Abstract
Bacterial two-hybrid analysis identified the Staphylococcus aureus RNA degradosome-like complex to include RNase J1, RNase J2, RNase Y, polynucleotide phosphorylase (PNPase), enolase, phosphofructokinase, and a DEAD box RNA helicase. Results also revealed that the recently recognized RNase RnpA interacts with the S. aureus degradosome and that this interaction is conserved in other Gram-positive organisms.
TEXT
Escherichia coli bulk RNA degradation is mediated by a holoenzyme complex, the RNA degradosome, which includes the ribonucleases RNase E and polynucleotide phosphorylase (PNPase), an RNA helicase, RhlB, and the glycolytic enzyme enolase (3, 20). RNase E is considered the key component of the E. coli degradosome; it catalyzes the initiation of mRNA degradation and serves as a scaffold for the assembly of the other degradosome components (2). By comparison, most Gram-positive bacteria do not contain an RNase E amino acid ortholog and much less is known about their mRNA degradation machinery. Nonetheless, recent studies suggest that the Gram-positive organism Bacillus subtilis produces two RNase E functional orthologs, ribonucleases J1 (RNase J1) and J2 (RNase J2) (17). Using bacterial two-hybrid analyses, both enzymes have been shown to interact with an RNA degradosome-like complex consisting of PNPase, enolase, and a DEAD box RNA helicase (CshA), as well as phosphofructokinase and RNase Y (6).
Staphylococcus aureus is a Gram-positive bacterial pathogen of immense health care concern that is evolutionarily similar, but divergent, from the Bacillus genus. Herein we used bacterial two-hybrid and biochemical analyses to measure interactions between putative members of the S. aureus RNA degradosome and compare the results to those with B. subtilis. Results revealed that although many interactions are conserved between both bacterial species, differences are likely to exist. Moreover, we show that the RNase RnpA interacts with components of the S. aureus RNA degradosome and that this interaction is conserved within B. subtilis.
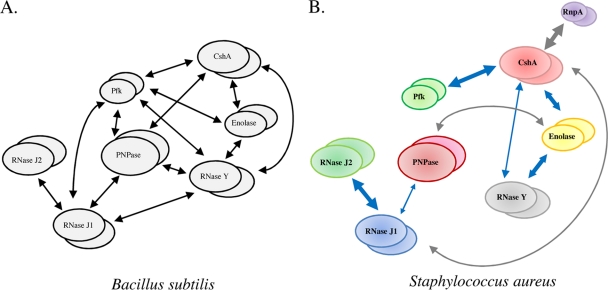
Using bacterial two-hybrid analyses, interactions between putative members of the B. subtilis RNA degradosome were recently measured and used to develop the first model of the Gram-positive holoenzyme complex (Fig. 1A) (6, 13). According to the model, the E. coli RNase E orthologs RNase J1 and RNase J2 interact with one another; RNase J1 also interacts with two additional ribonucleases, PNPase and RNase Y, as well as with the glycolytic enzyme phosphofructokinase, each of which binds to the RNA helicase CshA (Fig. 1A). CshA also binds to a second glycolytic enzyme, enolase. Although the mechanism of B. subtilis cellular RNA degradation has yet to be established, RNases J1 and J2 are bifunctional ribonucleases with endonuclease and 5′→3′ exoribonuclease activities (8, 16, 24). In complex, the enzymes act synergistically and may initiate 5′ mRNA digestion and/or generate endonucleolytic products that could subsequently be digested by RNA helicase-facilitated 3′→5′ PNPase exoribonuclease activity (14, 17). RNase Y is hypothesized to act upon highly structured RNA fragments that are not effectively degraded by RNase J1 (21) and to interact with enolase to modulate the mRNA turnover of Streptococcus pyogenes virulence factors (10). It is not clear what direct role, if any, phosphofructokinase plays in the RNA degradation process.
Fig. 1.
B. subtilis and S. aureus mRNA degradosome-like complexes. (A) B. subtilis RNA degradosome-like complex (6). (B) Deduced S. aureus RNA degradosome-like complex. Blue arrows represent conserved interactions between S. aureus and B. subtilis proteins. Arrow thickness indicates the strength of interactions as determined by quantitative β-galactosidase activity results (see Table S2 in the supplemental material).
S. aureus contains orthologs of each putative B. subtilis RNA degradosome subunit, suggesting that the RNA degradation complexes of these two bacterial species may be highly conserved. To evaluate this possibility, the bacterial adenylate cyclase-based two-hybrid (BACTH; Euromedex, Inc., France) system was used to measure all possible combinations of interactions between S. aureus enolase (SAR0832), phosphofructokinase (SAR1777), DEAD box RNA helicase (SAR2168), PNPase (SAR1250), RNase J1 (SAR1063), RNase J2 (SAR1251), and RNase Y (SAR1262), as well as the recently recognized S. aureus RNase RnpA (SAR2799) (19). Accordingly, the coding sequence of each gene was amplified from S. aureus strain UAMS-1 and cloned into the pCRII vector (Invitrogen, CA) (see Tables S1 and S2 in the supplemental material). Genes were subsequently digested with BamHI or XbaI and KpnI and cloned in frame into the bacterial two-hybrid expression vectors pKNT25, pKT25, pUT18, and pUT18C to create N- and C-terminal fusions to the catalytic domains T25 and T18 of Bordetella pertussis adenylate cyclase CyaA (see Table S2 in the supplemental material). All combinations of plasmid pairs were transformed into E. coli DHM1 (Δcya) cells. Cellular interactions between T25- and T18-fused proteins were measured as a function of their ability to reconstitute CyaA activity and, consequently, cellular β-galactosidase levels due to cyclic AMP-mediated activation of catabolic operons (11). DHM1 cells transformed with the BACTH system kit-supplied plasmids pKT25-zip and pUT18C-zip or with the empty vectors pKT25 and pUT18C were used as positive and negative controls, respectively.
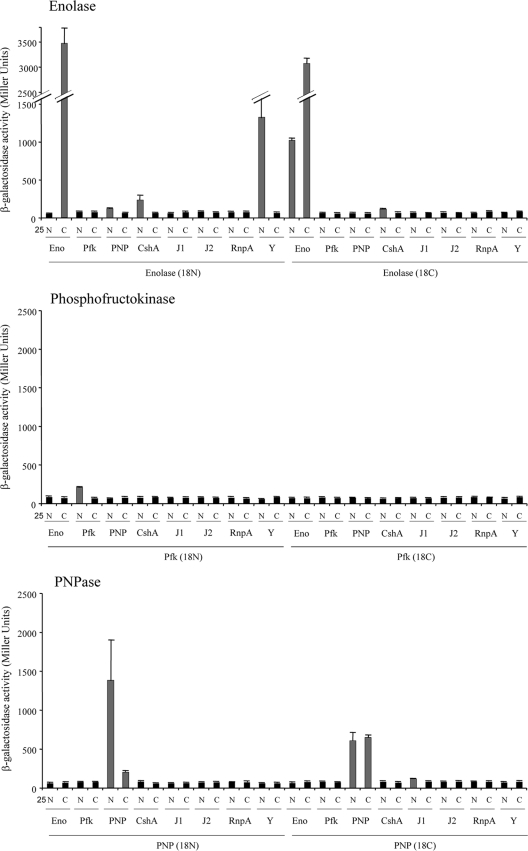
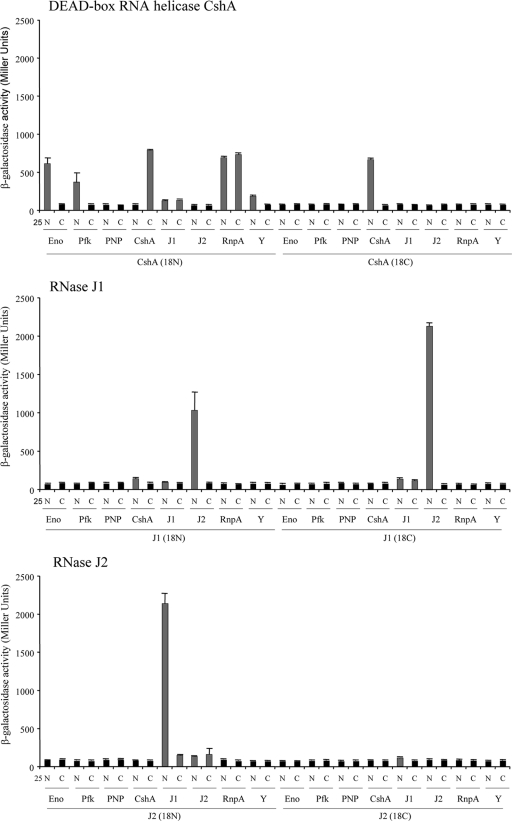
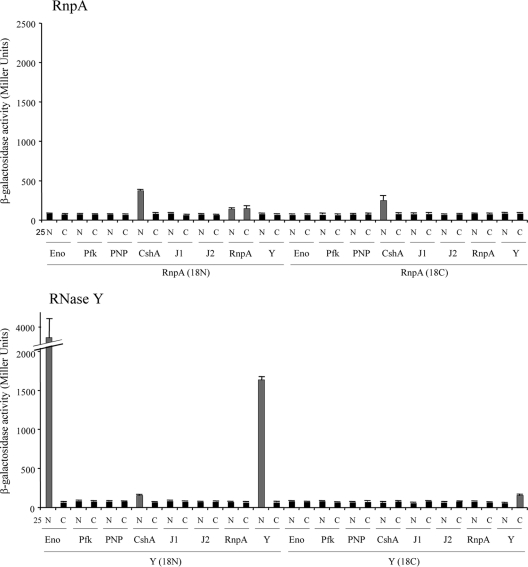
To measure protein-protein interactions, the β-galactosidase levels of a total of 256 transformants, collectively representing all possible plasmid pairs, were independently measured (see Table S3 in the supplemental material). Transformants were grown statically overnight at 30°C in Luria-Bertani (LB) medium supplemented with 100 μg/ml ampicillin, 50 μg/ml kanamycin, and 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). A 500-μl aliquot was added to 2 ml of M63 minimal medium [15 mM (NH4)2SO4, 100 mM KH2PO4, 1.8 μM FeSO47H2O, 3 μM vitamin B1, pH 7.0], and the optical density at 600 nm was measured. Cells were subsequently permeabilized with 30 μl of chloroform and 30 μl of 0.1% SDS, and an aliquot (500 μl) was mixed with 500 μl of PM2 buffer (70 mM Na2HPO4-H2O, 30 mM NaHPO4-H2O, 1 mM MgSO4, 0.2 mM MnSO4, 100 mM β-mercaptoethanol). For β-galactosidase measurements, 250 μl of o-nitrophenol-β-galactoside (ONPG; 4 mg/ml) was added and reactions were stopped after 15 min of incubation at 30°C by adding 500 μl of 1 M Na2CO3. The absorbance was read at 420 nm (cleavage of ONPG) and 550 nm (cellular debris). All interactions were tested in triplicate and are expressed in Miller units (18). Transformants judged to have interacting proteins exhibited a significant increase (Student's t test; P value ≤ 0.01) in β-galactosidase activity in comparison to negative-control cells. Results are presented in Fig. 2 and also in Table S3 in the supplemental material.
Fig. 2.
β-Galactosidase-deduced protein-protein interactions. β-Galactosidase activities of transformants with the indicated S. aureus proteins fused to the N terminus (N) or C terminus (C) of the T25 and T18 catalytic fragments of CyaA. Gray bars represent statistically significant, positive interactions of proteins/domains, whereas black bars represent negative interactions. S. aureus proteins tested included enolase (Eno), phosphofructokinase (Pfk), PNPase (PNP), DEAD box RNA helicase CshA, RNase J1 (J1), RNase J2 (J2), RnpA, and RNase Y (Y).
Taken together, results revealed that orthologs of the putative B. subtilis RNA degradosome subunits are also capable of interacting to form an S. aureus degradosome, suggesting a high degree of conservation of the RNA degradosome among Gram-positive bacteria. However, there were several noted differences between individual protein interactions (compare Fig. 1A and B). For instance, as observed for B. subtilis, our results indicate that S. aureus ribonucleases RNase J1 and RNase J2 interacted the most strongly and that RNase J1 also interacts with PNPase (6). However, we did not detect significant interactions between S. aureus RNase J1 and RNase Y or phosphofructokinase. Rather, we found that the only other RNase J1-interacting protein is the DEAD box RNA helicase CshA. In Gram-negative bacteria, RNase E interacts with both PNPase and an RNA helicase (3, 12) and initiates RNA degradation by cleaving substrate molecules, which are subsequently degraded in a 3′→5′ direction by the concerted activities of RNA helicase and PNPase. Accordingly, the interactions between S. aureus RNase J1 (an RNase E functional ortholog) and both CshA and PNPase suggest a conserved mechanism of RNA degradation in Staphylococcus. Consistent with that possibility, we have previously shown that PNPase contributes to bulk S. aureus mRNA decay (1). Furthermore, the B. subtilis DEAD box RNA helicase CshA interacts with phosphofructokinase, enolase, PNPase, and RNase Y (Fig. 1A) (6, 21). Similarly, we observed that S. aureus CshA interacts with phosphofructokinase, enolase, and RNase Y. However, S. aureus DEAD box RNA helicase does not appear to interact with PNPase (Fig. 1B).
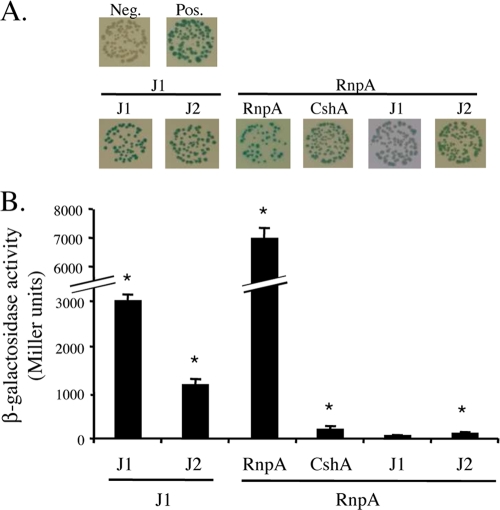
We recently reported that the protein subunit of S. aureus RNase P, RnpA, is involved in bulk RNA degradation and hypothesized that the enzyme is a component of the organism's RNA turnover machinery (19). Consistent with that prediction, results revealed that S. aureus CshA interacts with RnpA, suggesting that RNA helicase may augment cellular RnpA RNase activity. To investigate whether the observed interaction between S. aureus RnpA and CshA is conserved among Gram-positive bacteria, the bacterial two-hybrid studies were expanded to include B. subtilis RnpA (BSU41050) and CshA (BSU04580). Accordingly, both B. subtilis genes were cloned in frame into the bacterial two-hybrid expression vectors for quantitative measurement of β-galactosidase activity (as described above). Interactions were also qualitatively assessed by color change on agar plates containing 0.5 mM IPTG and 40 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside. B. subtilis RNase J1 (BSU14530) and RNase J2 (BSU16780) were used as positive controls, whereas the vector alone was used as a negative control (6). As shown in Fig. 3, our results confirmed that B. subtilis RNase J1 dimerizes and interacts with RNase J2. Further, as observed for S. aureus, B. subtilis RnpA dimerized and interacted with CshA but not with RNase J1 (see Table S4 in the supplemental material). Collectively, these data suggest that RnpA interacts with the DEAD box RNA helicase CshA and that this interaction is conserved among Gram-positive bacteria.
Fig. 3.
Bacterial two-hybrid analyses of selected B. subtilis proteins. (A) Qualitative analysis of the β-galactosidase activities of the indicated protein pairs. (B) Corresponding quantitative analysis of β-galactosidase activity. * indicates a P value of ≤0.01 (see Table S4 in the supplemental material). B. subtilis proteins tested were RNase J1 (J1), RNase J2 (J2), CshA, and RnpA.
The interaction of the S. aureus DEAD box RNA helicase CshA with three ribonucleases, RNase J1, RNase Y, and RnpA, suggests that the helicase is a central component of the organism's RNA degradosome. Nonetheless, it is intriguing to consider that CshA could also facilitate the activity of endo- and exoribonucleases independently of their participation in the degradosome complex. Indeed, E. coli PNPase can degrade RNA in the presence of the DEAD box RNA helicase independently of PNPase and RNA helicase binding to RNase E (15). Similarly, our results indicate that each protein examined can multimerize, consistent with previous observations. Indeed, each of these molecules has been shown to form homodimers (DEAD box RNA helicase, enolase, RNase Y, and RnpA [4, 6, 9, 15]), trimers (PNPase [22]), tetramers (phosphofructokinase and RNase J1/J2 [7, 23]), or combinations of multimer populations (RNase J1 and J2 form homodimers and homotetramers [17]). While the goal of the current study is to genetically evaluate the potential interacting partners of the S. aureus degradosome, it is clear that such work must be biochemically complemented to fully recognize the stoichiometry of the RNA degradosome subunits and to observe whether subset populations of proteins exist within the cell (such as helicase/RNase combinations).
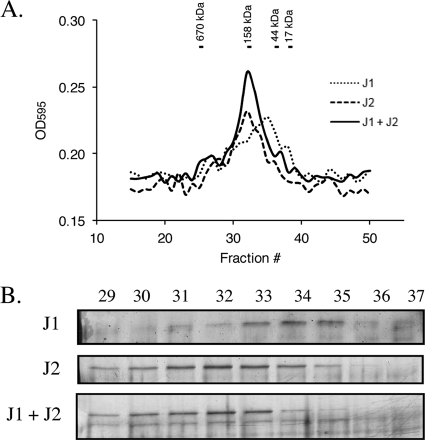
As a preliminary means of validating our bacterial two-hybrid analyses, we biochemically confirmed that purified RNase J1 and RNase J2 interact in vitro; both are hypothesized to be essential S. aureus enzymes (5) and, thus, may play major roles in mRNA decay. For purification, the RNase J1 gene was cloned into the pTrcHis2A expression vector (Invitrogen) and the RNase J2 gene was cloned into pET-30 (Novagen, WI). Proteins were expressed within E. coli BL21(DE3) cells following the addition of 0.25 mM IPTG for 2 h at 30°C or 0.5 mM IPTG for 3 h at 37°C for RNase J1 and RNase J2, respectively. Cultures were centrifuged and cell pellets were resuspended in lysis buffer (50 mM Tris-HCl [pH 7.5], 10% glycerol, 300 mM NaCl, 10 mM imidazole) supplemented with protease inhibitor cocktail (1/30, P8849; Sigma, MO), 5 μg/ml lysozyme (Sigma, MO), and Benzonase nuclease according to the manufacturer's instructions (Novagen, WI). Cells were disrupted by three cycles of pressure in a French press (14,000 lb/in2), cellular debris was removed by centrifugation for 30 min at 13,000 rpm, and proteins were purified using HisPur cobalt columns according to the manufacturer's instructions (Pierce, IL). To assess whether S. aureus RNase J1 and RNase J2 interact, 75 μg of each purified protein was incubated individually or in combination for 30 min at 37°C and was subjected to gel filtration on a Superose 6 HR 10/30 column (GE Life Sciences) in phosphate-buffered saline (PBS) buffer (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.47 mM KH2PO4, pH 7.4). Resulting fractions (25 μl) were loaded on a 10% SDS-PAGE gel, and proteins were detected with silver stain according to the manufacturer's instructions (Bio-Rad Laboratories, CA). As shown in Fig. 4A and B, RNase J1 eluted predominantly as monomers (collected in later fractions; migrated to around 67 kDa). Conversely, RNase J2 formed predominantly dimers (collected in earlier fractions; migrated to around 140 kDa). Following coincubation of both RNases, we observed a shift in the elution and gel migration of RNase J1. Further, the detection of RNase J1 in earlier fractions coincided with a loss of RNase J2 monomers. Taken together, these results suggest that S. aureus RNase J1 and RNase J2 interact under these experimental conditions.
Fig. 4.
In vitro interaction of purified S. aureus RNase J1 and RNase J2. (A) Gel filtration analysis of RNase J1, RNase J2, and RNase J1 plus RNase J2, as measured by determining the optical density at 595 nm (OD595). Molecular mass size standards are shown above the chromatogram. (B) Silver-stained 10% SDS-PAGE of eluted RNase J1, RNase J2, and RNase J1 plus RNase J2. Elution fractions are indicated above the gels.
The bacterial two-hybrid system has been an important tool in identifying components of the putative B. subtilis RNA degradosome. For this study, we also used this system to determine and draw comparisons between the S. aureus and B. subtilis RNA degradosome-like complexes. Results revealed that most interactions are conserved, but significant differences are likely to exist. Further, we show that RnpA interacts with the DEAD box RNA helicase in both S. aureus and B. subtilis.
Supplementary Material
Acknowledgments
We thank Gregory Tombline for technical assistance and John Morrison for critical reading of the manuscript.
This research was supported by NIH, NIAID award 1R01 AI073780-01 (P.M.D.).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 15 July 2011.
REFERENCES
- 1. Anderson K. L., Dunman P. M. 2009. Messenger RNA turnover processes in Escherichia coli, Bacillus subtilis, and emerging studies in Staphylococcus aureus. Int. J. Microbiol. 2009:525491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carpousis A. J. 2002. The Escherichia coli RNA degradosome: structure, function and relationship in other ribonucleolytic multienzyme complexes. Biochem. Soc. Trans. 30:150–155 [PubMed] [Google Scholar]
- 3. Carpousis A. J., Van Houwe G., Ehretsmann C., Krisch H. M. 1994. Copurification of E. coli RNase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell 76:889–900 [DOI] [PubMed] [Google Scholar]
- 4. Chandran V., Luisi B. F. 2006. Recognition of enolase in the Escherichia coli RNA degradosome. J. Mol. Biol. 358:8–15 [DOI] [PubMed] [Google Scholar]
- 5. Chaudhuri R. R., et al. 2009. Comprehensive identification of essential Staphylococcus aureus genes using transposon-mediated differential hybridisation (TMDH). BMC Genomics 10:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Commichau F. M., et al. 2009. Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing. Mol. Cell. Proteomics 8:1350–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Evans P. R., Farrants G. W., Hudson P. J. 1981. Phosphofructokinase: structure and control. Philos. Trans. R. Soc. Lond. B Biol. Sci. 293:53–62 [DOI] [PubMed] [Google Scholar]
- 8. Even S., et al. 2005. Ribonucleases J1 and J2: two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res. 33:2141–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fang X. W., et al. 2001. The Bacillus subtilis RNase P holoenzyme contains two RNase P RNA and two RNase P protein subunits. RNA 7:233–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kang S. O., Caparon M. G., Cho K. H. 2010. Virulence gene regulation by CvfA, a putative RNase: the CvfA-enolase complex in Streptococcus pyogenes links nutritional stress, growth-phase control, and virulence gene expression. Infect. Immun. 78:2754–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Karimova G., Pidoux J., Ullmann A., Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U. S. A. 95:5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee K., Cohen S. N. 2003. A Streptomyces coelicolor functional orthologue of Escherichia coli RNase E shows shuffling of catalytic and PNPase-binding domains. Mol. Microbiol. 48:349–360 [DOI] [PubMed] [Google Scholar]
- 13. Lehnik-Habrink M., et al. 2010. The RNA degradosome in Bacillus subtilis: identification of CshA as the major RNA helicase in the multiprotein complex. Mol. Microbiol. 77:958–971 [DOI] [PubMed] [Google Scholar]
- 14. Lin P. H., Lin-Chao S. 2005. RhlB helicase rather than enolase is the beta-subunit of the Escherichia coli polynucleotide phosphorylase (PNPase)-exoribonucleolytic complex. Proc. Natl. Acad. Sci. U. S. A. 102:16590–16595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liou G. G., Chang H. Y., Lin C. S., Lin-Chao S. 2002. DEAD box RhlB RNA helicase physically associates with exoribonuclease PNPase to degrade double-stranded RNA independent of the degradosome-assembling region of RNase E. J. Biol. Chem. 277:41157–41162 [DOI] [PubMed] [Google Scholar]
- 16. Mathy N., et al. 2007. 5′-to-3′ exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell 129:681–692 [DOI] [PubMed] [Google Scholar]
- 17. Mathy N., et al. 2010. Bacillus subtilis ribonucleases J1 and J2 form a complex with altered enzyme behaviour. Mol. Microbiol. 75:489–498 [DOI] [PubMed] [Google Scholar]
- 18. Miller J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 19. Olson P. D., et al. 2011. Small molecule inhibitors of Staphylococcus aureus RnpA alter cellular mRNA turnover, exhibit antimicrobial activity, and attenuate pathogenesis. PLoS Pathog. 7:e1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Py B., Causton H., Mudd E. A., Higgins C. F. 1994. A protein complex mediating mRNA degradation in Escherichia coli. Mol. Microbiol. 14:717–729 [DOI] [PubMed] [Google Scholar]
- 21. Shahbabian K., Jamalli A., Zig L., Putzer H. 2009. RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J. 28:3523–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shi Z., Yang W. Z., Lin-Chao S., Chak K. F., Yuan H. S. 2008. Crystal structure of Escherichia coli PNPase: central channel residues are involved in processive RNA degradation. RNA 14:2361–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shirakihara Y., Evans P. R. 1988. Crystal structure of the complex of phosphofructokinase from Escherichia coli with its reaction products. J. Mol. Biol. 204:973–994 [DOI] [PubMed] [Google Scholar]
- 24. Yao S., Sharp J. S., Bechhofer D. H. 2009. Bacillus subtilis RNase J1 endonuclease and 5′ exonuclease activities in the turnover of ΔermC mRNA. RNA 15:2331–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.