Abstract
Despite the emerging impact of serogroup 11 serotypes in Streptococcus pneumoniae epidemiology, the structures of serogroup 11 capsule types have not been fully elucidated, particularly the locations of O-acetyl substitutions. Here, we report the complete structures of the serotype 11B, 11C, and 11F polysaccharides and a revision to the serotype 11A capsular polysaccharide using nuclear magnetic resonance (NMR). All structures shared a linear, tetrasaccharide backbone with a pendant phosphopolyalcohol. Three of four saccharides are conserved in all serotypes. The individual serotype capsules differed in the identity of one saccharide, the pendant phosphopolyalcohol, and the O-acetylation pattern. Though the assigned locations of O-acetate substitutions in this study differed from those of previous reports, our findings were corroborated with strong correlations to serology and genetics. We examined the binding of serotyping sera to serogroup 11 polysaccharides by using flow cytometry and an inhibition-type enzyme-linked immunosorbent assay (ELISA) and found that de-O-acetylation of capsular polysaccharides by mild hydrolysis decreases its immunoreactivity, supporting the crucial role of O-acetylation in the antigenicity of these polysaccharides. Due to strong correlations between polysaccharide structures and capsule biosynthesis genes, we were able to assign target substrates for the O-acetyltransferases encoded by wcwC, wcwR, wcwT, and wcjE. We identified antigenic determinants for serogroup 11 serotyping sera and highlight the idea that conventional serotyping methods are not capable of recognizing all putative variants of S. pneumoniae serogroup 11.
INTRODUCTION
Streptococcus pneumoniae is a significant opportunistic pathogen whose transmissibility and virulence are increased by its ability to express a protective polysaccharide (PS) capsule (1). There are over 90 distinct S. pneumoniae PS capsules which are structurally and antigenically distinguishable (5, 10, 16). Each capsule is composed of unique PS repeat subunits that are synthesized, exported, and polymerized by enzymes encoded in the capsular synthesis locus (23). Current S. pneumoniae vaccines target the PS capsule of historically predominant serotypes and have been effective in reducing the occurrence of disease (17, 25). However, vaccination efforts have resulted in epidemiological replacement by nonvaccine serotypes. For example, S. pneumoniae serogroup 11 members, serotypes 11F (“F” denotes the first described member of a serogroup), 11A, 11B, 11C, 11D, and 11E, have become more prevalent in pediatric populations over the last few years following the introduction of the pneumococcal pediatric conjugated vaccine Prevnar, which does not target serogroup 11 serotypes (9, 11, 21). The emergence of serogroup 11 is mostly driven by serotype 11A, which makes up the vast majority of all serogroup 11 isolates despite being targeted by the adult polysaccharide vaccine, Pneumovax. However, other serogroup 11 members have also been identified as globally emerging serotypes (8, 18), and therefore, understanding the relation between the biochemical structures, antigenic qualities, and biosynthesis genes of all serogroup 11 PS capsules is essential for designing future interventions against this highly adaptable pathogen.
All members of serogroup 11 characteristically react with group 11 reference antiserum, and each serotype is identified according to its reactivity with polyclonal factor sera, which are named according to the four indeterminate factors that they bind: 11b, 11c, 11f, and 11g (Table 1). To further discriminate between serotype (ST) 11A and the newly identified ST 11E, which are indistinguishable according to factor sera binding, a monoclonal antibody (MAb) targeting an O-acetylation-dependent epitope is employed (5). Except for ST 11D, the capsular PS structures of serogroup 11 have been reported (19, 20, 27). All capsules share similar linear tetrasaccharide repeat units with a phospholinked polyalcohol branch and various O-acetyl substitutions, some of which were not definitively assigned (Table 2). However, previous structural analyses of these capsule types were not corroborated with a comprehensive understanding of the genetic basis of capsule synthesis (19, 20) or with comparative analysis of all serogroup 11 capsules (27), thus limiting the usefulness of these reported structural assignments.
Table 1.
Expected and observed reactivity of serogroup 11 members with serotyping antibodies
| Serotype | Resulta for polyclonal antiserum in indicated assay |
MAb Hyp11AM2 result in FCSA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Group 11 |
11b |
11c |
11f |
11g |
|||||||
| Q | FCSA | Q | FCSA | Q | FCSA | Q | FCSA | Q | FCSA | ||
| 11A | + | 108.1 | − | 12.6 | + | 308.2 | − | 7.0 | − | 3.5 | 53.3 |
| 11B | + | 52.0 | + | 937.6 | − | 27.6 | + | 214.9 | + | 146.0 | 3.5 |
| 11C | + | 74.1 | + | 652.5 | + | 190.0 | + | 100.2 | − | 19.7 | 3.4 |
| 11D | + | 172.5 | + | 403.4 | + | 353.7 | − | 6.7 | − | 11.5 | 55.8 |
| 11E | + | 75.9 | − | 28.1 | + | 302.5 | − | 15.0 | − | 2.4 | 3.3 |
| 11F | + | 152.5 | + | 306.4 | − | 28.2 | − | 4.2 | + | 382.2 | 65.8 |
Q, expected reactivity according to conventional Quellung assay; FCSA, flow cytometric serotyping assay. MFI values are shown. Boldface and shading indicate a strong signal (>50 MFI), boldface alone indicates a moderate signal (50 to 5 MFI), and lightface indicates no signal (<5 MFI).
Table 2.
Previously reported structures of serogroup 11 PS repeat unitsa
| Serotype | Carbohydrate core structureb |
O-acetylation site (mol eq) |
Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Core 1 | Core 2 | Core 3 | Core 4 | Phosphopolyalcohol pendant | Total | 1 | 2 | 3 | 4 | Unplaced | ||
| 11A | (1→6) α-Glc | (1→4) α-Gal | (1→3) β-Gal | (1→4) β-Glc | Gro | 2.6 | α-Glc H-2 (0.6) | α-Glc H-3 (0.5) | β-Gal H-4 (1.0) | Gro H-3 (0.5) | 27 | |
| 11B | (1→6) α-GlcNAc | (1→4) α-Gal | (1→3) β-Gal | (1→4) β-Glc | Rib | 1.4 | α-GlcNAc H-3 (0.9) | (0.5) | 19 | |||
| 11C | (1→6) α-GlcNAc | (1→4) α-Gal | (1→3) β-Gal | (1→4) β-Glc | Gro | 1.0 | α-GlcNAc H-3 (1.0) | 19 | ||||
| 11E | (1→6) α-Glc | (1→4) α-Gal | (1→3) β-Gal | (1→4) β-Glc | Gro | 1.7 | α-Glc H-2 (1.0) | α-Glc H-3 (0.4) | β-Gal H-4 (0.3) | 27 | ||
| 11F | (1→6) α-GlcNAc | (1→4) α-Gal | (1→3) β-Gal | (1→4) β-Glc | Rib | 2.0 | α-GlcNAc H-3 (0.5) | α-Gal H-2 (1.0) | (0.5) | 19 | ||
Serotype 11D PS structure has not been reported.
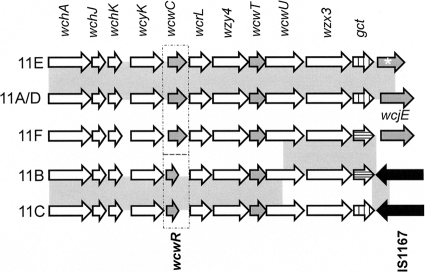
The capsular polysaccharide synthesis (cps) loci of serogroup 11 are homologous (5, 14), but three evident divergences between cps genotypes may be responsible for antigenic differences between the serotypes (Fig. 1), as follows: (i) ST 11B and ST 11C cps loci contain the putative O-acetyltransferase (OAcT) wcwR, which is replaced by the unrelated OAcT wcwC in ST 11A, ST 11D, ST 11E, and ST 11F cps loci; (ii) ST 11A, ST 11F, and ST 11D cps loci contain putatively intact OAcT wcjE, while ST 11E cps loci contain putatively inactivated wcjE alleles (5) and ST 11B and ST 11C cps loci completely lack the gene (2); and (iii) ST 11F and 11B cps loci contain a gct allele with a frameshift mutation putatively resulting in an inactive gct gene product (14). Putative synthesis roles of serogroup 11 cps glycosyltransferases, the polyalcohol phosphate transferase wcwU, and the CDP-glycerol synthetase gct were assigned according to comparative analysis of S. pneumoniae cps loci and known capsule structures (14). However, since the descriptions of PS structures upon which the analysis relied are incomplete, namely, in the assignment of O-acetyl substitution locations, the biosynthetic roles of the putative cps OAcTs wcwC, wcwR, wcwT, and wcjE could not be conclusively resolved.
Fig. 1.
Alignment of serogroup 11 cps genes involved in capsule synthesis. Regulatory genes wzg, wzh, wzd, and wze are not shown. Highlighted genes include putative O-acetyltransferase genes (gray fill) and the CDP-glycerol synthetase, gct (vertical lines), which contains a frameshift mutation in the ST 11F and ST 11B cps loci (horizontal lines). The transposable element IS1167 is represented with arrows with black fill. Regions that share greater than 99% homology between cps loci are highlighted with gray boxes. Dotted boxes show a divergent region which is the result of the replacement of wcwC with wcwR. The ST 11A and 11D cps loci are 99.9% homologous. The ST 11E cps locus differs from ST 11A due to a disruptive mutation to wcjE (white asterisk). The 11A and 11E cps sequences are according to reference 5. The 11F, 11B, 11C, and 11D sequences are according to reference 2.
Here, we report a revised and completed structural characterization of ST 11A, ST 11B, ST 11C, and ST 11F capsule polysaccharides (PS), including the location of all previously unassigned O-acetate substitutions, using modern nuclear magnetic resonance (NMR) methods (6). Using a novel serotyping assay, we quantified and evaluated the specificity of reference antisera and MAb binding to bacterial surfaces, and through in vitro immunological assays, we show that all these epitopes are dependent on PS acetylation. By comparative analysis, we identified serogroup 11 serotyping serum antigenic determinants and the putative biosynthesis targets of all serogroup 11 OAcT gene products. These findings demonstrate the important role of acetylation in serogroup 11 antigenicity.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Serogroup 11 standard strains were obtained from Statens Serum Institut (SSI, Copenhagen, Denmark): ST 11B, SSISP 11B/2; ST 11C, SSISP 11C/1; ST 11D, SSISP 11D/1; and ST 11F, SSISP 11F/2. For ST 11A and ST 11E, we used the previously characterized clinical isolates MNZ272 and MNZ264 (5, 27), respectively. Unless otherwise specified, strains were cultured on blood agar plates or Todd Hewitt liquid medium (BD Biosciences, San Jose, CA) plus 0.5% yeast extract (THY). All cultures were grown at 37°C in 5% CO2.
PS purification and de-O-acetylation.
To analyze the biochemical structures of the capsule of serogroup 11 serotypes, capsular PS was purified as previously described (27). Briefly, each culture was grown overnight at 37°C in chemically defined medium (JRH Biosciences, Lenexa, KS) (24) supplemented with choline chloride (1 g/liter), sodium bicarbonate (2.5 g/liter), and cysteine-HCl (0.73 g/liter). Culture lysates were obtained by treatment with 0.05% deoxycholate, and cell debris was removed by centrifugation. Precipitation with 50% ethanol per volume and centrifugation were used to remove amino acids and nucleic acids from lysates. Subsequently, 75% ethanol precipitation was used to recover PS. Precipitated PS was dissolved in 0.2 M NaCl, dialyzed in water at 4°C, and purified by ion exchange on a column of DEAE-Sepharose (GE Healthcare Bio-Sciences, United Kingdom) where PS eluted with a salt gradient. PS was then dialyzed, lyophilized, and fractionated on a column of Sephacryl S-300 HR (Sigma Chemical Company, ST. Louis, MO). Fractions containing PS according to reactivity with anthrone were pooled (15). To de-O-acetylate PS samples, purified PS was incubated in 0.2 M NaOH at room temperature for 3 h and desalted by dialysis in water. NMR analysis (see below) confirmed that mild hydrolysis with NaOH results in complete loss of O-acetate substitutions.
NMR analysis of PS.
All NMR data were acquired on a Bruker Avance operating at 700 MHz (Bruker Biospin, Billerica, MA) as previously described (27). Spectra were acquired at 35°C or 49°C in 99.998% deuterium oxide with deuterated dimethyl sulfoxide (DMSO-d6) and 4,4-dimethyl-4-silapentane-1-sulfonic acid–d6 (DSS-d6) as internal standards. Spectra were referenced to DSS at 0 ppm or DMSO at 2.71 ppm (1H) and 39.56 ppm (13C). All data were processed using MNova (Mestrelab Research).
Antibodies.
Hyp11AM2 mouse monoclonal antibodies (MAb) were produced as described previously (13), and hybridoma culture supernatants were used. Group 11 antiserum and factor sera 11b, 11c, 11f, and 11g are rabbit polyclonal sera purchased from SSI.
Flow cytometry.
To quantify epitope expression on bacterial surfaces, we examined the binding of polyclonal antisera or anti-11A PS MAb Hyp11AM2 to bacterial cells with flow cytometry. Cells were stained with a 1:1,000 dilution of polyclonal antisera or a 1:100 dilution of Hyp11AM2 hybridoma culture supernatant. Bound antibodies were detected with goat anti-rabbit Ig fluorescein isothiocyanate (FITC)-conjugated antibodies (Southern Biotechnology Associates, Inc., Birmingham, AL) or with rabbit anti-mouse IgM phycoerythrin (PE)-Cy7-conjugated antibodies (Southern Biotechnology). Bacteria were analyzed using a FACSCalibur (BD Biosciences, San Jose, CA). Data analysis was performed with FCS Express (De Novo Software, Los Angeles, CA) and binding of antibodies to bacteria was graded according to mean fluorescent intensity (MFI). MFI values of >50, between 50 and 5, and <5 were graded as “strong,” “moderate,” and “negative” binding, respectively. Samples stained with secondary antibody alone were used as negative controls. Although all factor sera displayed some moderate binding to nontarget serotypes, only strong signals correlate with Quellung reaction results (Table 1), consistent with reports of serogroup 6 factor sera binding to bacteria expressing ST 6A, 6B, 6C, and 6D PS capsules (4).
Inhibition ELISA.
To examine whether serotyping antibodies target O-acetate-dependent epitopes, we investigated the ability of native and de-O-acetylated PS to competitively inhibit antibody binding, similar to assays performed previously (5). To maximize the sensitivity of our assays, we detected the binding of rabbit polyclonal antisera to native PS from serotypes that displayed the lowest “strong” MFI value according to a flow cytometric serotyping assay (FCSA), i.e., ST 11B for factor serum 11g (1:2,000), ST 11C for factor sera 11c (1:2,000) and 11f (1:2,000), and ST 11F for factor serum 11b (1:1,000). For additional analysis, inhibition was also evaluated for factor serum 11b (1:8,000) binding to 11C PS. Since all strong Hyp11AM2 MFI values were relatively similar, ST 11F was also used to test inhibition of Hyp11AM2 binding. Antibodies were coincubated with soluble native or de-O-acetylated PS (ranging from 20 ng/ml to 200 μg/ml) in enzyme-linked immunosorbent assay (ELISA) wells coated with 2.5 μg/ml of purified native capsular PS in phosphate-buffered saline. After washing, bound antibodies were detected with alkaline phosphatase-conjugated anti-mouse or anti-rabbit antibody. The optical density at 405 nm (OD405) was recorded for each well. Native PS purified from a serotype 9A standard strain (SSISP 9A/1) was used as a control displaying no competitive binding, i.e., 0% inhibition. If a PS sample achieved 50% inhibition of binding compared to that of the negative control, it was graded positive for inhibition.
RESULTS
SSISP 11F/2 (11F) PS contains O-acetate substitutions at β-Gal H-6 and H-4 and at α-GlcNAc H-3.
The core (de-O-acetylated PS) of SSISP 11F/2 (herein referred to as ST 11F) is very similar to the core of MNZ272 (herein referred to as ST 11A) PS (27), as evidenced by the high degree of overlap in the 1H-13C heteronuclear single-quantum correlation spectroscopy (HSQC) spectrum (see Fig. S1 in the supplemental material). One key difference is in the anomeric region, where an ∼2 ppm δC shift in the anomeric resonance at ∼4.9 ppm δH is observed (see Fig. S1, inset). A peak typical of 2-acetamido-2-deoxy residues was observed at ∼54 ppm δC and was correlated to the anomeric signal at 4.99 ppm δH by 1H-13C heteronuclear multiple-bond correlation spectroscopy (HMBC) and 1H-13C HSQC-total correlation spectroscopy (HSQC-TOCSY). The remaining resonances for this residue were assigned by standard methods. Taken together, these resonances were assigned as α-N-acetylglucosamine (α-GlcNAc) replacing the α-glucose (α-Glc) seen in ST 11A. The remaining resonances in the repeat unit—α-galactose (α-Gal), β-glucose (β-Glc), β-galactose (β-Gal), and ribitol—were confirmed by a combination of gradient-selected correlation spectroscopy (gCOSY), TOCSY, HMBC, and HSQC-TOCSY experiments, and 1H-31P HMBC was used to confirm the linkage of a phosphoribitol (pRib) to the α-GlcNAc via a 1-4 linkage (see Table S1 in the supplemental material). In summary, our analysis of ST 11F confirms that the core carbohydrate structure is identical to the previously reported ST 11F PS core structure (Table 2) (19).
It was reported that ST 11F PS contains approximately 2 equivalents (eq) of O-acetate per mol of PS, of which 0.5 eq was not assigned to a specific residue (19). Since the cps loci of ST 11F and ST 11A both contain the putative O-acetyltransferases wcwC, wcwT, and wcjE, we hypothesized that the O-acetylation pattern of 11F PS would be identical to that of ST 11A PS. Comparison of the ST 11F core HSQC data to the native (O-acetylated PS) HSQC data (see Fig. S2 in the supplemental material), correlation to C′ resonances (∼174 ppm δC), and correlations from the acetate methyls in the 1H-13C HMBC data (data not shown) indicate that there are three sites of O-acetylation: 5.57, 5.24, and 4.26/4.15 [-CH2-] ppm δH. The first two sites of O-acetylation in 11F PS are β-Gal H-4 (5.57 ppm) and α-GlcNAc H-3 (5.24 ppm) (see Table S2 in the supplemental material), identical to ST 11A PS. This is in contrast to the previously reported 11F PS structure (19), which reported α-Gal H-2 O-acetate instead of β-Gal H-4 O-acetate as seen in this study. We initially interpreted the proton pair at 4.25/4.16 ppm δH as O-acetyl-phosphoribitol based on the high similarity to corresponding signals in the ST 11A assigned as O-acetyl-phosphoglycerol (pGro) (27). However, close inspection of the HMBC data for ST 11F showed that this O-acetylated methylene group has a strong cross-peak to the H-4 O-acetate substitution on β-Gal. The placement of the pRib on the α-GlcNAc in the ST 11F core structure precludes the 4.25/4.16 cross-peak from being pRib. In ST 11F PS, this carbon chemical shift is unambiguous, as opposed to that in ST 11A (see below). An HSQC-TOCSY assay (100 ms isotropic mixing time [τm]) confirmed the connectivity from this O-acetyl-CH2 to β-Gal H-4. These two pieces of data (from HSQC-TOCSY and HMBC) taken together led to the assignment of the O-acetylated CH2 at 4.26/4.16 as β-Gal H-6 instead of pRib.
MNZ272 (11A) PS contains an O-acetate group on carbon 6 of β-Gal which is absent in MNZ264 (11E) PS.
The unexpected finding that ST 11F PS did not contain an O-acetylated polyalcohol prompted us to reexamine the structure of ST 11A PS. HSQC-TOCSY data were acquired for ST 11A PS (50 ms τm) and compared to the data from ST 11F PS; the data showed weak but real peaks from the β-Gal H-4 O-acetate to the O-acetylated CH2 group at 4.26/4.15, supporting the hypothesis that these protons are in the same residue (β-Gal). This prompted us to reexamine the original ST 11A TOCSY and HMBC data (27). The 1H-1H TOCSY data showed a very weak (i.e., just above the noise) cross-peak from 5.59 to 4.26. In the HMBC, there is a cross-peak from 4.26 to 67.1 ppm δC, which was interpreted as Gro H-1 to Gro H-3 (3.88/3.80) (27). However, an alternate assignment could be to the β-Gal H-4 O-acetate (5.59 ppm δH). This assignment was originally considered but was ruled out due to the strong correlations in the pGro spin system and because of the lack of other supporting data at the time (27, 28). In light of the ST 11F results, the new data, and the reexamination of previous data, we need to revise our assignment of the pGro O-acetate: the correct interpretation of the ST 11A PS structure is that it does not contain O-acetate on pGro but instead contains an H-6 O-acetate substitution on β-Gal. Other than minor differences in the level of O-acetate substitutions at different sites, the only major distinction between the HSQC of native MNZ264 (herein referred to as ST 11E PS) and native ST 11A PS was the absence of signals at 4.26/4.15 ppm δH in 11E PS (27), which should now be interpreted as ST 11E PS being identical to 11A PS except for the lack of β-Gal H-6 O-acetylation.
SSISP 11B/2 (11B) and SSISP 11C/1 (11C) PS contain an O-acetate group on the 2 carbon of α-Gal.
The SSIPS11C/1 (herein referred to as ST 11C) core PS was reported to be similar to ST 11A, differing only by a substitution of α-GlcNAc (ST 11C) for α-Glc (ST 11A) (19, 27), similar to ST 11F. Indeed, the HSQC of ST 11C core and ST 11A core were highly similar (see Fig. S3 in the supplemental material). The shifted peaks in ST 11C PS were identical to the α-GlcNAc peaks in ST 11F PS (see Table S3 in the supplemental material), confirming the replacement of the α-Glc by α-GlcNAc. Also, the presence of pendant pGro on the α-GlcNAc in ST 11C core PS was confirmed. As expected (19), the core structure of SSISP 11B/2 (herein referred to as ST 11B PS) was exactly the same as the ST 11F core (see Fig. S4) and the assignments of the 11B PS core structure was straightforward (see Table S4).
ST 11B and ST 11C cps loci contain wcwT and the uncharacterized putative O-acetyltransferase wcwR (2), and thus, we hypothesized that ST 11B and ST 11C would share similar O-acetylation patterns. To identify the location of the O-acetate substitutions in these capsule types and gain insight into the role of wcwR in capsule synthesis, we compared the NMR spectra of native ST 11C and ST 11B PS to the spectra of native ST 11A and 11F, serotypes that do not contain wcwR in their cps loci. In contrast to ST 11A and ST 11F PS, ST 11C PS has only two sites of O-acetylation based upon 1H-13C HMBC correlations, 5.27, 72.63 and 5.05, 71.56. The signal at 5.27 ppm δH in ST 11C PS was congruent to the 5.25 ppm δH O-acetylated peak in ST 11F PS (α-GlcNAc H-3); however, the signal at 5.06 ppm δH was on the entirely different residue α-Gal H-2 (see Table S5 in the supplemental material), indicating O-acetyl substitution at this site on 11C PS.
The majority of peaks in native ST 11B PS were assigned using the same methods as described above (see Table S6 and Fig. S5 in the supplemental material). There is remarkable similarity between native ST 11C and ST 11B, and this was used to assign the vast majority of peaks in the ST 11B spectrum. Two peaks with significant intensities in the 1H-13C HSQC were not completely assigned (5.26, 96.52 and 4.98, 98.35). The peak at 5.26 is assigned as the nonacetylated α-Gal H-1 based upon comparison to PS structures that do not contain α-Gal O-acetate but was not further analyzed due to the extreme overlap with the α-GlcNAc H-3 O-acetate resonance in the 1H dimension. The peak at 4.98 was assigned to the α-N-acetylgalactosamine of cell wall PS based upon data acquired on purified cell wall PS (data not shown). In summary, similar to ST 11C PS, ST 11B PS contained two sites of O-acetylation: α-GlcNAc H-2 and α-Gal H-2.
Serogroup 11 serotypes display variable binding by polyclonal antisera.
As these assignments of O-acetyl substitutions differ from previously reported structures (19, 27), we corroborated our findings with serological evidence. It is commonly thought that serotypes that cross-react to a factor serum share a structural determinant that is absent in nonreactive serotypes. Conventional Quellung and latex agglutination assays do not allow us to quantify and compare the binding of sera to bacteria, so we analyzed the reactivity of each serotype to all available serogroup 11-specific polyclonal antisera using FCSA (Table 1). The mean fluorescent intensity (MFI) values of serogroup 11 bacteria stained with secondary antibody alone (i.e., without anticapsule antibodies) ranged between 2 and 5 (data not shown). ST 33F bacteria stained with serogroup 11-specific factor sera also yielded MFI values of <5 (data not shown), confirming that all antibody binding was specific to capsule antigens.
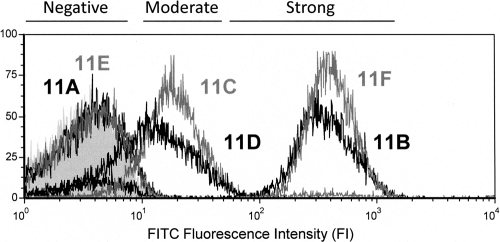
When comparing the binding of factor sera 11g and 11f to bacteria, MFI values fell into three categories: strong signals (MFI > 50), moderate signals (MFI between 50 and 5), and negative binding (MFI < 5). For example, of the bacteria stained with factor serum 11g, ST 11B and ST 11F displayed strong MFI values, ST 11D and ST 11C displayed moderate MFI values, and ST 11A and ST 11E displayed negative MFI values (Fig. 2). Factor serum 11f staining yielded strong MFI values for serotypes 11B and 11C, moderate MFI values for 11A, 11D, and 11E, and a negative MFI value for 11F. Factor serum 11b staining resulted in strong MFI values for serotypes 11B, 11C, 11D, and 11F and moderate MFI values for 11A and 11E. Factor serum 11c staining presented strong MFI values for 11A, 11C, 11D, and 11E and moderate MFI values for 11B and 11F. As expected, all serotypes displayed strong signals for group 11 antiserum staining, though the MFI values ranged from 52 for ST 11B to 172.5 for ST 11D. It should be noted that 11A and 11E behaved similarly with the conventional factor sera, in concordance with previous observations using the Quellung assay (5). Although all factor sera displayed moderate binding to nontarget serotypes, only strong signals correlate with Quellung reaction results (Table 1); therefore, we only considered strong FCSA signals for our final antigenic determinant analyses (see Discussion).
Fig. 2.
Staining intensity curves fall into three categories according to FCSA. Flow cytometry histogram displaying the fluorescence intensity (FI) curves of factor serum 11g binding to all serogroup 11 serotypes. Serotypes yielding each binding curve are matched by shade next to their corresponding curves. The gray-filled curve corresponds to serotype 11B stained with secondary antibody only (negative control). Bands above the diagram depict the FI ranges corresponding to signal grading.
Hyp11AM2 clearly binds or does not bind serogroup 11 members.
It was previously determined that Hyp11AM2 MAb binds to ST 11A, ST 11D, and ST 11F PS (26). To determine whether Hyp11AM2 also reacts to other serogroup 11 members, we quantified Hyp11AM2 binding to all serogroup 11 serotypes using FCSA (Table 1). Staining with this antibody resulted in comparably strong signals for serotypes 11A, 11D, and 11F (i.e., MFI values of 53.3, 55.8, and 65.8, respectively) and MFI values of less than 5 for serotypes 11B, 11C, and 11E. Notably, no moderate signal was observed when staining bacteria with this MAb.
De-O-acetylation of PS affects its capacity to competitively bind antibody.
Detecting direct binding of serotyping antibodies to bacterial surfaces did not reveal whether these antibodies bound O-acetate-dependent epitopes, and it is unclear whether a factor serum bound to a single structural determinant shared by cross-reactive serotypes. To confirm that serotyping sera target single epitopes and to show that these antigenic determinants are dependent on PS O-acetylation, we tested the ability of native and de-O-acetylated PS to competitively bind polyclonal antisera and MAb by inhibition ELISA (Table 3). The binding of all antibodies analyzed was not inhibited by native PS purified from the unrelated serotype 9A control. Factor serum 11b binding to ST 11C PS was inhibited only by native 11C and 11B PS, but de-O-acetylated PS was incapable of achieving inhibition. Remarkably, 11b binding to 11F PS was inhibited by both native and de-O-acetylated PS from serotypes 11B, 11C, 11D, and 11F. Factor serum 11c binding to 11C PS was inhibited by native 11A, 11C, 11D, and 11E PS, 11f binding to 11C PS was inhibited by native 11C and 11B PS, and 11g binding to 11B PS was inhibited by native 11B and 11F PS. No other native or de-O-acetylated PS samples were able to inhibit the binding of polyclonal antisera to target PS. Hyp11AM2 MAb binding to 11F PS was inhibited only by native 11A, 11D, and 11F PS and not by any other PS sample.
Table 3.
Inhibition of antibody binding by native and de-O-acetylated PS
| Inhibition PS | Bindinga of indicated group 11 antiserum or MAb to target PS (antiserum, target PSb) |
||||||
|---|---|---|---|---|---|---|---|
| 11b, 11C | 11b, 11F | 11c, 11C | 11f, 11C | 11g, 11B | Hyp11AM2 11F | ||
| 11A | Native | − | − | + | − | − | + |
| deOAcc | − | − | − | − | − | − | |
| 11B | Native | + | + | − | + | + | − |
| deOAc | − | + | − | − | − | − | |
| 11C | Native | + | + | + | + | − | − |
| deOAc | − | + | − | − | − | − | |
| 11D | Native | − | + | + | − | − | + |
| deOAc | − | + | − | − | − | − | |
| 11E | Native | − | − | + | − | − | − |
| deOAc | − | − | − | − | − | − | |
| 11F | Native | − | + | − | − | + | + |
| deOAc | − | + | − | − | − | − | |
+ inhibition, PS is capable of competitively inhibiting antibody binding to target PS (≤50% signal compared to control); − inhibition, PS is unable to competitively inhibit antibody binding (>50% signal compared to control).
Native PS used to coat ELISA plates.
deOAc, PS de-O-acetylated by mild alkali hydrolysis.
DISCUSSION
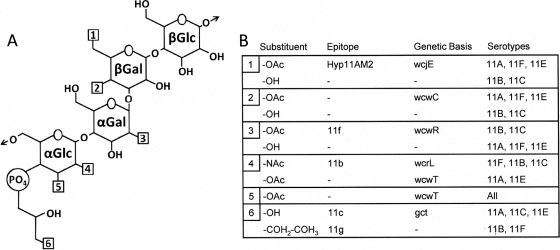
Recent studies have highlighted the increasing prevalence of S. pneumoniae serogroup 11 members, primarily serotype 11A (9, 11, 21), yet the capsule PS structures of most of these serotypes have not been addressed in over 20 years (19). As the results of recent investigations concerning serogroup 11 (5, 27) were inconsistent with previously reported PS capsule structures (19), we readdressed the structures of ST 11A, ST 11B, ST 11C, and ST 11F PS capsules. With newer NMR technology and a better knowledge of the capsule synthesis genes than when these PS structures were initially investigated, we were able to completely elucidate the O-acetylation patterns of serogroup 11 PS capsules (Table 4). Further supporting the PS structures reported here, we identified strong correlations between the revised models of serogroup 11 PS structures, the reactivities of the PS with serotyping sera, and the O-acetyltransferase genes putatively involved in capsule biosynthesis (Fig. 3).
Table 4.
Revised assignment of O-acetate substitutions
| Serotype | O-acetylation site (mol eq): |
||||
|---|---|---|---|---|---|
| Total | 1 | 2 | 3 | 4 | |
| 11A | 2.6 | α-Glc H-2 (0.6) | α-Glc H-3 (0.5) | β-Gal H-4 (1.0) | β-Glc H-6 (0.5) |
| 11B | 1.2 | α-GlcNAc H-3 (0.8) | α-Gal H-2 (0.4) | ||
| 11C | 1.2 | α-GlcNAc H-3 (0.9) | α-Gal H-2 (0.3) | ||
| 11E | 1.6 | α-Glc H-2 (1.0) | α-Glc H-3 (0.3) | β-Gal H-4 (0.3) | |
| 11F | 2.4 | α-GlcNAc H-3 (1.0) | β-Gal H-4 (0.8) | β-Glc H-6 (0.6) | |
Fig. 3.
Correlation of PS structure, antibody binding, and cps genes for serogroup 11. (A) Depiction of the core carbohydrate shared by all serogroup 11 capsules. Arrows represent the bonds across which PS subunits are polymerized. Hydroxyl substitutions that differ among serotypes are denoted with numbered boxes. (B) Correlation of characteristics at each numbered location (structure, epitope, genetic basis, and relevant serotypes). The substituent structures are as follows: OH, hydroxyl group; OAc, submolar O-acetate substitution; NAc, N-acetate substitution. “Epitope” gives the epitope that is dependent on the presence of each substituent. “Genetic Basis” gives the cps gene whose integrity is correlated with each structure. Note that factor serum 11b contains two subpopulations of antibodies that target separate epitopes, one of which clearly binds a GlcNAc-associated epitope (see text).
Consistent with previous reports that antibodies against various bacterial species often bind capsule PS in O-acetylation-dependent manners (3, 7, 12, 22), we observed a decrease in the competitive binding of each serogroup 11 factor serum to de-O-acetylated PS. The binding of factor serum 11f to ST 11C PS was inhibited by native ST 11C and ST 11B PS but was not inhibited by any other PS sample, evidence that factor 11f is associated with O-acetylation of α-Gal H-2, the only O-acetate substitution detected exclusively in the 11f-positive serotypes ST 11B and ST 11C (Fig. 3). Unambiguously, factor serum 11c binds to serotypes containing pGro (ST 11A, ST 11E, and ST 11C) and factor serum 11g binds to serotypes containing pRib (ST 11B and ST 11F). However, since competitive binding of these factor sera by capsule PS was abolished following mild alkali hydrolysis and α-Glc or α-GlcNAc is the only site of O-acetylation conserved among all serogroup 11 capsules, it is likely that factors 11c and 11g are conformational epitopes created by corresponding polyalcohols and proximal O-acetyl groups on the adjacent α-Glc or α-GlcNAc.
Identifying factor 11b presents a greater challenge because the serum displayed two distinct binding patterns. A high dilution (i.e., 1:8,000) of factor serum clearly bound 11C PS, a reaction which could only be inhibited by ST 11B or ST 11C PS and not by any other native or de-O-acetylated sample. This suggests that factor 11b is associated with α-Gal O-acetylation. In contrast, the binding of a low dilution (i.e., 1:1,000) of factor serum 11b to ST 11F-coated plates was competitively inhibited by ST 11B, ST 11C, and ST 11F PS even after mild alkali hydrolysis but not by ST 11A or ST 11E PS, indicating that these antibodies bind the de-O-acetylated backbone of repeat units containing α-GlcNAc. We propose that the dominant antibodies in this polyclonal antiserum bind capsule PS containing an O-acetylation pattern characteristic of ST 11B and 11C capsule, while other antibodies in the serum bind the PS containing α-GlcNAc in an O-acetate-independent manner. A heterogeneous antibody population also explains the ∼2-fold higher MFI values for factor serum 11b staining of serotypes 11B and 11C compared to that of serotypes 11D and 11F. Thus, there is no single “factor 11b” structure that mediates reactivity to this factor serum.
Correlation of the native capsule PS of these serotypes to the published serogroup 11 cps loci (2, 5) supports our structural models and permits us to assign biosynthetic roles to wcjE, wcwR, wcwC, and wcwT (Fig. 3). For example, the examined serotypes that contain putatively functional wcjE genes, i.e., ST 11A and ST 11F, contain an O-acetate substitution on β-Gal H-6, a feature which the PS of the wcjE-null ST 11B, ST 11C, and ST 11E lack. This finding is consistent with the results of recent molecular studies which clearly established the role of wcjE in the antigenic disparity between ST 11A and ST 11E (5). Prior to the present study, it was unclear whether wcwR performed any function in ST 11B and ST 11C capsule synthesis. Here, we show that the replacement of wcwC, present in ST 11A, ST 11F, and ST 11E cps loci, with wcwR in ST 11B and ST 11C (Fig. 1) strongly correlates to loss of β-Gal H-4 O-acetylation and the gain of α-Gal H-2 O-acetylation, revealing candidate enzymatic targets for each of these OAcTs. The α-glucosyl residue and wcwT are the only O-acetylation site and OAcT gene shared by all serogroup 11 members. Therefore, we propose that the wcwT gene product is a promiscuous O-acetyltransferase capable of recognizing either α-Glc or α-GlcNAc as a substrate. The wcwT gene product O-acetylates α-Glc at either the H-2 and H-3 carbons (ST 11A and ST 11E) or the H-3 carbon of α-GlcNAc (ST 11B, ST 11C, and ST 11F). The lack of identification of a wcwT-null serogroup 11 variant could be due to the lack of a factor serum that recognizes PS without this O-acetate substitution. However, the possibility that a wcwT-null variant is clinically nonviable cannot be dismissed.
It should be noted that NMR analysis of ST 11D capsule yielded ambiguous results (data not shown), and a conclusive PS structure will be presented pending further analyses. However, since the ST 11D cps locus is nearly identical to the ST 11A cps locus (2, 5) and both these serotypes are bound by Hyp11AM2 and factor serum 11c but not by factor serum 11f or 11g, it can be predicted that ST 11D PS is very similar to ST 11A PS in terms of O-acetate placement and the identity of the polyalcohol pendant. Antigenic divergences between the two may be due to the single amino acid polymorphism between ST 11A and ST 11D wcrL (Fig. 1), whose gene product putatively transfers either an α-Glc or α-GlcNAc to the capsule subunit (14). Indeed, since ST 11D is bound by factor serum 11b, i.e., an antigenic feature strongly correlated to N-acetylation of the α-Glc (Fig. 3), we postulate that ST 11D PS contains α-GlcNAc.
Our findings highlight a need to reevaluate conventional serogroup 11 serotyping tools, as limitations in these methods may obscure the actual epidemiology of this increasingly prevalent serogroup (11, 21). For example, serogroup 11 antiserum staining often showed moderate cross-reactivity to multiple serotypes, and at least one factor serum (11b) contains antibodies targeting clearly distinct antigenic determinants. Furthermore, in accordance with previous reports on ST 11A and ST 11E (5), we observed that no factor serum recognizes the wcjE-associated β-Gal H-6 O-acetate substitution. We recently found that inactivation of wcjE in the serotype 9V cps locus results in serotype 9A expression (5a), supporting the existence of multiple instances of wcjE-mediated dichotomies between serotypes. Conceivably, an ST 11C variant with intact wcjE expression or an ST 11F variant that loses wcjE expression would express antigenically divergent capsule types but would be indistinguishable from the current ST 11C or ST 11F standards by conventional serotyping means. Hyp11AM2 displayed clear positive or negative staining and recognizes wcjE-associated structural determinants. The use of Hyp11AM2 and other similar MAbs in investigating unidentified serovariants in serogroup 11 and other potentially heterogeneous S. pneumoniae serogroups is currently being explored.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants AI093103 to J.J.C. from the National Institutes of Health, AI31473 to M.H.N. from the National Institutes of Health, and GM008361 to the UAB Medical Scientist Training Program from the National Institutes of Health.
The University of Alabama at Birmingham owns the intellectual property rights to some of the reagents described in this work. J.J.C. and M.H.N. are employees of the University of Alabama at Birmingham.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 29 July 2011.
REFERENCES
- 1. Avery O. T., Dubos R. 1931. The protective action of a specific enzyme against type III pneumococcus infection in mice. J. Exp. Med. 54:73–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bentley S. D., et al. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Berry D. S., Lynn F., Lee C. H., Frasch C. E., Bash M. C. 2002. Effect of O acetylation of Neisseria meningitidis serogroup A capsular polysaccharide on development of functional immune responses. Infect. Immun. 70:3707–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bratcher P. E., Nahm M. H. 2010. Cross-reactivity of current serogroup 6 factor sera from Statens Serum Institut with the recently described pneumococcal serotype 6d. J. Clin. Microbiol. 48:3044–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Calix J. J., Nahm M. H. 2010. A new pneumococcal serotype, 11E, has variably inactivated wcjE gene. J. Infect. Dis. 202:29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a. Calix J. J., et al. Streptococcus pneumoniae serotype 9A isolates contain diverse mutations to wcjE that result in variable expression of serotype 9V-specific epitope. J. Infect. Dis., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Duus J., Gotfredsen C. H., Bock K. 2000. Carbohydrate structural determination by NMR spectroscopy: modern methods and limitations. Chem. Rev. 100:4589–4614 [DOI] [PubMed] [Google Scholar]
- 7. Fattom A. I., Sarwar J., Basham L., Ennifar S., Naso R. 1998. Antigenic determinants of Staphylococcus aureus type 5 and type 8 capsular polysaccharide vaccines. Infect. Immun. 66:4588–4592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Flasche S., et al. 2011. Effect of pneumococcal conjugate vaccination on serotype-specific carriage and invasive disease in England: a cross-sectional study. PLoS Med. 8:e1001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang S. S., et al. 2009. Continued impact of pneumococcal conjugate vaccine on carriage in young children. Pediatrics 124:e1–e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jin P., et al. 2009. First report of putative Streptococcus pneumoniae serotype 6D among nasopharyngeal isolates from Fijian children. J. Infect. Dis. 200:1375–1380 [DOI] [PubMed] [Google Scholar]
- 11. Kellner J. D., et al. 2008. Effects of routine infant vaccination with the 7-valent pneumococcal conjugate vaccine on nasopharyngeal colonization with streptococcus pneumoniae in children in Calgary, Canada. Pediatr. Infect. Dis. J. 27:526–532 [DOI] [PubMed] [Google Scholar]
- 12. Kennedy D. A., Buchanan J. G., Baddiley J. 1969. The type-specific substance from Pneumococcus type 11A(43). Biochem. J. 115:37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lin J., et al. 2006. Validation of a multiplex pneumococcal serotyping assay with clinical samples. J. Clin. Microbiol. 44:383–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mavroidi A., et al. 2007. Genetic relatedness of the Streptococcus pneumoniae capsular biosynthetic loci. J. Bacteriol. 189:7841–7855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Morris D. L. 1948. Quantitative determination of carbohydrates with Dreywood's anthrone reagent. Science 107:254–255 [DOI] [PubMed] [Google Scholar]
- 16. Park I. H., et al. 2007. Discovery of a new capsular serotype (6C) within serogroup 6 of Streptococcus pneumoniae. J. Clin. Microbiol. 45:1225–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Poehling K. A., et al. 2006. Invasive pneumococcal disease among infants before and after introduction of pneumococcal conjugate vaccine. JAMA 295:1668–1674 [DOI] [PubMed] [Google Scholar]
- 18. Reyna J., Limon A. E. 2008. High prevalence of serotype 11B of Streptococcus pneumoniae isolated in the nasopharynx of Mexican children. Arch. Med. Res. 39:629–630 [DOI] [PubMed] [Google Scholar]
- 19. Richards J. C., Perry M. B., Kniskern P. J. 1985. The structure of the specific capsular polysaccharide of Streptococcus pneumoniae type 11F (American type 11). Can. J. Biochem. Cell Biol. 63:953–968 [DOI] [PubMed] [Google Scholar]
- 20. Richards J. C., Perry M. B., Moreau M. 1988. Elucidation and comparison of the chemical structures of the specific capsular polysaccharides of Streptococcus pneumoniae groups 11 (11F, 11B, 11C, and 11A), p. 595–596 In Wu A. M. (ed.), The molecular immunology of complex carbohydrates. Plenum, New York, NY [Google Scholar]
- 21. Richter S. S., et al. 2009. Changing epidemiology of antimicrobial-resistant Streptococcus pneumoniae in the United States, 2004-2005. Clin. Infect. Dis. 48:e23–e33 [DOI] [PubMed] [Google Scholar]
- 22. Theilacker C., et al. 2003. Construction and characterization of a Pseudomonas aeruginosa mucoid exopolysaccharide-alginate conjugate vaccine. Infect. Immun. 71:3875–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Trzcinski K., Thompson C. M., Lipsitch M. 2003. Construction of otherwise isogenic serotype 6B, 7F, 14, and 19F capsular variants of Streptococcus pneumoniae strain TIGR4. Appl. Environ. Microbiol. 69:7364–7370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van de Rijn I., Kessler R. E. 1980. Growth characteristics of group A streptococci in a new chemically defined medium. Infect. Immun. 27:444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Whitney C. G., et al. 2006. Effectiveness of seven-valent pneumococcal conjugate vaccine against invasive pneumococcal disease: a matched case-control study. Lancet 368:1495–1502 [DOI] [PubMed] [Google Scholar]
- 26. Yu J., et al. 2005. Rapid multiplex assay for serotyping pneumococci with monoclonal and polyclonal antibodies. J. Clin. Microbiol. 43:156–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zartler E. R., et al. 2009. Structure of the capsular polysaccharide of pneumococcal serotype 11A reveals a novel acetylglycerol that is the structural basis for 11A subtypes. J. Biol. Chem. 284:7318–7329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zartler E. R., Porambo R. J., Anderson C. L., Yu J., Nahm M. H. 2009. Identification of 3-O-acetylglycerol, a novel structural element in bacterial polysaccharides. Carbohydr Res. 344:2586–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.