Abstract
Aeromonas hydrophila polar-flagellum class I gene transcription is σ70 dependent, which is consistent with the fact that the A. hydrophila polar flagellum is constitutively expressed. In contrast to other bacteria with dual flagellar systems such as Vibrio parahaemolyticus, the A. hydrophila LafK protein does not compensate for the lack of the polar-flagellum regulator FlrA (V. parahaemolyticus FlaK homologue). This is consistent with the fact that the A. hydrophila FlrA mutation abolishes polar-flagellum formation in liquid and on solid surfaces but does not affect inducible lateral-flagellum formation. The results highlight that the polar- and lateral-flagellum interconnections and control networks are specific and that there are differences between the dual flagellar systems in A. hydrophila and V. parahaemolyticus. Furthermore, our results indicate that the A. hydrophila polar-flagellum transcriptional hierarchy (also in class II, III, and IV genes) shares some similarities with but has many important differences from the transcriptional hierarchies of Vibrio cholerae and Pseudomonas aeruginosa. The A. hydrophila flhF and flhG genes are essential for the assembly of a functional polar flagellum because in-frame mutants fail to swim in liquid medium and lack the polar flagellum. In Vibrio and Pseudomonas flhG disruption increases the number of polar flagella per cell, and Pseudomonas flhF disruption gives an aberrant placement of flagellum. Here, we propose the gene transcriptional hierarchy for the A. hydrophila polar flagellum.
INTRODUCTION
Flagellum motility represents an important advantage for bacteria in moving toward favorable conditions or in avoiding detrimental environments, and it allows flagellated bacteria to successfully compete with other microorganisms (14). Flagellum morphogenesis is a complex cascade of events that requires coordinate expression of more than 50 genes encoding structural subunits, regulatory proteins, and chemo-sensor machinery. These genes have been categorized in relation to their temporal requirement during the assembly process into three groups: early, middle, and late genes (1, 10). Early genes encode regulatory proteins that control the expression of the entire regulon. Middle genes include structural components of the hook, the basal body, the export apparatus, and regulatory proteins that couple late-gene expression; and late genes include the filament, motor force generators, and chemotactic proteins. The expression of these genes is an energetically expensive process for the bacterium, and all flagellar systems are highly regulated. Regulation of flagellum biogenesis involves a combination of transcriptional, translational, and posttranslational mechanisms (1). In relation to their transcriptional hierarchy, the flagellar clusters of different bacterial species are transcribed from three different promoter classes, whose differential expression is coordinated by the activity of transcriptional regulators which include alternative sigma factors and anti-sigma factors (10, 25). The coordinated expression of these promoters cluster gene transcription in three or four levels of hierarchy: classes I to III or I to IV. In peritrichous flagellated bacteria, such as Escherichia coli and Salmonella, three levels of hierarchy have been described (22). Transcription of class I and II genes requires the housekeeping sigma factor 70 (σ70). The single class I promoter responds to a number of global regulatory factors (23) and transcribes the genes for the FlhDC master activator, required for expression of all class II σ70-dependent promoters (43). A class II promoter transcribes the gene for the sigma factor 28 (σ28), which directs transcription of class III genes (31). Class III promoters are negatively regulated by the anti-sigma factor FlgM (9). However, inducible peritrichous flagella (lateral flagella) of Vibrio parahaemolyticus and Aeromonas hydrophila do not posses an FlhDC master regulator and are sigma factor 54 (σ54) dependent (8, 39). Polar flagellated Gammaproteobacteria, such as Vibrio and Pseudomonas, show four transcriptional levels, where classes II and III are σ54 dependent and class IV is σ28 dependent (12, 33). At the top of the hierarchy is a σ54-associated transcriptional activator (FlrA of Vibrio cholerae and FleQ of Pseudomonas aeruginosa) which activates class II σ54-dependent promoters. Class II promoters encode a two-component signal-transducing system (FlrBC of V. cholerae and FleSR in P. aeruginosa) whose regulator (FlrC/FleR) activates class III σ54-dependent promoters. Moreover, in Vibrio spp. class II promoters also encode the σ28 factor which activates transcription of class IV genes (11, 12, 33).
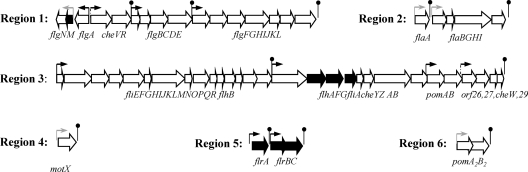
Mesophilic Aeromonas is a ubiquitous aquatic microorganism that constitutively expresses a single polar flagellum although about 60% of strains most commonly associated with diarrhea (20) are also able to express many lateral flagella when grown in viscous environments or on surfaces (37). The Aeromonas polar-flagellum genes are organized in different clusters distributed in six chromosomal regions (Fig. 1). These genes encode structural, regulatory, and chemotaxis proteins, as well as enzymes involved in flagellin glycosylation (2, 7, 44). The regulatory genes are localized in region 1, 3, and 5 (2, 7). Region 1 contains the gene encoding the anti-σ28 transcription factor FlgM. Region 3 contains three regulatory genes: the σ28 transcription factor (fliA) and two genes found only in polar-flagellum systems, flhF and flhG, which encode proteins that play a regulatory role in V. cholerae and P. aeruginosa flagellum biosynthesis (11, 30). Region 5 encodes three proteins homologous to the FlrA transcriptional activator and the FlrBC two-component signal-transducing system of V. cholerae. In addition, outside the polar-flagellum regions is RpoN (σ54) that is essential for polar- and lateral-flagellum system transcription (8). Given the critical role these genes play in regulating polar-flagellum expression in different bacterial species (11, 12, 30, 33), we investigated the aeromonad polar-flagellum transcriptional hierarchy by two techniques: measurement of the β-galactosidase activity of promoter-lacZ fusions in several mutant backgrounds and reverse transcription-PCR (RT-PCR) assays.
Fig. 1.
Organization of A. hydrophila AH-3 polar-flagellum chromosomal regions. Arrows indicate the direction of transcription and the extent of coding sequence for each gene. Black arrows indicate regulatory genes. Small arrows indicate in silico predicted promoters (black, σ54 promoters; gray, σ28 promoters; dotted line, undetermined promoter). Lollipops indicate predicted rho-independent transcriptional terminators.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were grown on Luria-Bertani (LB) Miller broth and LB Miller agar at 37°C, while Aeromonas strains were grown in either tryptic soy broth (TSB) or agar (TSA) at 30°C. When required, ampicillin (50 μg/ml), kanamycin (50 μg/ml), rifampin (100 μg/ml), spectinomycin (50 μg/ml), chloramphenicol (25 μg/ml), gentamicin (10 μg/ml), and tetracycline (20 μg/ml) were added to the medium.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and/or descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| A. hydrophila | ||
| AH-3 | A. hydrophila wild type, serogroup O:34 | 26 |
| AH-405 | AH-3, spontaneous Rifr | 2 |
| AH-5502 | AH-405 rpoN::Kmr | 8 |
| AH-4443 | AH-405 fliA::Kmr | 7 |
| AH-3 flrA | AH-405; flrA::Kmr | This work |
| AH-3 flrBC | AH-405 flrB::pSF, Kmr | This work |
| AH-3 ΔflhF | AH-405 with in-frame ΔflhF | This work |
| AH-3 ΔflhG | AH-405 with in-frame ΔflhG | This work |
| E. coli | ||
| DH5α | λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | 16 |
| MC1061λpir | thi thr1 leu6 proA2 his4 argE2 lacY1 galK2 ara14 xyl5 supE44 λ pir | 34 |
| Plasmids | ||
| pGEMT-easy | Cloning vector, Apr | Promega |
| pRK2073 | Helper plasmid, Spr | 34 |
| pDM4 | Suicide plasmid, pir dependent with sacAB genes, oriR6K, Cmr | 28 |
| pDM-FlhF | pDM4 with ΔflhF fragment, Cmr | This work |
| pDM-FlhG | pDM4 with ΔflhG fragment, Cmr | This work |
| pDM-FlrAKm | pDM4 with AH-3 flrA::Km, Cmr Kmr | This work |
| pFS100 | pGP704 suicide plasmid, pir dependent, Kmr | 34 |
| pFS-FlrB | pFS100 with a AH-3 flrB internal fragment, Kmr | This work |
| pBAD33 | Arabinose-induced expression vector, orip15 PBAD Cmr | 15 |
| pBAD33-FLHF | pBAD33 with AH-3 flhF gene, Cmr | This work |
| pBAD33-FLHG | pBAD33 with AH-3 flhG gene, Cmr | This work |
| pBAD33-Gm | pBAD33 arabinose-induced expression vector with Gmr | |
| pBAD33Gm-FLIA | pBAD33 with AH-3 fliA gene, Gmr | This work |
| pBAD33Gm-FLRA | pBAD33 with AH-3 flrA gene, Gmr | This work |
| pACYC184 | Plasmid vector; Cmr Tcr | 35 |
| pACYC-RPON | pACYC184 with AH-3 rpoN gene, Tcr | 8 |
| pACYC-FLR1 | pACYC184 with AH-3 flrBC genes, Tcr | 7 |
| pDN19 lacΩ | Promoterless lacZ fusion vector; Spr Smr Tcr | 42 |
| pDNlac-flaAp | flaA promoter-lacZ fusion in pDN19lacΩ, Tcr | This work |
| pDNlac-flaBp | flaB promoter-lacZ fusion in pDN19lacΩ, Tcr | This work |
| pDNlac-flgAp | flgA promoter-lacZ fusion in pDN19lacΩ, Tcr | This work |
| pDNlac-flgBp | flgB promoter-lacZ fusion in pDN19lacΩ, Tcr | This work |
| pDNlac-flgFp | flgF promoter-lacZ fusion in pDN19lacΩ, Tcr | This work |
| pDNlac-flgMp | flgM promoter-lacZ fusion in pDN19lacΩ, Tcr | This work |
| pDNlac-flhAp | flhA promoter-lacZ fusion in pDN19lacΩ, Tcr | This work |
| pDNlac-fliEp | fliE promoter-lacZ fusion in pDN19lacΩ, Tcr | This work |
| pDNlac-flrAp | flrA promoter-lacZ fusion in pDN19lacΩ, Tcr | This work |
| pDNlac-flrBp | flrB promoter-lacZ fusion in pDN19lacΩ, Tcr | This work |
| pDNlac-lafKp | lafK promoter-lacZ fusion in pDN19lacΩ, Tcr | This work |
| pDNlac-motXp | motX promoter-lacZ fusion in pDN19lacΩ, Tcr | This work |
| pDNlac-pomAp | pomA promoter-lacZ fusion in pDN19lacΩ, Tcr | This work |
| pDNlac-pomA2p | pomA2 promoter-lacZ fusion in pDN19lacΩ, Tcr | This work |
Tcr, tetracycline resistant; Kmr, kanamycin resistant; Apr, ampicillin resistant; Rifr, rifampin resistant; Cmr, chloramphenicol resistant; Spr, spectinomycin resistant; Smr, streptomycin resistant; Gmr, gentamicin resistant.
Motility assays (swarming and swimming).
Freshly grown bacterial colonies were transferred with a sterile toothpick into the center of swarm agar (1% tryptone, 0.5% NaCl, 0.5% agar) or swim agar (1% tryptone, 0.5% NaCl, 0.25% agar). The plates were incubated face up for 16 to 24 h at 25°C, and motility was assessed by examining the migration of bacteria through the agar from the center toward the periphery of the plate. Moreover, swimming motility was assessed by light microscopy observations in liquid medium.
TEM.
Bacterial suspensions were placed on Formvar-coated grids and negatively stained with a 2% solution of uranyl acetate, pH 4.1. Preparations were observed on a Hitachi 600 transmission electron microscope (TEM).
DNA techniques.
DNA manipulations were carried out essentially according to standard procedures (35). DNA restriction endonucleases and E. coli DNA polymerase Klenow fragment were obtained from Promega. T4 DNA ligase and alkaline phosphatase were obtained from Invitrogen and GE Healthcare, respectively. PCR was performed using BioTaq DNA polymerase (Ecogen) in a Gene Amplifier PCR System 2400 thermal cycler (Perkin Elmer).
Nucleotide sequencing and computer sequence analysis.
Plasmid DNA for sequencing was isolated by a Qiagen plasmid purification kit (Qiagen, Inc. Ltd.) as recommended by the suppliers. Double-strand DNA sequencing was performed by using the Sanger dideoxy-chain termination method (36) with a BigDye Terminator, version 3.1, cycle sequencing kit (Applied Biosystems). Custom-designed primers used for DNA sequencing were purchased from Sigma-Aldrich. The DNA sequences were inspected in the GenBank and EMBL databases at the National Center for Biotechnology Information (NCBI) (3). The Terminator search program in the GCG Wisconsin package was used to search for factor-independent transcriptional terminators. The Neural Network Promoter Prediction, PromScan (40), and PRODORIC (29) programs were used to search promoter sequences.
Total RNA extraction and RT-PCR.
Total RNA was isolated, by means of an RNA Protect Bacteria Reagent (Qiagen) and an RNeasy Mini Kit (Qiagen), from A. hydrophila AH-3, and flrA, flrBC, flhF, flhG, and fliA mutants were grown in liquid (TSB) or solid agar (TSA) medium. To ensure that RNA was devoid of contaminating DNA, the preparation was treated with RNase-free Turbo DNase I (Ambion). First-strand cDNA synthesis was carried out with Moloney murine leukemia virus (M-MuLV) reverse transcriptase (New England BioLabs) and random oligonucleotides (Promega) on 5 μg of total RNA, DNase digested. The reaction mixtures were incubated at 25°C for 10 min, 37°C for 120 min, and 75°C for 15 min. Control reactions lacking reverse transcriptase were performed to confirm that RNA samples were not contaminated with genomic DNA (RT negative controls). PCR, second-strand synthesis, and subsequent DNA amplification were carried out using the Accuprime TaqDNA polymerase (Invitrogen) and specific oligonucleotides. Amplicons were analyzed by agarose gel electrophoresis with ethidium bromide staining. A. hydrophila ribosomal 16S primers were used as a control for cDNA template. RT-PCR amplifications were performed at least twice, with total RNA preparations obtained from a minimum of two independent extractions.
Mapping the A. hydrophila AH-3 flrA and pomA2 transcription start sites by 5′ RACE PCR.
Amplification of the A. hydrophila AH-3 flrA and pomA2 cDNA 5′ ends was performed using a 5′ RACE (random amplification of cDNA ends) System, version 2.0 (Invitrogen). Total RNA extraction from A. hydrophila AH-3 was performed as mentioned above. First-strand cDNA was synthesized using the entire volume of DNase-digested total RNA (5 μg), the flrA internal primer GSP1-FlrA (5′-GAGAGAGCTCGTGAAT-3′), or the pomA2 internal primer GSP1-PomA2 (5′-GCGCCATACAGAGTA-3′) and the Thermoscript RT-PCR system (Invitrogen) at 45°C for 45 min. Reverse transcriptase was deactivated at 85°C for 5 min, and 1 μl of RNase H was then added and incubated at 37°C for 20 min. Purification of cDNA with S.N.A.P. columns (Invitrogen Life Technologies), as well as tailing of purified cDNA using terminal deoxynucleotidyl transferase and dCTP, was done according to 5′ RACE System, version 2.0 instructions. Confirmation of cDNA was performed after each step by PCR with nested primers. Tailed cDNA was amplified by primary PCR using a 10 μM concentration of each primer, the 5′ RACE abridged anchor primer (AAP) that binds to the tailed cDNA sequence, and GSP2-FlrA (5′-CCTGACAGAAGTGCAGATG-3′) or GSP2-PomA2 (5′-TTTCATGAAGGCATTTGG-3′) that binds to an internal gene sequence. The PCR program applied was 94°C for 1 min, followed by 35 cycles of 94°C for 45 s, 55°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 5 min. Nested PCR was performed for pomA2 transcription start site amplification using the primary PCR product diluted 1:100 as a template and 10 mM (each) nested primer abridged by a universal amplification primer (AUAP) and GSP3-PomA2 (5′-GGCATTTGGCACTTCG-3′). The PCR program applied was 94°C for 1 min, followed by 35 cycles of 94°C for 45 s, 55°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 5 min. PCR products were analyzed by agarose gel electrophoresis, and amplified bands were excised from the gel, purified, and sequenced with GSP2-FlrA or GSP3-PomA2 primer.
Construction of defined mutants.
The chromosomal in-frame flhF and flhG deletion mutants, A. hydrophila AH-3 ΔflhF and AH-3 ΔflhG, respectively, were constructed by allelic exchange as described by Milton et al. (28). Briefly, DNA regions upstream (fragment AB) and downstream (fragment CD) of the flhF and flhG genes were independently amplified using two sets of asymmetric PCRs for each gene to amplify DNA fragments of 807 (FlhF-AB) and 570 (FlhF-CD) bp for the flhF in-frame deletion and of 641 (FlhG-AB) and 576 (FlhG-CD) bp for the flhG in-frame deletion. DNA fragment FlhF-AB contains 687 bp upstream of flhF and the first 39 codons of flhF. DNA fragment FlhF-CD contains from the first base in codon 448 of flhF to 496 bp downstream of flhF. DNA fragment FlhG-AB contains 583 bp upstream flhG and the first 18 codons of flhG. DNA fragment FlhG-CD contains from the first base in codon 270 of flhG to 502 bp downstream of flhG. DNA fragments AB and CD of each gene were annealed at the overlapping regions provided by the primers B and C and amplified as a single fragment using primers A and D (Table 2). The fusion products were purified, BglII digested (the BglII site is present in primers A and D), ligated into BglII-digested and phosphatase-treated pDM4 vector (27), electroporated into E. coli MC1061 (λpir), and plated on chloramphenicol plates at 30°C to obtain pDM-FlhF and pDM-FlhG plasmids. Plasmids with mutated genes were transferred into an A. hydrophila AH-405 rifampin-resistant (Rifr) strain by triparental matings using the E. coli MC1061 (λpir) containing the insertion constructs and the mobilizing strain HB101/pRK2073. Transconjugants were selected on plates containing chloramphenicol and rifampin. PCR analysis confirmed that the vector had integrated correctly into the chromosomal DNA. After sucrose treatment, transconjugants that were rifampin resistant (Rifr) and chloramphenicol sensitive (Cms) were chosen and confirmed by PCR.
Table 2.
Primers used in the construction of chromosomal in-frame flhF and flhG deletion mutant
| Target and primer | Primer sequencea | Amplified fragment |
|---|---|---|
| flhF mutant | ||
| A primer | 5′-AGATCTCCCAGACCCTCGGTTATAC-3′ | FlhF-AB |
| B primer | 5′-CCCATCCACTAAACTTAAACATATCTCCACCCCACCTGTG-3′ | |
| C primer | 5′-TGTTTAAGTTTAGTGGATGGGGAGCGGGAAACAGAAGAAC-3′ | FlhF-CD |
| D primer | 5′-AGATCTGATGGAGGTAGGCTCATCG-3′ | |
| flhG mutant | ||
| A primer | 5′-AGATCTCGGTTTGCCATGAAGTATG-3′ | FlhG-AB |
| B primer | 5′-CCCATCCACTAAACTTAAACAGTTTTGACGCATTTTGCG-3′ | |
| C primer | 5′-TGTTTAAGTTTAGTGGATGGGGGCGGTCATCTCGAATTTT-3′ | FlhG-CD |
| D primer | 5′-AGATCTGTTCGTCAAACCCGCTGT-3′ |
Underlining indicates overlapping regions. BglII restriction sites are shown in boldface.
To obtain the A. hydrophila AH-3 flrA mutant, the flrA gene was amplified by PCR with 5′-GCTCTAGATTGTCCTCGTTGCGATG-3′ and 5′-GCTCTAGACCTTGAACAAAGGCGTCA-3′ (XbaI sites are underlined), ligated into the vector pGEM-T Easy (Promega), and transformed into E. coli DH5α (16). The Tn5-derived kanamycin resistance cartridge (nptll) from pUC4-KIXX was obtained by SmaI digestion (5), and the cassette was inserted into the SmaI restriction internal site of flrA. The presence of a single BglII site in the SmaI-digested cassette allowed its orientation to be determined. Constructs containing the mutated genes were ligated into the XbaI-digested and phosphatase-treated pDM4 suicide vector (28), electroporated into E. coli MC1061 (λpir), and plated on chloramphenicol and kanamycin plates at 30°C to obtain the pDM-FlrAKm plasmid. Introduction of pDM-FlrAKm plasmid into A. hydrophila AH-405 was performed as previously described, and transconjugants were selected on plates containing chloramphenicol, kanamycin, and rifampin. PCR analysis confirmed that the vector had integrated correctly into the chromosomal DNA. After sucrose treatment, transconjugants that were Rifr, kanamycin resistant (Kmr), and Cms were chosen and confirmed by PCR.
To obtain the A. hydrophila AH-3 flrBC mutant, we used a single defined insertion in the flrB gene by a method based on suicide plasmid pFS100 (34). Briefly, an internal fragment of flrB was amplified by PCR (5′-CTGACCGAAACCCGCAAAC-3′ and 5′-GAACGACAGGGTAAAGCAG-3′), ligated into pGEM-T Easy (Promega), and transformed into E. coli DH5α (16). The DNA insert was recovered by EcoRI restriction digestion and was ligated into EcoRI-digested and phosphatase-treated pFS100 (pFS-FlrB). Ligation was transformed into E. coli MC1061 (λpir) and selected for kanamycin resistance (Kmr). Triparental mating with the mobilizing strain HB101/pRK2073 was used to transfer recombinant plasmid into the A. hydrophila AH-405 rifampin-resistant (Rifr) strain to obtain defined insertion mutants, selecting for Rifr and Kmr.
Plasmid constructions.
Plasmid pBAD33-FLHF and pBAD33-FLHG containing the complete flhF and flhG genes of A. hydrophila AH-3, respectively, under the arabinose promoter (PBAD) on pBAD33 (15) and plasmids pBAD33Gm-FLIA and pBAD33Gm-FLRA containing the complete fliA and flrA genes of A. hydrophila AH-3, respectively, under the arabinose promoter (PBAD) on pBAD33-Gm (17) were obtained. Oligonucleotides 5′-TCCCCCGGGTGCCTGATGACAAGCAA-3′ and 5′-GCTCTAGACAGGACATGGGAGAGGTTG-3′ generated a band of 1,723 bp containing the flhF gene, and oligonucleotides 5′-GATATCGAGCGGGAAACAGAAGAAC-3′ and 5′-GCTCTAGACCTCGGTATCACGAGCAT-3′ generated a band of 1,303 bp (the SmaI site is in boldface, the XbaI site is underlined, and the EcoRV site is in italics) containing the flhG gene. The amplified bands were digested with SmaI and XbaI or with EcoRV and XbaI and ligated into SmaI- and XbaI-digested pBAD33 vector to construct the pBAD33-FLHF and pBAD33-FLHG recombinant plasmids. Oligonucleotides 5′-GGAATTCCGGTCATCTCGAATTTTTCC-3′ and 5′-TCCCCCGGGTTGCGGATTATGCCTTAGAG-3′ generate a band of 840 bp containing the fliA gene, and oligonucleotides 5′-GGAATTCCGTGGCTAGACCACAGATC-3′ and 5′-CCCAAGCTTCTGGCTATTGGGTCAGGTT-3′ generate a band of 1,580 bp containing the flrA gene (the EcoRI site is underlined, the SmaI site is in boldface, and the HindIII site is in italics). The amplified bands were digested with EcoRI and SmaI or EcoRI and HindIII and ligated into EcoRI- and SmaI-digested or EcoRI- and HindIII-digested pBAD33-Gm vector (17) to construct the pBAD33Gm-FLIA and pBAD33Gm-FLRA recombinant plasmids. Plasmids were independently introduced into the E. coli DH5α (16) and sequenced.
Construction of flagellar promoter-lacZ fusions.
Oligonucleotide primer pairs for the A. hydrophila AH-3 promoter regions of the flaA, flaB, flgB, flgF, flgM, flhA, fliE, flrA, flrB, pomA, motX, pomA2, and lafK genes (2, 7, 8, 44) are listed in Table 3. Primers were designed to amplify a fragment of 216 to 1,732 bp that encompassed regions both upstream and downstream of the predicted start codon. Restriction sites were added to some primers for cloning purposes. Promoter fragments were PCR amplified from A. hydrophila AH-3 genomic DNA, ligated into pGEM-T Easy (Promega), and transformed into E. coli DH5α (16). DNA inserts containing flaA, flgM, flgB, fliE, flrA, motX, pomA, and pomA2 promoters were recovered by EcoRI/BamHI restriction digestion; inserts containing flaB, flgF, flhA, and flrB promoters were recovered by EcoRI/BglII restriction digestion; and the insert containing the flgA promoter was recovered by NotI-blunt ended/BamHI restriction digestion. The BamHI restriction site in the flrA insert is 86 bp downstream from the flrA start codon, and the BglII restriction sites in the flaB and flrB inserts are 146 and 223 bp downstream from the flaB and flrB start codons, respectively. The EcoRI or NotI restriction site comes from the pGEM-T Easy plasmid. Digested fragments were ligated into plasmid pDN19lacΩ EcoRI/BamHI-digested or EcoRI-blunt ended/BamHI (42), transformed into E. coli DH5α (16), and selected for tetracycline resistance (Tcr). The final constructs were confirmed by DNA-sequencing.
Table 3.
Primers used for flagellar promoter-lacZ fusion constructions
| Promoter | Primer sequencea | Siteb |
|---|---|---|
| flaAp | 5′-AATGGCTGCCTGCAAAAG-3′ | −357 flaA |
| 5′-CGGGATCCCAGGCGGGTGTAGGAAGTA-3′ | +96 flaA | |
| flaBp | 5′-GCTGCATCGACCATACTGT-3′ | −650 flaB |
| 5′-TCCGCAGTCTGAGCAACA-3′ | +235 flaB | |
| flgAp | 5′-AAGAATCGTCTGCCACCAG-3′ | −705 flgA |
| 5′-CGGGATCCGTGAGAAATACCGCAAAA-3′ | +52 flgA | |
| flgBp | 5′-CTTTCGCCCTTGATGACTC-3′ | −781 flgB |
| 5′-CGGGATCCAACACTCAGCGCGTATTGA-3′ | +53 flgB | |
| flgFp | 5′-AAGATGATCACGCCGACTAC-3′ | −679 flgF |
| 5′-GGAAGATCTATGCTGCAGGTTCTGACC-3′ | +203 flgF | |
| flgMp | 5′-TGACTATCTCAGCGATCCG-3′ | −360 flgM |
| 5′-CGGGATCCGAGGTCGCTGGTTTGGTAT-3′ | +89 flgM | |
| flhAp | 5′-AGCTCAAGATGACCAAGCAG-3′ | −586 flhA |
| 5′-GGAAGATCTGAAAGCGCAATATTGAAGGA-3′ | +160 flhA | |
| fliEp | 5′-TTGTCGCATGGTACTGCTC-3′ | −1010 fliE |
| 5′-CGGGATCCGACCATCTTGTTACGCACC-3′ | +281 fliE | |
| flrAp | 5′-CCCTCTGTTGCTCGATTG-3′ | −579 flrA |
| 5′-CCAGCTCGTTCTCGACTATC-3′ | +120 flrA | |
| flrBp | 5′-TACTATCGCCTCAATGTCTTCC-3′ | −670 flrB |
| 5′-TCATCTTCCACGACCAGAAT-3′ | +1062 flrB | |
| lafKp | 5′-GGGCAAGTTGGGCCAATAT-3′ | −997 lafK |
| 5′-CGGGATCCTTTCCATTTGATAACGCAGG-3′ | +98 lafK | |
| motXp | 5′-GCCACTCTGAAAGCCGATA-3′ | −172 motX |
| 5′-CGGGATCCGTGGTCAGCAAACAAGCAA-3′ | +44 motX | |
| pomAp | 5′-CGGGTCAAGGAAATATCGC-3′ | −746 pomA |
| 5′-CGGGATCCAGGATCAGGCCAAACAT-3′ | +24 pomA | |
| pomA2p | 5′-ATGGTTTCCAGCTCTTCCA-3′ | −283 pomA2 |
| 5′-CGGGATCCCAGCACTATGCCAAT-3′ | +37 pomA2 |
BamHI restriction sites are underlined; BglII restriction sites are in boldface.
Nucleotide position from the gene start codon.
β-Galactosidase assays.
The promoter-lacZ fusion plasmids described above were introduced into several A. hydrophila strains (Table 1). The cultures were grown in TSB medium at 25°C to an optical density of 0.4 to 0.8 at 600 nm. Bacterial cells were permeabilized with chloroform and sodium dodecyl sulfate (SDS) overnight and assayed for β-galactosidase activity as described by Miller (27). All experiments were performed at least three separate times.
Statistical analysis.
The data obtained for the β-galactosidase assays were analyzed by the t test using Microsoft Excel software.
RESULTS
The polar-flagellum σ28 factor (FliA) does not control pomA2B[r]2 stator motor transcription.
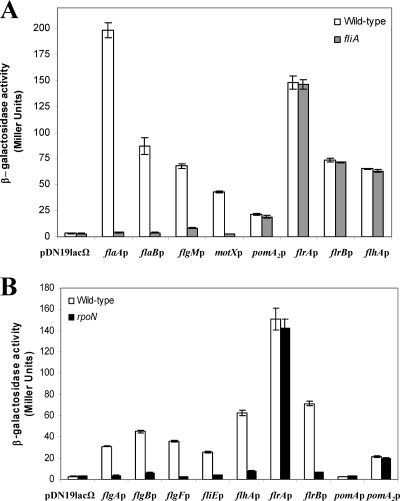
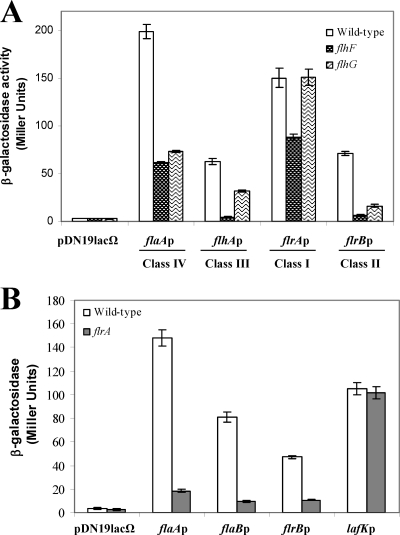
Transcription of the V. parahaemolyticus and P. aeruginosa polar-flagellum late genes (class IV) are σ28 dependent (11, 12, 33). In A. hydrophila mutation of FliA abolishes polar-flagellum formation (7), and both polar flagella and swimming motility were restored by complementation with the pBAD33Gm-FLIA plasmid in the presence of 0.2% l-arabinose. In silico sequence analysis of the A. hydrophila AH-3 polar-flagellum regions show putative σ28 promoter sequences upstream of flaA, flaB, flgM, motX (7), and pomA2 (44), which is redundant to pomA located in polar-flagellum region 3. In order to study whether any of these A. hydrophila promoters are σ28 dependent, we independently transferred the promoter-lacZ fusion plasmids pDNlac-flaAp (flaAp-lacZ), pDNlac-flaBp (flaBp-lacZ), pDNlac-flgMp (flgMp-lacZ), pDNlac-motXp (motXp-lacZ), and pDNlac-pomA2p (pomA2p-lacZ) into A. hydrophila AH-405 (AH-3 rifampin-resistant) and the fliA mutant (AH-4443) by triparental mating. Transconjugants that were Rifr Tcr or Rifr Kmr Tcr were chosen, and β-galactosidase activity was measured. Transcription from the flaA, flaB, flgM, and motX promoters in the fliA mutant background shows a 97, 95, 87, and 93% reduction in activity, respectively, in comparison to the A. hydrophila AH-405 value. However, pomA2 expression exhibited comparable values in both strains (Fig. 2A). Furthermore, total RNA from A. hydrophila AH-3 and the fliA mutant was used to amplify internal fragments of flaA, flaB, flgM, motX, and pomA2 transcripts, but no flaA, flaB, flgM, and motX amplicons were obtained in the fliA mutant, whereas an amplicon was observed for pomA2 (data not shown). These results suggest that flaA, flaB, flgM, and motX transcription is σ28 dependent but that pomA2 transcription is not.
Fig. 2.
Analysis of β-galactosidase activity. (A) pDNlac-flaAp, pDNlac-flaBp, pDNlac-flgMp, pDNlac-motXp, pDNlac-pomA2p, pDNlac-flrAp, pDNlac-flrBp, and pDNlac-flhAp plasmids in A. hydrophila wild-type (AH-405) and fliA mutant (AH-4443) after growth in TSB at 25°C. (B) pDNlac-flgAp, pDNlac-flgBp, pDNlac-flgFp, pDNlac-fliEp, pDNlac-flhAp, pDNlac-flrAp, pDNlac-flrBp, pDNlac-pomA, and pDNlac-pomA2p plasmids in A. hydrophila wild type (AH-405) and rpoN mutant (AH-5502) after growth in TSB at 25°C. As a control we also measured the pDN19lacΩ promoterless plasmid. The results shown are representative of three independent experiments. Bars represent standard deviations.
A. hydrophila AH-3 encodes an alternative σ54 sigma factor (RpoN) which is essential for both polar- and lateral-flagellum expression (8). We analyzed the possibility that pomA2 could be transcribed from a σ54-dependent promoter. The pDNlac-pomA2p plasmid was transferred into the A. hydrophila rpoN mutant (AH-5502), and β-galactosidase activity was measured. Similar activity was detected in the rpoN mutant and the wild type (Fig. 2B).
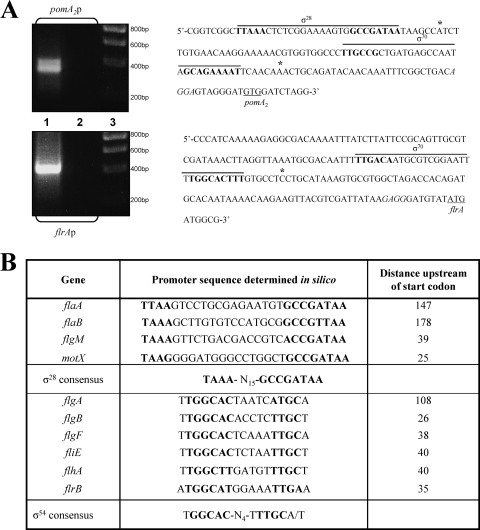
To identify the pomA2 promoter region, amplification of the A. hydrophila AH-3 pomA2 cDNA 5′ end was performed using 5′ RACE as described in Materials and Methods. Primary PCR of tailed cDNA using primers AAP (abridged anchor primer) and GSP2-PomA2 give two very faint bands, but nested PCR using primers AUAP (abridged universal amplification primer) and GSP3-PomA2 showed two DNA bands of approximately 350 and 400 bp (Fig. 3). The smaller band shows enhanced intensity compared to the larger band on the agarose gel, suggesting that pomA2B[r]2 is more actively transcribed from the promoter region closer to the pomA2 start codon. DNA sequences of the amplified bands indicate that both were tailed with G residues, and therefore pomA2B[r]2 is transcribed from two promoter regions. The pomA2 transcription starts were located −41 nucleotides (nt) and −107 nt upstream from the pomA2 translation start site. DNA sequence upstream of the −41-nt transcription start contains a σ70 promoter sequence (TTGCCG-N14-GCAGAAAAT), and sequence upstream of the −107-nt transcription start contains a σ28 promoter sequence (TAAA-N14-GCCGATAA) (Fig. 3). Using the same techniques, we were able to identify a σ70 promoter sequence and upstream of it a σ28 promoter sequence in strains A. hydrophila ATCC 7966T and A. caviae Sch3N (data not shown).
Fig. 3.
(A) Amplification of the A. hydrophila AH-3 pomA2 and flrA cDNA 5′ end performed using the 5′ RACE System, version 2.0 (Invitrogen). An amplicon was obtained by nested PCR using primers AUAP (abridged universal amplification primer) and GSP3-PomA2 (pomA2p) and by primary PCR using primers AAP (abridged anchor primer) and GSP2-FlrA (flrAp). Lanes 1, primary PCR template; lanes 2, PCR negative control; and lanes 3, molecular size standard (Ecogen). Underlined sequences show start codons, italics indicate the ribosome binding sites, asterisks show locations of the transcriptional start sites, and bold nucleotides show potential consensus sequences. (B) Alignment in silico of σ28 and σ54 promoter elements in A. hydrophila polar-flagellum promoters. The consensus σ28 sequence is from Kutsukake (22). The consensus σ54 sequence is from Barrios et al. (4).
RpoN is not involved in flrA transcription in A. hydrophila AH-3.
RpoN is involved in A. hydrophila polar- and lateral-flagellum formation (8), and in silico analysis of A. hydrophila AH-3 polar-flagellum regions shows putative σ54 promoter sequences upstream of flgA, flgB, flgF, flhA, fliE, flrA, flrB, and pomA genes (2, 7). Furthermore, transcription of Vibrio and Pseudomonas polar-flagellum class II and III genes, which include structural components of the hook, the basal body, the export apparatus, and regulatory proteins, is σ54 dependent (12, 33). Therefore, we investigated the effect of rpoN mutation on the transcription of different Aeromonas polar-flagellum promoters. β-Galactosidase activity of A. hydrophila wild type and the rpoN mutant (AH-5502) carrying the polar-flagellum gene promoter-lacZ fusion plasmids pDNlac-flgAp, pDNlac-flgBp, pDNlac-flgFp, pDNlac-fliEp, pDNlac-flhAp, pDNlac-flrAp, pDNlac-flrBp, and pDNlac-pomAp was measured. No significant β-galactosidase activity was detected from pDNlac-flgAp, pDNlac-flgBp, pDNlac-flgFp, pDNlac-flhAp, pDNlac-fliEp, and pDNlac-flrBp in the rpoN mutant background in comparison to the wild-type activity levels, suggesting that they are σ54-dependent promoters (Fig. 2B). However, β-galactosidase activity from pDNlac-flrAp plasmid is not affected by loss of σ54 factor, as no reduction in activity in comparison to the wild type was observed (Fig. 2B). Surprisingly, no β-galactosidase activity was detected from the in silico predicted pomA promoter in either the wild type or the mutant strain tested. To determine whether pomA is flhA cotranscribed, total RNA from A. hydrophila AH-3 was subjected to RT-PCR using primer pairs which amplified the flhA-flhF, flhF-fliA, fliA-cheZ, cheZ-cheA, cheA-cheB, and cheB-pomA genes. Amplicons were obtained with all primer pairs used (data not shown), suggesting that pomA transcription is under the control of the flhA promoter.
Given that flrA transcription was σ54 independent, we performed 5′ RACE, as described in Materials and Methods, to further analyze the flrA promoter region. Primary PCR of tailed cDNA using primers AAP (abridged anchor primer) and GSP2-FlrA gave a unique band of approximately 400 bp (Fig. 3). DNA sequence of the amplified band indicates that it was tailed with G residues. The flrA transcription start was located −68 nt upstream from the flrA translation start site, and DNA sequence upstream of the transcription start contains a σ70 promoter sequence (TTGACA-N14-TGGCACTTT) (Fig. 3). Using the same techniques, we were able to identify a σ70 promoter sequence upstream of flrA in strains A. hydrophila ATCC 7966T and A. caviae Sch3N (data not shown).
Identification of σ54 A. hydrophila promoters that are FlrA or FlrC dependent.
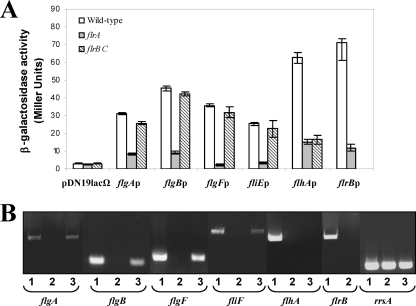
Promoters recognized by the σ54 holoenzyme require specialized enhancer-binding proteins, which bind specific sequences located in a relatively remote position from the transcription start site (6). Two σ54-dependent regulators are required to direct polar-flagellum class II and III gene transcription in V. cholerae and P. aeruginosa (12, 33). In A. hydrophila the mutation of flrA or flrC, which encodes σ54 enhancer-binding proteins homologous to V. cholerae FlrA/FlrC and P. aeruginosa FleQ/FleR (7), abolishes polar-flagellum formation, and the ability of the mutants to synthesize polar-flagellum formation and swimming motility was restored by complementation with the pBAD33Gm-FLRA plasmid in the presence of 0.2% l-arabinose and plasmid pACYC-FLR1 (7), respectively. To investigate which of the A. hydrophila σ54 polar-flagellum promoters are FlrA or FlrC dependent, β-galactosidase activity of the A. hydrophila wild type and the mutant strains AH-3 flrA and AH-3 flrBC carrying the promoter-lacZ fusion plasmids pDNlac-flgAp, pDNlac-flgBp, pDNlac-flgFp, pDNlac-fliEp, pDNlac-flhAp, and pDNlac-flrBp was measured. Transcription from flgA, flgB, flgF, fliE, and flrB promoters appeared to be affected by the flrA mutation, showing a 73, 79, 93, 86, and 83% reduction of β-galactosidase activity, respectively, in the AH-3 flrA strain compared to the wild type, and no significant variations were obtained in the AH-3 flrBC mutant (Fig. 4A). However, transcription from the flhA promoter showed β-galactosidase activity reduction in both mutant strains (76% in AH-3 flrA and 74% in AH-3 flrBC), suggesting that the flhA promoter is FlrC dependent. Furthermore, RT-PCRs to compare flgA, flgB, flgF, fliF, and flhA gene transcription levels in the wild-type as well as in the flrA and flrBC mutants produced flgA, flgB, flgF, and fliF amplicons in the wild type and the flrB mutant, whereas the flhA amplicon was found only in the wild type (Fig. 4B). Transcription of flrB was tested in the wild type and the flrA mutant, detected amplicons in the wild type only (Fig. 4B).
Fig. 4.
(A) Analysis of β-galactosidase activity of pDNlac-flgAp, pDNlac-flgBp, pDNlac-flgFp, pDNlac-fliEp, pDNlac-flhAp, and pDNlac-flrBp plasmids in A. hydrophila wild-type (AH-405) and the AH-3 flrA or AH-3 flrBC mutant after growth in TSB at 25°C. As a control we also measured the pDN19lacΩ promoterless plasmid. The results shown are representative of three independent experiments. (B) RT-PCR amplification of flgA, flgB, flgF, fliF, flhA, and flrB from cDNA of the AH-3 (lanes 1) strain and the AH-3 flrA (lanes 2) and AH-3 flrBC (lanes 3) mutants. A. hydrophila ribosomal 16S (rrsA) amplification was used as a control for cDNA template. RT-PCR amplifications were performed at least twice, with total RNA preparations obtained from a minimum of two independent extractions.
A. hydrophila flhF and flhG are involved in polar-flagellum biosynthesis and regulation.
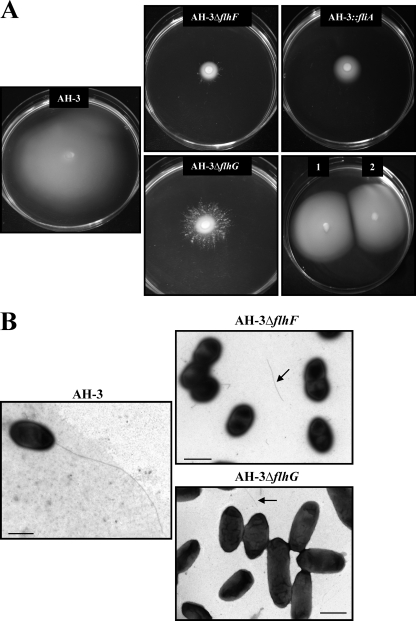
It has been reported that FlhF and FlhG are unique to polar-flagellated bacteria and regulate the number and distribution of flagella in Vibrio and Pseudomonas (11, 21, 32). A. hydrophila contains within the polar-flagellum flhA-cheW gene cluster two genes whose encoded proteins showed 51% and 70% identity to FlhF and FlhG of Vibrio spp. (Fig. 1) (7). A. hydrophila AH-3 flhF and flhG in-frame mutants were constructed, as described in Materials and Methods, and transcription of the downstream gene fliA was investigated by RT-PCR (data not shown). Both in-frame mutants showed expression of fliA, which is located downstream of flhG, and they also exhibited growth in TSB similar to that of the wild type. Unlike the wild type, both mutants failed to swim in liquid medium, as determined by light microscopy observation, and they also showed a reduced spread in semisolid agar plates, similar to that observed for polar-flagellum mutants such as flaH, flhA, or fliA (7). However, the spread of these two mutants in semisolid plates did not constitute homogeneous concentric rings, especially in the flhG mutant, but clotted and branching rings were observed (Fig. 5). Electron microscopic observations of the flhF and flhG mutants grown in liquid medium revealed that both mutants lacked a polar flagellum although approximately 10% of bacteria showed flagellum fragments on their surface, and some flagellum structures were observed in the medium (Fig. 5). Swimming motility was restored by complementation of the flhF and flhG mutants with the pBAD33-FLHF and pBAD33-FLHG plasmids, respectively, in the presence of 0.2% l-arabinose (Fig. 5). These results indicate that flhF and flhG are required for A. hydrophila assembly of a functional polar flagellum.
Fig. 5.
Phenotypes of FlhF and FlhG mutants. (A) Swarming motility observed for A. hydrophila AH-3, AH-3 ΔflhF, AH-3 ΔflhG, and AH-3 fliA (AH-4443), as well as complemented AH-3 ΔflhF (1) and AH-3 ΔflhG (2) strains with the pBAD33-FLHF and pBAD33-FLHG plasmids, respectively, in the presence of 0.2% l-arabinose. (B) Transmission electron microscopy of A. hydrophila AH-3, AH-3 ΔflhF, and AH-3 ΔflhG grown at 25°C on liquid medium. Arrows show flagellum fragments. Bacteria were gently placed onto Formvar-coated copper grids and negatively stained using 2% uranyl acetate. Scale bar, 1 μm.
To determine whether A. hydrophila FlhF and FlhG affect polar-flagellum gene transcription, plasmids containing promoters belonging to different classes of the flagellum hierarchy fused to lacZ were transferred into the A. hydrophila flhF and flhG mutants. Analysis of the β-galactosidase activity of pDNlac-flaAp, pDNlac-flhAp, pDNlac-flrAp, and pDNlac-flrBp plasmids in the flhF mutant showed reduced activity of 68, 92, 41, and 91% from the flaA, flhA, flrA, and flrB promoters, respectively, in comparison to the wild-type values, suggesting that FlhF positively regulates all classes of A. hydrophila polar-flagellum genes (Fig. 6A). The flhG mutant showed an flrA promoter transcription level similar to that of the wild type and a significant reduction of 63, 48 and 77% in the activity of the flaA, flhA, and flrB promoters versus the wild-type values, respectively. The data suggest that FlhG positively regulates A. hydrophila polar-flagellum gene classes II, III, and IV (Fig. 6A).
Fig. 6.
Analysis of β-galactosidase activity. (A) pDNlac-flaAFp, pDNlac-flhAp, pDNlac-flrAp, and pDNlac-flrBp plasmids in A. hydrophila wild type (AH-405) and AH-3 ΔflhF and AH-3 ΔflhG mutants after growth in TSB at 25°C. (B) pDNlac-lafKp, pDNlac-flaAp, pDNlac-flaBp, and pDNlac-flrBp plasmids in A. hydrophila wild-type (AH-405) after growth in plates at 25°C. As a control we also measured the pDN19lacΩ promoterless plasmid. The results shown are representative of three independent experiments.
FlrA cannot be substituted for the lateral-flagellum regulator LafK.
A. hydrophila as well as V. parahaemolyticus has dual flagellum systems (polar and lateral flagella) which do not share structural or regulatory genes, and both contribute to motility in semisolid plates. One open reading frame of lateral-flagellum clusters of both species encodes a σ54-dependent response regulator, LafK, essential for lateral-flagellum generation (8, 39). V. parahaemolyticus LafK is able to compensate for mutation of the polar-flagellum σ54-dependent response regulator, FlaK (19). The A. hydrophila FlrA mutation abolished polar-flagellum formation but did not affect lateral-flagellum-dependent swarming motility on semisolid medium; furthermore an A. hydrophila LafK mutation that abolished lateral-flagellum formation did not affect polar-flagellum biogenesis (7, 8). β-Galactosidase activity of pDNlac-lafKp (lafKp-lacZ) was measured after growth in semisolid plates (inducing conditions for lateral flagella) to ensure that LafK was transcribed. Analyses show that lafK was transcribed similarly in the wild-type and the flrA mutant strains in semisolid medium. We measured the β-galactosidase activity of pDNlac-flaAp, pDNlac-flaBp, and pDNlac-flrBp plasmids in the A. hydrophila wild type and the flrA mutant after growth on semisolid medium. β-Galactosidase values were reduced in the flrA mutant versus the wild-type background (Fig. 6B). These results suggest that LafK cannot supplement FlrA function and that FlrA is required for polar-flagellum production and swimming motility.
DISCUSSION
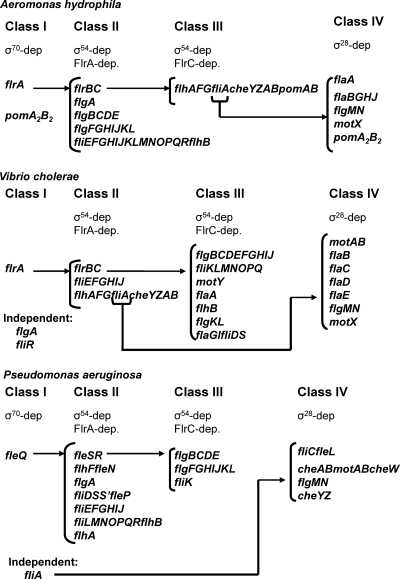
Mesophilic Aeromonas produces a single polar flagellum constitutively expressed in both liquid and solid media. In addition, 50 to 60% of strains also have an inducible lateral-flagellum system that is expressed in high-viscosity medium. Polar-flagellum genes are organized in different clusters distributed in six chromosomal regions (Fig. 1). The measurement of promoter-lacZ fusion activities and RT-PCR assays in defined flagellum-regulatory mutants allowed us to analyze the A. hydrophila polar-flagellum cluster transcription hierarchy. A. hydrophila polar-flagellum expression, as in other mono-flagellated bacteria, seems to be organized in four transcriptional levels (classes I to IV), where each level contains the activator for the subsequent transcriptional level.
Class I.
A. hydrophila polar-flagellum-regulatory cascade class I seems to include only the flrA gene since its mutation reduced the β-galactosidase activity of most promoter-lacZ fusions tested (Fig. 4). A. hydrophila FlrA looks like the master regulator of the polar-flagellum regulon, and the analysis of flrAp-lacZ fusion in the A. hydrophila rpoN and fliA mutants (AH-5502 and AH-4443, respectively) showed β-galactosidase activity levels similar to the level determined in the wild type (Fig. 2), suggesting that flrA is transcribed in a σ54- and σ28-independent manner. Amplification of the flrA cDNA 5′ ends by 5′ RACE allowed us to obtain a DNA sequence upstream of the flrA transcription start which contained a σ70 promoter sequence (TTGACA-N14-TGGCACTTT) (Fig. 3). This result indicated that A. hydrophila polar-flagellum class I gene transcription is σ70 dependent in several strains (Fig. 7) and is consistent with the fact that the A. hydrophila polar flagellum is constitutively expressed, which is in contrast to other bacteria with dual flagellum systems, such as V. parahaemolyticus (24). Our data are similar to those for flagellum class I gene transcription of P. aeruginosa (13) although the σ54 factor is possibly involved in the transcription regulation of other pseudomonads (38). In Vibrio spp. class I gene transcription is σ54 and σ28 independent, but no specific motifs for σ70 binding were found in their promoter sequences (33).
Fig. 7.
Comparative proposed A. hydrophila, V. cholerae (41), and P. aeruginosa (12) polar-flagellum gene transcription hierarchies.
V. parahaemolyticus and some A. hydrophila strains express dual flagellum systems, and LafK is the lateral-flagellum master regulator (8, 39). In V. parahaemolyticus LafK is able to compensate for the mutation of the polar-flagellum σ54-dependent response regulator FlaK (19). Data obtained are consistent with the fact that the A. hydrophila FlrA mutation abolishes polar-flagellum formation in liquid and solid media but does not affect inducible lateral-flagellum formation (7). These results suggest that despite the 57% similarity between A. hydrophila LafK and FlrA, their C-terminal domains might recognize different DNA binding regions which do not coexist in polar-flagellum class II promoter sequences. It is tempting to suggest that V. parahaemolyticus polar-flagellum class II promoter sequences should contain binding sites for LafK and FlaK. Therefore, results highlight that polar- and lateral-flagellum interconnections and control networks are specific and different for the dual flagellum systems of A. hydrophila and V. parahaemolyticus.
Class II and III.
Transcription of the Pseudomonas and V. cholerae polar-flagellum genes included in the class II and III levels, as well as the dually flagellated V. parahaemolyticus, is σ54 dependent. Their transcriptions require specialized enhancer-binding proteins (FleQ, FlrA, and FlaK, respectively) involved in class II and class III (FleS, FlrC, and FlaM, respectively) transcription (12, 18, 33, 41). Transcription of A. hydrophila polar-flagellum promoters, whose sequences in silico contain conserved nucleotides that could represent σ54-dependent consensus regions, was analyzed in the A. hydrophila wild type and the AH-5502 (rpoN mutant) (8), AH-3 flrA, and AH-3 flrBC mutants by promoter-lacZ fusions and RT-PCR. Each of these three mutants restored swimming motility after being complemented with the mutated gene. A previous study suggested that A. hydrophila pomAB, in polar-flagellum region 3 (Fig. 1), constituted an independent transcriptional unit, and a putative σ54-dependent promoter upstream of pomA was found (7); however, no β-galactosidase activity was detected in the pomAp-lacZ fusion in either the wild type or mutant strains tested (Fig. 2B). The data obtained by both techniques indicate that pomAB are not independently transcribed and constitute a unique transcript from an flhA promoter previously identified although this fact does not completely eliminate the possibility of additional promoters. The data obtained suggest that flgA, flgB, flgF, fliE, and flrB promoters are σ54 and FlrA dependent, whereas the flhA promoter is σ54 and FlrC dependent. Furthermore, analysis of flrBp-lacZ and flhAp-lacZ fusions representing class II and class III promoters in the A. hydrophila fliA mutant showed β-galactosidase activities similar to the activity determined in the wild type (Fig. 2).
Therefore, the A. hydrophila polar-flagellum class II transcription level includes the following: flgA, which is necessary for P-ring addition; flgB-E (operon comprising flgB and flgE and the genetic material between the two genes) and flgF-L, which encode proteins involved in basal body, L and P rings, and hook formation; fliE-flhB, which encode the export/assembly apparatus, the MS ring-switch complex, and the hook length control protein; and flrBC, which encode regulatory proteins. All A. hydrophila polar-flagellum genes included in the class III transcription level are localized in a unique cluster, flhA-pomB, whose genes encode proteins involved in export/assembly, motor, and chemotaxis, as well as the σ28 factor and two flagellum regulators (Fig. 7).
Although A. hydrophila, V. cholerae, and P. aeruginosa polar-flagellum clusters are quite conserved, they differ in their chromosomal distributions and in the transcription hierarchies (12, 25, 33). While genes homologous to flrBC and fliE-J are transcribed as class II in the three species, greater diversity in the transcriptional hierarchy is shown in the remaining polar-flagellum class II/III genes. A. hydrophila flgB-E, flgF-L, and fliK are class II, but in P. aeruginosa and V. cholerae they are class III. A. hydrophila flhA-G are class III, P. aeruginosa and V. cholerae homologous genes are class II. A. hydrophila and P. aeruginosa fliL-flhB and flgA are class II, whereas V. cholerae fliL-flhB are class III and flgA is σ54- and σ28-independently transcribed. The genes transcribed in a more variable hierarchical order are fliA, cheY-B, and pomAB. Thus, P. aeruginosa fliA is σ54 independent, and cheY-B pomAB are class IV genes, whereas A. hydrophila and V. cholerae fliA cheY-B are transcribed from the flhA promoter, being σ54 and FlrA dependent in V. cholerae (class II) and σ54 and FlrC dependent in A. hydrophila (class III). A. hydrophila pomAB are also transcribed from the flhA promoter, but in V. cholerae these two genes constituted a transcriptionally independent, σ28-dependent unit (class IV) located in a different chromosomal region.
Class IV.
A. hydrophila fliA encodes a σ28 homologue that is required for polar-flagellum formation (7). FliA from Gammaproteobacteria is involved in transcription of late flagellar genes, such as flagellins, the anti-sigma factor FlgM, and motor components (12, 33). Analysis of β-galactosidase activity of flaAp-lacZ, flaBp-lacZ, flgMp-lacZ, motXp-lacZ, and pomA2p-lacZ fusions in the A. hydrophila wild type and the fliA mutant showed that all promoter-lacZ fusions tested, with the exception of pomA2p-lacZ, have a low level of transcription in the fliA mutant, suggesting that they are σ28 dependent (Fig. 2A). The pomA2 promoter exhibited a similar level of transcription in both strains, as well as in the rpoN mutant, suggesting that it is σ54 and σ28 independent (Fig. 2B). Analysis of pomA2 cDNA 5′ ends by 5′ RACE showed two different transcription start sites at −41 and −107 nt. The DNA fragment upstream of the −41-nt transcription start contained a σ70 promoter sequence, and a fragment upstream of the −107-nt transcription start contained a σ28 promoter sequence (Fig. 3). Since A. hydrophila has a redundant set of motor proteins involved in polar-flagellum rotation, PomA2B2 and PomAB, with PomA2B2 being more sensitive to low-level sodium ion variations and having a lower level of transcription than PomAB (44), the results obtained suggest that transcription of these redundant motor proteins is specifically regulated and expressed at different levels of the flagellar hierarchy. Thus, while pomAB is transcribed from the flhA promoter, which is σ54 and FlrC dependent, pomA2B[r]2 is transcribed independently of the flagellum hierarchy although conditions that increase its transcription remain to be determined (Fig. 7). Homologous motor proteins of Vibrio (PomAB) and Pseudomonas (MotAB), as well as MotX, are transcribed from σ28-dependent promoters (12, 33, 41).
A. hydrophila, P. aeruginosa, and V. cholerae polar-flagellum class IV genes include flgMN and most flagellin genes, as well as motX. Genes that encode the filament length control (flaG), the filament cap protein (flaH), and a chaperone (flaJ) are more diversely transcribed in the hierarchies of these three species. A. hydrophila flaGHJ homologues and P. aeruginosa flaG homologues are class IV genes, P. aeruginosa flaHJ homologues are class II, and V. cholerae flaGHJ are class III (Fig. 7). On the other hand, although flgMN constitutes a σ28-dependent transcription unit in these three species, in P. aeruginosa and V. cholerae these genes are also transcribed with flgA, whereas in A. hydrophila no mRNA containing flgA and flgM was found (data not shown).
FlhF and FlhG structural and regulatory implications.
It has been reported that FlhF and FlhG act together to regulate flagellum placement and number in Vibrio alginolyticus and Pseudomonas species. V. cholerae and V. alginolyticus flhF disruption abolishes flagellum formation (11, 21), and Pseudomonas flhF disruption gives an aberrant placement of flagella (30, 32). In Vibrio and Pseudomonas, flhG disruption increases the number of polar flagella per cell (11, 12, 21, 30). Our results indicate that A. hydrophila FlhF and FlhG are essential for the assembly of a functional polar flagellum because in-frame mutants fail to swim in liquid medium and lack polar flagella although some flagellum fragments were observed in the medium (Fig. 5). Furthermore, the irregular motility in a semisolid plate (Fig. 5), especially of the flhG mutant, together with the presence of some polar-flagellum fragments in the medium, might suggest that some cells assemble an unstable polar flagellum, which allows them to swim for a short time and then constitute a colony after loosing the flagellum.
A. hydrophila FlhF positively regulates all polar-flagellum transcription levels (classes I to IV), whereas FlhG positively regulates classes II, III, and IV. Because of the hierarchical nature of flagellum transcription, the reduction in the transcription of the master regulator FlrA may be the cause for the general reduction in all of the classes of flagellum promoters, suggesting that FlhF exerts its influence by positively regulating the transcription of flrA. Furthermore, FlhG may exert its function through the regulation of the activity of FlrA. Among Aeromonas, Vibrio, and Pseudomonas species, FlhF and FlhG homologues do not play the same role in flagellum gene regulation, but they regulate the expression of the majority of flagellum genes directly or indirectly. FlhF increases the expression of Vibrio class III genes only (11), whereas there was an increase in expression of all Aeromonas polar-flagellum gene classes. A. hydrophila FlhG is a positive regulator of class II genes, whereas in Vibrio it is a repressor of class I genes, and in Pseudomonas it is a repressor of class II genes (11, 13).
Our results indicate that the A. hydrophila polar-flagellum transcription hierarchy shares some similarities but many important differences with the transcription hierarchies of V. cholerae and P. aeruginosa (12, 33, 41).
ACKNOWLEDGMENTS
This work was supported by Plan Nacional de I + D (Ministerio de Educación, Ciencia y Deporte and Ministerio de Sanidad, Spain) and from Generalitat de Catalunya (Centre de Referència en Biotecnologia). R.M. has a predoctoral fellowship from Ministerio de Educación, Ciencia y Deporte.
We thank Maite Polo for her technical assistance and the Servicios Científico-Técnicos from the University of Barcelona.
Footnotes
Published ahead of print on 22 July 2011.
REFERENCES
- 1. Aldridge P., Hughes K. T. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5:160–165 [DOI] [PubMed] [Google Scholar]
- 2. Altarriba A., et al. 2003. A polar flagella operon (flg) of Aeromonas hydrophila contains genes required for lateral flagella expression. Microb. Pathog. 34:249–259 [DOI] [PubMed] [Google Scholar]
- 3. Altschul F. S., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acid Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barrios H., Valderrama B., Morett E. 1999. Compilation and analysis of σ54-dependent promoter sequences. Nucleic Acid Res. 27:4305–4313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bott M., Meyer M., Dimroth P. 1995. Regulation of anaerobic citrate metabolism in Klebsiella pneumoniae. Mol. Microbiol. 18:533–546 [DOI] [PubMed] [Google Scholar]
- 6. Buck M., Gallegos M. T., Studholme D. J., Guo Y., Gralla J. D. 2000. The bacterial enhancer-dependent sigma 54 transcription factor. J. Bacteriol. 182:4129–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Canals R., et al. 2006. Polar flagellum biogenesis in Aeromonas hydrophila. J. Bacteriol. 188:542–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Canals R., et al. 2006. Analysis of the lateral flagellar gene system of Aeromonas hydrophila AH-3. J. Bacteriol. 188:852–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chadsey M. S., Karlinsey J. E., Hughes K. T. 1998. The flagellar anti-sigma factor FlgM actively dissociates Salmonella typhimurium σ28 RNA polymerase holoenzyme. Genes Dev. 12:3123–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chilcott G. S., Hughes K. T. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Correa N. E., Peng F., Klose K. E. 2005. Roles of the regulatory proteins FlhF and FlhG in the Vibrio cholerae flagellar transcription hierarchy. J. Bacteriol. 187:6324–6332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dasgupta N., et al. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50:809–824 [DOI] [PubMed] [Google Scholar]
- 13. Dasgupta N., Ferrell E. P., Kanack K. J., West S. E. H., Ramphal R. 2002. fleQ, the gene encoding the major flagellar regulator of Pseudomonas aeruginosa, is σ70 dependent and is downregulated by Vfr, a homolog of Escherichia coli cyclic AMP receptor protein. J. Bacteriol. 184:5240–5250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fenchel T. 2002. Microbial behavior in a heterogeneous world. Science 296:1068–1071 [DOI] [PubMed] [Google Scholar]
- 15. Guzman L.-M., Belin D., Carson M. J., Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 17. Jimenez N., et al. 2009. Genetics and proteomics of Aeromonas salmonicida lipopolysaccharide core biosynthesis. J. Bacteriol. 191:2228–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim Y. K., McCarter L. L. 2000. Analysis of the polar flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 182:3693–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim Y. K., McCarter L. L. 2004. Cross-regulation in Vibrio parahaemolyticus: compensatory activation of polar flagellar genes by the lateral flagellar regulator LafK. J. Bacteriol. 186:4014–4018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kirov S. M., et al. 2002. Lateral flagella and swarming motility in Aeromonas species. J. Bacteriol. 184:547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kusumoto A., et al. 2008. Collaboration of FlhF and FlhG to regulate polar-flagella number and localization in Vibrio alginolyticus. Microbiology 154:1390–1399 [DOI] [PubMed] [Google Scholar]
- 22. Kutsukake K., Ohya Y., Iino T. 1990. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J. Bacteriol. 172:741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kutsukake K. 1997. Autogenous and global control of the flagellar master operon, flhDC, in Salmonella typhimurium. Mol. Gen. Genet. 254:440–448 [DOI] [PubMed] [Google Scholar]
- 24. McCarter L. L., Hilmen M., Silvermanm M. 1988. Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell 54:345–351 [DOI] [PubMed] [Google Scholar]
- 25. McCarter L. L. 2001. Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 65:445–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Merino S., Camprubi S., Tomas J. M. 1991. The role of lipopolysaccharide in complement-killing of Aeromonas hydrophila strains of serotype O:34. J. Gen. Microbiol. 137:1583–1590 [DOI] [PubMed] [Google Scholar]
- 27. Miller J. H. 1992. A short course in bacterial genetics, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 28. Milton D. L., O'Toole R., Horstedt P., Wolf-Watz H. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178:1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Münch R., et al. 2005. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 21:4187–4189 [DOI] [PubMed] [Google Scholar]
- 30. Murray T. S., Kazmierczak B. I. 2006. FlhF is required for swimming and swarming in Pseudomonas aeruginosa. J. Bacteriol. 188:6995–7004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ohnishi K., Kutsukake K., Suzuki H., Iino T. 1990. Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol. Gen. Genet. 221:139–147 [DOI] [PubMed] [Google Scholar]
- 32. Pandza S., et al. 2000. The G-protein FlhF has a role in polar flagellar placement and general stress response induction in Pseudomonas putida. Mol. Microbiol. 36:414–423 [DOI] [PubMed] [Google Scholar]
- 33. Prouty M. G., Correa N. E., Klose K. E. 2001. The novel sigma54 and sigma28 dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39:1595–1609 [DOI] [PubMed] [Google Scholar]
- 34. Rubires X., et al. 1997. A gene (wbbL) from Serratia marcescens N28b (O4) complements the rfb-50 mutation of Escherichia coli K-12 derivatives. J. Bacteriol. 179:7581–7586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 36. Sanger F., Nicklen S., Coulson A. R. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74:5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shimada T., Sakazaki R., Suzuki K. 1985. Peritrichous flagella in mesophilic strains of Aeromonas. Jpn. J. Med. Sci. Biol. 38:141–145 [DOI] [PubMed] [Google Scholar]
- 38. Soutourina O. A., Semenova E. A., Parfenova V. V., Danchin A., Bertin P. 2001. Control of bacterial motility by environmental factors in polarly flagellated and peritrichous bacteria isolated from Lake Baikal. Appl. Environ. Microbiol. 67:3852–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stewart B. J., McCarter L. L. 2003. Lateral flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 185:4508–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Studholme D. J., Dixon R. 2003. Domain architectures of σ54-dependent transcriptional activators. J. Bacteriol. 185:1757–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Syed K. A., et al. 2009. The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors. J. Bacteriol. 191:6555–6570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Totten P. A. A., Lory S. 1990. Characterization of the type a flagellin gene from Pseudomonas aeruginosa PAK. J. Bacteriol. 172:7188–7199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang S., Fleming R. T., Westbrook E. M., Matsumura P., McKay D. B. 2006. Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription. J. Mol. Biol. 355:798–808 [DOI] [PubMed] [Google Scholar]
- 44. Wilhelms M., et al. 2009. Two redundant sodium-driven stator motor proteins are involved in Aeromonas hydrophila polar flagellum rotation. J. Bacteriol. 191:2206–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]