonella. PLoS One 4:e5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gopaul D. N., Duyne G. D. 1999. Structure and mechanism in site-specific recombination. Curr. Opin. Struct. Biol. 9:14–20 [DOI] [PubMed] [Google Scholar]
- 10. Hisert K. B., et al. 2005. A glutamate-alanine-leucine (EAL) domain protein of Salmonella controls bacterial survival in mice, antioxidant defence and killing of macrophages: role of cyclic diGMP. Mol. Microbiol. 56:1234–1245 [DOI] [PubMed] [Google Scholar]
- 11. Hughes K. T., Gillen K. L., Semon M. J., Karlinsey J. E. 1993. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science 262:1277–1280 [DOI] [PubMed] [Google Scholar]
- 12. Ikebe T., Iyoda S., Kutsukake K. 1999. Structure and expression of the fliA operon of Salmonella typhimurium. Microbiology 145:1389–1396 [DOI] [PubMed] [Google Scholar]
- 13. Ikebe T., Iyoda S., Kutsukake K. 1999. Promoter analysis of the class 2 flagellar operons of Salmonella. Genes Genet. Syst. 74:179–183 [DOI] [PubMed] [Google Scholar]
- 14. Iyoda S., Kamidoi T., Hirose K., Kutsukake K., Watanabe H. 2001. A flagellar gene fliZ regulates the expression of invasion genes and virulence phenotype in Salmonella enterica serovar Typhimurium. Microb. Pathog. 30:81–90 [DOI] [PubMed] [Google Scholar]
- 15. Iyoda S., Kutsukake K. 1995. Molecular dissection of the flagellum-specific anti-sigma factor, FlgM, of Salmonella typhimurium. Mol. Gen. Genet. 249:417–424 [DOI] [PubMed] [Google Scholar]
- 16. Jonas K., et al. 2010. Complex regulatory network encompassing the Csr, c-di-GMP and motility systems of Salmonella Typhimurium. Environ. Microbiol. 12:524–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kage H., Takaya A., Ohya M., Yamamoto T. 2008. Coordinated regulation of expression of Salmonella pathogenicity island 1 and flagellar type III secretion systems by ATP-dependent ClpXP protease. J. Bacteriol. 190:2470–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koop A. H., Hartley M. E., Bourgeois S. 1987. A low-copy-number vector utilizing β-galactosidase for the analysis of gene control elements. Gene 52:245–256 [DOI] [PubMed] [Google Scholar]
- 19. Kutsukake K. 1994. Excretion of the anti-sigma factor through a flagellar substructure couples the flagellar gene expression with flagellar assembly in Salmonella typhimurium. Mol. Gen. Genet. 243:605–612 [DOI] [PubMed] [Google Scholar]
- 20. Kutsukake K. 1997. Autogenous and global control of the flagellar master operon, flhD, in Salmonella typhimurium. Mol. Gen. Genet. 254:440–448 [DOI] [PubMed] [Google Scholar]
- 21. Kutsukake K., Ide N. 1995. Transcriptional analysis of the flgK and fliD operons of Salmonella typhimurium which encode flagellar hook-associated proteins. Mol. Gen. Genet. 247:275–281 [DOI] [PubMed] [Google Scholar]
- 22. Kutsukake K., Iino T. 1994. Role of the FliA-FlgM regulatory system on the transcriptional control of the flagellar regulon and flagellar formation in Salmonella typhimurium. J. Bacteriol. 176:3598–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kutsukake K., Ikebe T., Yamamoto S. 1999. Two novel regulatory genes, fliT and fliZ, in the flagellar regulon of Salmonella. Genes Genet. Syst. 74:287–292 [DOI] [PubMed] [Google Scholar]
- 24. Kutsukake K., Iyoda S., Ohnishi K., Iino T. 1994. Genetic and molecular analyses of the interaction between the flagellum-specific sigma and anti-sigma factors in Salmonella typhimurium. EMBO J. 13:4568–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kutsukake K., Nakashima H., Tominaga A., Abo T. 2006. Two DNA invertases contribute to flagellar phase variation in Salmonella enterica serovar Typhimurium strain LT2. J. Bacteriol. 188:950–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kutsukake K., Nambu T. 2000. Bacterial flagellum: a paradigm for biogenesis of transenvelope supramolecular structures. Recent Res. Dev. Microbiol. 4:607–615 [Google Scholar]
- 27. Kutsukake K., Ohya Y., Iino T. 1990. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J. Bacteriol. 172:741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kutsukake K., Ohya Y., Yamaguchi S., Iino T. 1988. Operon structure of flagellar genes in Salmonella typhimurium. Mol. Gen. Genet. 214:11–15 [DOI] [PubMed] [Google Scholar]
- 29. Lanois A., Jubelin G., Givaudan A. 2008. FliZ, a flagellar regulator, is at the crossroads between motility, haemolysin expression and virulence in the insect pathogenic bacterium Xenorhabdus. Mol. Microbiol. 68:516–533 [DOI] [PubMed] [Google Scholar]
- 30. Liu X., Fujita N., Ishihama A., Matsumura P. 1995. The C-terminal region of the α subunit of Escherichia coli RNA polymerase is required for transcriptional activation of the flagellar level II operons by the FlhD/FlhC complex. J. Bacteriol. 177:5186–5188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu X., Matsumura P. 1994. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J. Bacteriol. 176:7345–7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miller J. 1972. Experiments in molecular genetics, p. 431-433 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 33. Ohnishi K., Kutsukake K., Suzuki H., Iino T. 1990. Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol. Gen. Genet. 221:139–147 [DOI] [PubMed] [Google Scholar]
- 34. Ohnishi K., Kutsukake K., Suzuki H., Iino T. 1992. A novel transcriptional regulation mechanism in the flagellar regulon of Salmonella typhimurium: an anti-sigma factor inhibits the activity of the flagellum-specific sigma factor, σF. Mol. Microbiol. 6:3149–3157 [DOI] [PubMed] [Google Scholar]
- 35. Pesavento C., et al. 2008. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 22:2434–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reese M. R. 2001. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 26:51–56 [DOI] [PubMed] [Google Scholar]
- 37. Saini S., Brown J. D., Aldridge P. D., Rao C. V. 2008. FliZ is a posttranslational activator of FlhD4C2-dependent flagellar gene expression. J. Bacteriol. 190:4979–4988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Saini S., et al. 2010. FliZ induces a kinetic switch in flagellar gene expression. J. Bacteriol. 192:6477–6481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saini S., Slauch J. M., Aldridge P. D., Rao C. V. 2010. Role of cross talk in regulating the dynamic expression of the flagellar Salmonella pathogenicity island 1 and type 1 fimbrial genes. J. Bacteriol. 192:5767–5777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vetcher A. A., et al. 2006. DNA topology and geometry in Flp and Cre recombination. J. Mol. Biol. 357:1089–1104 [DOI] [PubMed] [Google Scholar]
- 41. Wada T., et al. 2011. EAL domain protein YdiV acts as an anti-FlhD4C2 factor responsible for nutritional control of the flagellar regulon in Salmonella enterica serovar Typhimurium. J. Bacteriol. 193:1600–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang S., Fleming R. T., Westbrook E. M., Matsumura P., McKay D. B. 2006. Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription. J. Mol. Biol. 355:798–808 [DOI] [PubMed] [Google Scholar]
- 43. Wozniak C. E., Lee C., Hughes K. T. 2009. T-POP array identifies EcnR and PefI-SrgD as novel regulators of flagellar gene expression J. Bacteriol. 191:1498–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yamamoto S., Kutsukake K. 2006. FljA-mediated posttranscriptional control of phase 1 flagellin expression in flagellar phase variation of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:958–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yamamoto S., Kutsukake K. 2006. FliT acts as an anti-FlhD2C2 factor in the transcriptional control of the flagellar regulon in Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:6703–6708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yokoseki T., Kutsukake K., Ohnishi K., Iino T. 1995. Functional analysis of the flagellar genes in the fliD operon of Salmonella typhimurium. Microbiology 141:1715–1722 [DOI] [PubMed] [Google Scholar]