Abstract
During protein synthesis, translation elongation factor Tu (Ef-Tu) is responsible for the selection and binding of the cognate aminoacyl-tRNA to the acceptor site on the ribosome. The activity of Ef-Tu is dependent on its interaction with GTP. Posttranslational modifications, such as phosphorylation, are known to regulate the activity of Ef-Tu in several prokaryotes. Although a study of the Mycobacterium tuberculosis phosphoproteome showed Ef-Tu to be phosphorylated, the role of phosphorylation in the regulation of Ef-Tu has not been studied. In this report, we show that phosphorylation of M. tuberculosis Ef-Tu (MtbEf-Tu) by PknB reduced its interaction with GTP, suggesting a concomitant reduction in the level of protein synthesis. Overexpression of PknB in Mycobacterium smegmatis indeed reduced the level of protein synthesis. MtbEf-Tu was found to be phosphorylated by PknB on multiple sites, including Thr118, which is required for optimal activity of the protein. We found that kirromycin, an Ef-Tu-specific antibiotic, had a significant effect on the nucleotide binding of unphosphorylated MtbEf-Tu but not on the phosphorylated protein. Our results show that the modulation of the MtbEf-Tu–GTP interaction by phosphorylation can have an impact on cellular protein synthesis and growth. These results also suggest that phosphorylation can change the sensitivity of the protein to the specific inhibitors. Thus, the efficacy of an inhibitor can also depend on the posttranslational modification(s) of the target and should be considered during the development of the molecule.
INTRODUCTION
Elongation factor Tu (Ef-Tu) and its homologs play a pivotal role in protein biosynthesis in prokaryotes as well as in eukaryotes. Ef-Tu is a highly conserved protein that interacts with RNA, proteins, and nucleotides (43). It consists of three domains; of these, domain I exhibits a nucleotide-binding property (GTPase domain), which is highly conserved among all GTPases (2, 12). The 3′ end of tRNA resides in the crevice between domains I and II, while domain III interacts primarily with the 5′ acceptor arm of the tRNA. The overall effect of Ef-Tu activity depends largely on the interplay of all three domains (14). Protein translation in Escherichia coli is the most widely studied system in which Ef-Tu functions as a molecular switch that alternates between active and inactive states. In the active state, Ef-Tu binds to GTP, and the complex mediates accurate binding of aminoacyl-tRNA to the ribosome. This process is associated with the hydrolysis of GTP, after which Ef-Tu remains bound to GDP, becomes inactive, and is released from the ribosome (38, 39). E. coli Ef-Tu has a low dissociation rate for bound GDP, in comparison to GTP (19); as a result, Ef-Tu is found primarily in a GDP-bound state. Exchange of GDP with GTP is mediated by another elongation factor, Ef-Ts, which serves as the guanine nucleotide exchange factor. Subsequent to the exchange of GDP with GTP from Ef-Tu by Ef-Ts, Ef-Tu commences a new elongation cycle (1, 24).
Reversible phosphorylation, carried out by protein kinases and phosphatases, has evolved as a ubiquitous signaling mechanism that can alter the cellular metabolism in response to a changing environment. Phosphorylation of Ef-Tu has been demonstrated in several organisms, such as E. coli (28), Thermus thermophilus (28), Listeria monocytogenes (6), Streptococcus pneumoniae (50), Corynebacterium glutamicum (11), Bacillus subtilis (27), Mycoplasma pneumoniae (44), and Streptomyces collinus (30). The pathogenic bacterium Mycobacterium tuberculosis has 11 Ser/Thr protein kinases (STPKs), one Ser/Thr phosphatase (PstP), two Tyr phosphatases (16), and one Tyr kinase (9). PknB, an essential STPK of M. tuberculosis (42), has been associated with a large number of events relating to cell division and growth (18, 23, 25, 31), metabolism (8, 32, 33), cell wall synthesis (20, 51, 52), and stress response (10, 35). M. tuberculosis survives various stress conditions during disease establishment and has the ability to enter into dormancy. In order for M. tuberculosis to cope with the changing environment and become a successful pathogen, regulation of protein synthesis would be critical. However, the regulation of the components of protein synthesis in M. tuberculosis has not been explored so far. Most of the critical components of protein translation are also essential in M. tuberculosis, including elongation factor Tu (42). Although M tuberculosis Ef-Tu (MtbEf-Tu) has not been characterized, it has been found to be associated with the cell wall (40) and to be induced under anaerobic conditions (49) and with high-iron-containing media (56). MtbEf-Tu has also been implicated in binding to human plasminogen (57). Ef-Tu has been widely known to be regulated by multiple posttranslational modifications, including phosphorylation, acetylation, and methylation in various organisms, especially E. coli (4, 5, 22, 28). In E. coli, the phosphorylation of Ef-Tu has been suggested to facilitate its release from the ribosome, which may reduce the rate of protein synthesis (3).
In this report, we show that phosphorylation of MtbEf-Tu decreased the affinity of MtbEf-Tu with GTP. Reduced binding of GTP is expected to cause a reduction in the level of protein synthesis. Overexpression of PknB in Mycobacterium smegmatis showed that the level of protein synthesis was indeed decreased. Multiple residues of MtbEf-Tu are phosphorylated by PknB, and phosphorylated MtbEf-Tu is less sensitive to kirromycin, an antibiotic that inhibits the activity of the protein. Phosphorylation-mediated regulation of MtbEf-Tu may play a significant role in the dormancy of M. tuberculosis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli DH5α (Novagen) and BL21(DE3) (Stratagene) were used for cloning and protein expression. E. coli cells were grown and maintained with constant shaking (220 rpm) at 37°C in LB medium supplemented with 100 μg/ml ampicillin. Mycobacterial strains were grown in Middlebrook 7H9 broth supplemented with 0.5% glycerol, 0.05% Tween 80, 1% albumin-dextrose-catalase, and 50 μg/ml kanamycin at 37°C, with shaking (200 rpm).
Gene manipulation.
The genes coding for the cytosolic region of PknA (PknAc) (PknA amino acids [aa] 1 to 337) (rv0015c), PknBc (PknB aa 1 to 331) (rv0014c) (20), PknDc (PknD aa 1 to 378) (rv0931c), PknE aa 1 to 566 (rv1743), PknF aa 1 to 476 (rv1746), PknG aa 1 to 750 (rv0410c), PknHc (PknH aa 1 to 403) (rv1266c), PknJc (PknJ aa 1 to 320) (rv2088) (8), and PstPc (PstP aa 1 to 300) (rv0018c) (20) were PCR amplified using M. tuberculosis H37Rv genomic DNA. The amplicons thus generated were digested with the corresponding restriction enzymes and were ligated into vectors pProEx-HTc and/or pGEX-5X-3, previously digested with the same enzymes. The 1,188-bp gene coding for MtbEf-Tu (rv0685; tuf) was amplified from M. tuberculosis H37Rv genomic DNA and was cloned into the pProEx-HTc and pVV16 vectors. GST-Ef-Ts was constructed by a similar procedure using primers for amplification of the rv2889c (tsf) gene. HTc-MtbEf-Tu was subjected to site-directed mutagenesis to generate HTc-MtbEf-Tu-T118A using the QuikChange XL site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. For cloning into the dual-expression vector pETDuet-1 (Novagen), the gene coding for MtbEf-Tu was inserted in MCS1 with an N-terminal His6 tag, while the gene coding for PknB (aa 1 to 626) was cloned into MCS2 with an N-terminal maltose-binding protein (MBP) tag upstream of the kinase. The kinase-dead mutant of PknB, PknBK40M, cloned into MCS2 along with MtbEf-Tu in MCS1, served as a control. The details of PknB cloning in pETDuet-1 have been described previously (25). The genes encoding PknB and PknBK40M were also cloned into pSD5 for overexpression in M. smegmatis as described previously (46). The details of all the primers and clones are provided in Tables 1 and 2. The integrity of all clones was confirmed by DNA sequencing (TCGA, New Delhi, India).
Table 1.
Primers used in this study
| Primer name | Sequence (5′→3′)a |
|---|---|
| pknA FP | TGATCGAAGCCGGAATTCAGGGGGAACCATGA (EcoRI) |
| pknAc RP | AGCACCCCCGCGGCCGCGAGCAGCGCTCACTGACCGGAC (NotI) |
| pknD FP | TGACCTAGTGAAGGGAATTCGACGGTGAGC (EcoRI) |
| pknDc RP | GCAGCGCCGACGGCGGCCGCCTACTTCCGTTTGTTGC (NotI) |
| pknH FP | GCGGCAGCGGGTTGCTCGAGAGGATCAGCGATG (XhoI) |
| pknHc RP | GACGGCGGCGGCGGCGGCCGCTCAGGGCCACGGGTTG (NotI) |
| pknJ FP | GGTTGATGGGCGGCCGCGTGGCCCACGAGTTGAGTG (NotI) |
| pknJc RP | ATGCTGTCACCACGGCGAACCTCGAGCTGAGAACCTGGC (XhoI) |
| tuf FP | TGCTTTTATAAGCACCAACGGATCCAGGAGGACACA (BamHI) |
| tuf RP | CTTTTGCGTCTGGCGGCCGCTAGACCTACTTG (NotI) |
| tufT118A FP | GACGGCCCGATGCCCCAGGCCCGCGAGCACGTTCTGC |
| tufT118A RP | GCAGAACGTGCTCGCGGGCCTGGGGCATCGGGCCGTC |
| tsf FP | ACCAACCACAGATGGATCCTAGGAAAGGCT (BamHI) |
| tsf RP | GCGTGTACGCGCGGAATTCCCGGCTAAGCC (EcoRI) |
| pSD5 pknB FP | CGAGATAGCCATATGACCACCCCTTCCCACCTGTC (NdeI) |
| pSD5 pknB RP | CAACCGCTAACGCGTGCGAAGCTACTGGCCGAAC (MluI) |
| pknBK40M FP | CCGCGACGTTGCGGTCATGGTGCTGGCGCGCTGATG |
| pknBK40M RP | GATCAGCGCGCAGCACCATGACCGCAACGTCGCGG |
| pVV16 tuf FP | CAAGTCCAGGAGGACCATATGGTGGCGAAGGCGAAG (NdeI) |
| pVV16 tuf RP1 | TGCGTCTGGGTGGTGGTGCCGGTAGACCTTCTTGATGAT (His3) |
| pVV16 tuf RP2 | GTTGTTGTTAAGCTTGTGGTGGTGGTGGTGGTGCCGGTA (His6, HindIII) |
Restriction sites/stop codons/mutated sequences are underlined. Restriction enzymes and/or tags are given in parentheses after sequences.
Table 2.
Description of the plasmid constructs used
| Plasmid construct | Description | Reference or source |
|---|---|---|
| pProEx-HTc | E. coli expression vector containing an N-terminal His6 tag | Invitrogen |
| pProEx-HTc-pknBc | Expression of His6-PknB1-331 (cytosolic domain) | 20 |
| pProEx-HTc-pknAc | Expression of His6-PknA1-337 (cytosolic domain) | This study |
| pProEx-HTc-PstPc | Expression of His6-PstP1-300 (cytosolic domain) | 20 |
| pProEx-HTc-pknF | Expression of His6-PknF1-476 (full-length protein) | This study |
| pProEx-HTc-pknJc | Expression of His6-PknJ1-320 (cytosolic domain) | This study |
| pProEx-HTc-tuf | Expression of His6-MtbEf-Tu | This study |
| pProEx-HTc-tufT118A | pProEx-HTc-MtbEf-Tu with Thr118 mutated to Ala | This study |
| pGEX-5X-3 | E. coli expression vector containing an N-terminal glutathione S-transferase tag | GE Healthcare |
| pGEX-5X-3-pknBc | Expression of GST-PknB1-331 (cytosolic domain) | This study |
| pGEX-5X-3-pknE | Expression of GST-PknE1-566 (full-length protein) | This study |
| pGEX-5X-3-pknDc | Expression of GST-PknD1-378 (cytosolic domain) | This study |
| pGEX-5X-3-pknG | Expression of GST-PknG1-750 (full-length protein) | This study |
| pGEX-5X-3-pknHc | Expression of GST-PknH1-403 (cytosolic domain) | This study |
| pGEX-5X-3-tsf | Expression of GST-Ef-Ts | This study |
| pETDuet1 | E. coli dual-expression vector containing an N-terminal His6 tag in MCS1 and a C-terminal S tag in MCS2 | Novagen |
| pETDuet1-MtbEf-Tu/MBP-PknB | Expression of His6-MtbEf-Tu in MCS1 with MBP-tagged PknB in MCS2 | 25; this study |
| pETDuet1-MtbEf-Tu/MBP-PknBK40M | Expression of His6-MtbEf-Tu in MCS1 with MBP-tagged PknBK40M in MCS2 | This study |
| pSD5 | Mycobacterial expression vector with Kanr | 46 |
| pSD5-pknB | Expression of PknB1-626 (full-length protein) | This study |
| pSD5-pknBK40M | Expression of PknB kinase-dead mutant with Lys40 mutated to Met | This study |
| pVV16 | Mycobacterial expression vector with Kanr and a C-terminal His6 tag | TVTRMC,a Colorado State University |
| pVV16-tuf | Expression of His12-MtbEf-Tu (His6 of pVV16 + His6 added in the sequential pVV16 tuf reverse primers) | This study |
TVTRMC, Tuberculosis Vaccine Testing and Research Materials contract.
Growth of M. smegmatis and labeling of proteins with [35S]methionine.
Electrocompetent cells of M. smegmatis mc2155 were electroporated with 100 ng of plasmid DNA (pSD5-PknB or pSD5-PknBK40M) as described previously (34). For protein labeling, 5 ml of medium was inoculated with 1% of the primary culture, which was grown until the optical density at 600 nm (OD600) reached 1.0. Cells were harvested, resuspended in 1 ml of fresh medium containing 15 μCi of [35S]methionine (BRIT, Hyderabad, India), and incubated for 2 h. Cultures were harvested, sonicated, and centrifuged to remove intact cells. The supernatant was precipitated with 10% trichloroacetic acid (ice-cold), and the precipitate was then washed with cold acetone. The samples were dried and resuspended in 50 mM Tris-Cl buffer (pH 8.0). The total protein content of the lysate was estimated by a Bradford assay, and radioactive counts were measured using a liquid scintillation counter (Hidex). The relative loss in counts for the sample overexpressing PknB was calculated by considering the counts obtained for the sample overexpressing PknBK40M as 100%.
To assess the effect of kirromycin on the growth of M. smegmatis, cultures of transformed cells (5 ml) were grown in duplicate in the presence or absence of kirromycin (final concentration, 10 μM). The concentration of kirromycin was standardized (from 1 μM to 10 μM) such that it does not completely inhibit mycobacterial growth. The absorbance of cultures at 600 nm was taken at intervals for 48 h, and the generation time was calculated.
Purification of MtbEf-Tu from M. smegmatis and phosphoprotein enrichment.
His12-MtbEf-Tu was purified from M. smegmatis that had been electroporated with pVV16-MtbEf-Tu. M. smegmatis cells (4 liters) were cultured in 7H9 medium and were grown for 30 h, to log phase (OD600, 1.0); then they were harvested and lysed by sonication in sonication buffer (1× phosphate-buffered saline [PBS], 1 mM phenylmethylsulfonyl fluoride [PMSF], and protease inhibitor cocktail [Roche]). The lysate was centrifuged at 14,000 × g and 4°C for 30 min, and the resulting supernatant containing His12-MtbEf-Tu was incubated with Ni2+-nitrilotriacetic acid (NTA) resin (Qiagen). The resin was washed extensively with a buffer containing 1× PBS, 1 mM PMSF, 20 mM imidazole, and 10% glycerol, and proteins were eluted in the elution buffer (1× PBS, 1 mM PMSF, 500 mM imidazole, 10% glycerol). The purified protein was desalted using a PD-10 desalting column (GE Healthcare). Four hundred micrograms of purified MtbEf-Tu was further phosphoenriched by using a PhosphoProtein purification kit (Qiagen) according to the manufacturer's instructions. The enriched protein was concentrated and utilized for immunoblotting.
Protein purification and biochemical assays.
Proteins were expressed and purified from E. coli as described previously (8, 20, 25). In vitro phosphorylation of 5 μg MtbEf-Tu/MtbEf-TuT118A/Ef-Ts with PknB and/or other STPKs (0.5 to 3 μg) was carried out in kinase buffer [20 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 7.2), 5 mM MnCl2, and 5 mM MgCl2 for all the kinases and 25 mM Tris-Cl (pH 7), 1 mM dithiothreitol (DTT), 5 mM MgCl2, and 1 mM EDTA for PknE] containing 2μCi [γ-32P]ATP (BRIT, Hyderabad, India), followed by incubation at 25°C for 20 min. Reactions were terminated with 5× sodium dodecyl sulfate (SDS) sample buffer, followed by boiling at 100°C for 5 min. Proteins were separated by 12% SDS-polyacrylamide gel electrophoresis (PAGE) and were analyzed with a phosphorimager (FLA-2000; Fujifilm). The extent of loss of the phosphorylation signal in MtbEf-TuT118A was analyzed with Image Gauge software (Fujifilm), and the relative loss was calculated by considering the phosphorylation on native MtbEf-Tu as 100%.
To visualize the phosphorylation signal on His6 tag-cleaved MtbEf-Tu, the recombinant tag was removed by the addition of 2 μg tobacco etch virus (TEV) protease in cleavage buffer (Tris-Cl [pH 8.5], 5 mM EDTA, 300 mM NaCl, and 1 mM DTT) after the kinase reaction, followed by 2 h of incubation at 20°C. For phosphoamino acid analysis, MtbEf-Tu was phosphorylated by PknBc, cleaved with TEV protease, separated by SDS-PAGE, and electroblotted onto an Immobilon PVDF membrane (Millipore). Phosphoamino acid analysis by 2-dimensional thin-layer electrophoresis (2D-TLE) was performed as described previously (13, 20).
Immunoblotting with anti-phosphothreonine and anti-MtbEf-Tu antibodies.
To assess the phosphorylation status of purified MtbEf-Tu coexpressed in E. coli with either PknB or PknBK40M (phosphorylated MtbEf-Tu [MtbEf-Tuphos] or unphosphorylated MtbEf-Tu [MtbEf-Tuunphos], respectively) by Western blotting with anti-phosphothreonine (anti-pThr) antibodies, the samples were resolved by SDS-PAGE along with positive (purified PknBc) and negative (PstPc) controls and were transferred to a nitrocellulose membrane (Bio-Rad). Protocols for Western blotting similar to those described previously (20) were followed. The blots were developed using the SuperSignal West Pico chemiluminescent substrate kit (Pierce Protein Research Products) according to the manufacturer's instructions. The phosphorylation status of phosphoenriched MtbEf-Tu purified from M. smegmatis was also assessed in the same way. Polyclonal antibodies against MtbEf-Tu were generated in mice. To confirm that the protein bands were MtbEf-Tu, Western blotting was also performed using polyclonal antibodies generated against MtbEf-Tu. The identity of MtbEf-Tu was also confirmed by mass spectrometric analysis (TCGA, New Delhi, India).
Analysis of MtbEf-Tu by 2-dimensional PAGE and identification of the phosphorylation site(s).
Purified MtbEf-Tuphos and MtbEf-Tuunphos (30 μg each) were desalted by acetone precipitation. The proteins were separated by 2D-PAGE as described previously (25), except that 7-cm-long immobilized pH gradient (IPG) strips were used. After MtbEf-Tu was run in the second dimension, the gel was analyzed by immunoblotting using anti-pThr antibodies and anti-MtbEf-Tu antibodies.
For the identification of phosphorylation sites, MtbEf-Tuphos (3 μg) was run on a 12% SDS-PAGE gel, which was first stained with Coomassie brilliant blue and then destained, and the band corresponding to MtbEf-Tuphos was excised from the gel and was washed with MilliQ water. The samples were processed for the identification of phosphorylation sites by using a Thermo Finnigan LTQ electrospray instrument (Proteomics Core Facility, Children's Hospital, Boston, MA). The detailed protocol for sample processing has been described previously (41).
Interaction of MtbEf-Tu with BODIPY FL-GTP.
Unphosphorylated and phosphorylated MtbEf-Tu proteins were used in interaction studies with dipyrromethene boron difluoride (BODIPY) FL-GTP (Molecular Probes, Invitrogen) by a fluorimetric method. The reaction was initiated by the addition of MtbEf-Tu (1 to 2 μM) to fluorescence assay buffer F (20 mM Tris-Cl [pH 7.5], 150 mM NaCl, 1 mM DTT, 10 mM MgCl2, and 10% glycerol) containing 10 nM BODIPY FL-GTP. The emission spectra were recorded from 490 nm to 600 nm after excitation at 488 nm (Fluoromax-3 spectrofluorimeter; Jobin Yvon Horiba) with an integration time of 1 s. In a separate experiment, kirromycin (10 μM) was added to the reaction mixture, and spectra were recorded in order to investigate the role of kirromycin in the interaction of MtbEf-Tu with GTP. To investigate the role of Thr118, increasing concentrations (1 μM to 4 μM) of both MtbEf-Tu and MtbEf-TuT118A were used in the presence of a fixed concentration of BODIPY FL-GTP (200 nM). Spectra of buffer F containing BODIPY FL-GTP served as a control.
Dissociation of GDP from Ef-Tu.
Five-hundred-nanomolar concentrations of MtbEf-Tu (MtbEf-Tuunphos and MtbEf-Tuphos) were incubated with 100 nM BODIPY FL-GDP (Molecular Probes, Invitrogen) in buffer F for 30 min at 25°C, and the mixtures were passed through a Sephadex G-25 column to remove unbound GDP. Fluorescence stopped-flow measurements (SFA-20 Rapid Kinetics Accessories; Hi-Tech Scientific) were performed to assess the spontaneous dissociation of BODIPY FL-GDP from MtbEf-Tu by titrating the MtbEf-Tu–BODIPY FL-GDP complex with an excess of unlabeled GDP (500 nM) in buffer F at 25°C. BODIPY FL-GDP was excited at 488 nm, and emission was measured at 512 nm, with 5-nm-wide slits for both excitation and emission. Experiments were performed by rapidly mixing equal volumes (150 μl each) of the reactants and monitoring the fluorescence change with time. Data were evaluated by fitting to an exponential function using SigmaPlot software. The effect of Ef-Ts on the dissociation of BODIPY FL-GDP bound to MtbEf-Tu was assessed similarly by adding 200 nM GST-Ef-Ts to the reaction mixture. For steady-state measurements, the association of BODIPY FL-GTP with MtbEf-Tu was measured by mixing increasing concentrations (1 × 103 to 10 × 103 nM) of MtbEf-Tuunphos or MtbEf-Tuphos with 10 nM BODIPY FL-GTP in buffer F at 25°C. The saturation curve was plotted at a peak position of 515 nm and was fitted for single-ligand binding by using SigmaPlot to calculate the dissociation constant (Kd).
RESULTS
Effect of PknB expression on the protein synthesis and phosphorylation of MtbEf-Tu.
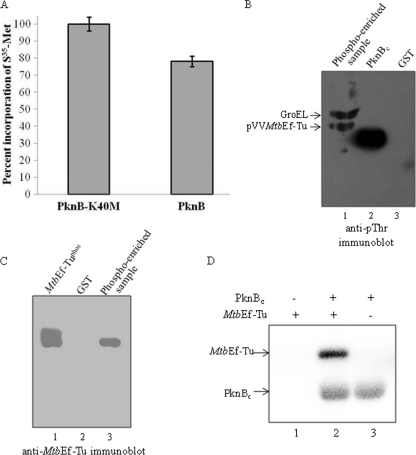
In several organisms, elongation factor Tu (Ef-Tu) has been shown to be regulated by phosphorylation (6, 11, 27, 28, 30, 44, 50). In many of these organisms where Ef-Tu was reported to be a substrate of STPKs, a homolog of PknB was also present (see Fig. S1 in the supplemental material). Ef-Tu is a well conserved protein among bacteria and has been shown to be essential for the survival of M. tuberculosis (42). However, the regulation of MtbEf-Tu has not been studied. Therefore, the effect of PknB-mediated phosphorylation on MtbEf-Tu was assessed in this study. Overexpression of PknB has been shown to slow the growth and alter the cell morphology of M. smegmatis (23); however, the factors leading to this growth inhibition are not well understood. Protein synthesis is an essential process required for the normal growth of all living organisms. The role of PknB-mediated phosphorylation in protein synthesis in the cell was determined by expressing PknB or its inactive mutant (PknBK40M) in M. smegmatis. The proteins in exponentially growing M. smegmatis cells were labeled with [35S]methionine for 2 h in order to evaluate protein synthesis. Overexpression of PknB resulted in approximately 25% less incorporation of [35S]methionine into proteins than that for the strain expressing PknBK40M (Fig. 1A). These results suggest that PknB can regulate cellular protein synthesis in M. smegmatis. Subsequently, we investigated the phosphorylation of MtbEf-Tu in M. smegmatis. To ascertain that MtbEf-Tu is phosphorylated in M. smegmatis, His12-MtbEf-Tu was expressed and purified by Ni2+-NTA affinity purification procedures using log-phase cells, and the purified protein was phosphoenriched. Since Ef-Tu is known to be phosphorylated on a conserved Thr residue in E. coli and other species (28, 44), the phosphoenriched protein was analyzed by immunoblotting using anti-pThr antibodies. We observed that MtbEf-Tu was indeed phosphorylated on Thr in M. smegmatis (Fig. 1B). The enriched sample also showed an additional protein band around 60 kDa as a copurified contaminant. Mass spectrometric analysis confirmed that the lower band was MtbEf-Tu, while the contaminant was identified as GroEL (Fig. 1B). Mycobacterial GroEL has been reported to be a substrate of STPKs (15), and in this study, GroEL was also observed to be phosphorylated. The identity of MtbEf-Tu was also confirmed by Western blotting with anti-MtbEf-Tu antibodies (Fig. 1C). Since PknB is expressed in log-phase cultures of mycobacteria (23), MtbEf-Tu was expected to be phosphorylated by PknB. In fact, MtbEf-Tu was phosphorylated by PknBc (the cytosolic domain of PknB [aa 1 to 331]) during an in vitro kinase assay (Fig. 1D). However, based on these experiments, we cannot rule out the possibility that MtbEf-Tu is phosphorylated by other STPKs. In vitro phosphorylation assays showed that MtbEf-Tu was also phosphorylated by several protein kinases of M. tuberculosis, including PknAc, PknDc, PknE, PknF, PknHc, and PknJc (see Fig. S2 in the supplemental material). The authenticity of phosphorylation on intact MtbEf-Tu but not on the recombinant His6 tag was confirmed by cleavage of the tag by TEV protease (data not shown).
Fig. 1.
Role of PknB in the regulation of protein synthesis and the phosphorylation of MtbEf-Tu in M. smegmatis. (A) Graphical representation of [35S]Met incorporation into proteins of M. smegmatis cells overexpressing PknB or PknBK40M. The PknB-overexpressing strain showed ∼25% less incorporation of [35S]Met than the PknBK40M-overexpressing strain when equal concentrations of the lysates were taken. The experiment was repeated four times; error bars represent standard deviations of four individual readings. (B) His12-MtbEf-Tu was purified from M. smegmatis and was phosphoenriched. The enriched protein was assessed for phosphorylation by immunoblotting with anti-pThr antibodies. Lane 1, phosphoenriched sample; lane 2, PknBc (positive control); lane 3, GST (negative control). (C) The phosphoenriched sample of MtbEf-Tu purified from M. smegmatis was also probed with anti-MtbEf-Tu antibodies. Lane 1, MtbEf-Tuphos (positive control); lane 2, GST (negative control); lane 3, phosphoenriched sample. (D) Autoradiogram showing the in vitro phosphorylation of MtbEf-Tu (5 μg) by PknBc (0.5 μg) in kinase buffer containing 2 μCi [γ-32P]ATP. Samples were separated by 12% SDS-PAGE and were analyzed with a phosphorimager.
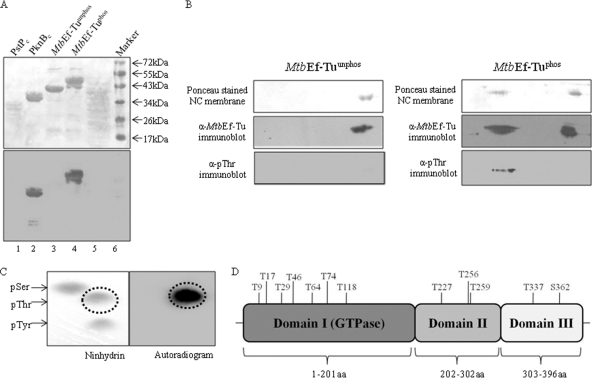
Phosphorylation of MtbEf-Tu was further validated by expressing MtbEf-Tu together with PknB or PknBK40M using a dual-expression system in the surrogate host E. coli. MtbEf-Tu was purified and assessed for phosphorylation by immunoblotting with anti-pThr antibodies. As expected, MtbEf-Tu was phosphorylated (MtbEf-Tuphos) when PknB was coexpressed, whereas no phosphorylation was observed in the presence of PknBK40M (MtbEf-Tuunphos) (Fig. 2A). Mass spectrometry and immunoblotting of purified MtbEf-Tu with mouse anti-MtbEf-Tu antibodies confirmed the proteins to be MtbEf-Tu (data not shown). The phosphorylated species of MtbEf-Tuphos and MtbEf-Tuunphos were separated by 2-dimensional PAGE on the basis of their isoelectric points. The 2D gels were subjected to Western blotting with anti-pThr and anti-MtbEf-Tu antibodies. The results showed that several isoforms were present in MtbEf-Tuphos (Fig. 2B, right), while no phosphorylated species were visible in MtbEf-Tuunphos (Fig. 2B, left). The presence of more than one species indicated that heterogeneous species of MtbEf-Tu were present, possibly due to differential phosphorylation of MtbEf-Tu. The blot was stripped and redeveloped using anti-MtbEf-Tu antibodies to confirm that all the spots corresponded to MtbEf-Tu (Fig. 2B).
Fig. 2.
Phosphorylation of MtbEf-Tu and identification of phosphorylation sites. (A) MtbEf-Tu was coexpressed with either PknB or PknBK40M in E. coli, and the phosphorylation status of purified MtbEf-Tu was analyzed by Western blotting using anti-pThr antibodies. (Top) Ponceau-stained nitrocellulose membrane; (bottom) immunoblot. Lane 1, PstPc (negative control); lane 2, PknBc (positive control); lane 3, MtbEf-Tu coexpressed with PknBK40M (MtbEf-Tuunphos)); lane 4, MtbEf-Tu coexpressed with PknB (MtbEf-Tuphos); lane 6, molecular size markers. (B) Phosphorylated species in MtbEf-Tuunphos (left) and MtbEf-Tuphos (right) were separated by bidirectional PAGE, and the proteins were transferred to a nitrocellulose membrane and were blotted with anti-pThr (α-pThr) and anti-MtbEf-Tu antibodies. Additional spots were probed for MtbEf-Tuphos, while no such spots were observed with MtbEf-Tuunphos. (C) Phosphoamino acid analysis of MtbEf-Tu phosphorylated by PknBc was performed by 2D-TLE and autoradiography. Mainly a Thr residue(s) was observed to be phosphorylated; no spot corresponding to pSer or pTyr was observed. (D) Diagrammatic representation of residues of MtbEf-Tuphos phosphorylated by PknB during coexpression in E. coli. A probable domain-wise distribution of phosphorylation sites is shown.
Identification of phosphorylated amino acids of MtbEf-Tu.
The phosphorylated amino acids of MtbEf-Tu were identified by phosphorylating MtbEf-Tu with PknBc and then analyzing the product by 2-dimensional thin-layer electrophoresis (2D-TLE). Phosphoamino acid analysis indicated that mainly Thr residues were phosphorylated (Fig. 2C), while no phosphorylation was observed on Ser or Tyr.
The sites of phosphorylation for MtbEf-Tuphos were identified by mass spectrometric analysis. Several Thr residues within domain I were phosphorylated (Thr9, Thr17, Thr29, Thr46, Thr64, Thr74, and Thr118). Residues Thr227, Thr256, and Thr259 of domain II and residues Thr337 and Ser362 of domain III were also phosphorylated (Fig. 2D). Since phosphoamino acid analysis by 2D-TLE is a less sensitive technique than mass spectrometry, phosphorylation on a Ser residue was not observed by 2D-TLE.
Effect of phosphorylation on the interaction of MtbEf-Tu with GTP.
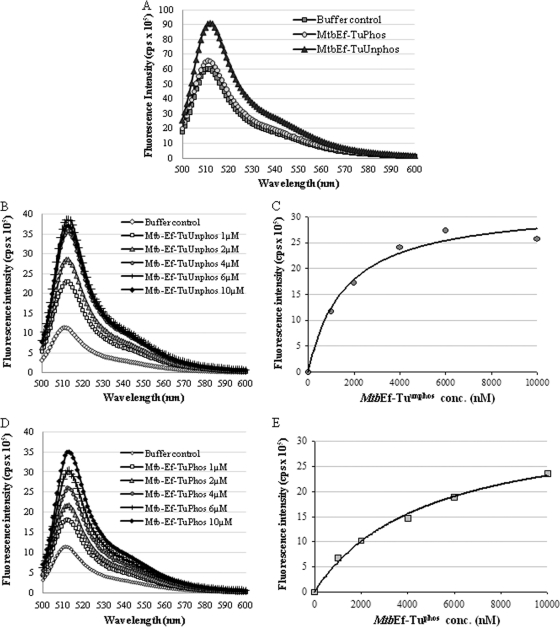
Since binding of GTP to Ef-Tu is critical for the activity of the enzyme, we tried to determine if the posttranslational modification identified had any effect on Ef-Tu–GTP interaction. The interactions of MtbEf-Tuphos and MtbEf-Tuunphos with GTP were measured by fluorimetric assays using the fluorescently tagged nucleotide BODIPY FL-GTP as described previously (29). BODIPY FL-GTP showed the characteristic maxima at 512 nm, and an increase in fluorescence intensity was associated with an increased interaction of BODIPY FL-GTP with MtbEf-Tu. The level of interaction of MtbEf-Tuunphos with GTP was higher than that of MtbEf-Tuphos (Fig. 3A), suggesting a negative impact of phosphorylation on GTP binding by MtbEf-Tu. The equilibrium constant of GTP binding to MtbEf-Tu was calculated through steady-state measurements using BODIPY FL-GTP. The Kd value obtained for MtbEf-Tuunphos was 1.6 × 10−6 M (standard error [SE], ±0.386 × 10−6), significantly lower than the Kd value for MtbEf-Tuphos, which was estimated to be 4.8 × 10−6 M (SE, ±0.698 × 10−6) (Fig. 3B, C, D, and E; Table 3). These results suggest that phosphorylation reduced the GTP-binding ability of MtbEf-Tu 3-fold.
Fig. 3.
Interaction of MtbEf-Tu with BODIPY FL-GTP. (A) The interactions of MtbEf-Tuphos and MtbEf-Tuunphos (2 μM each) with the fluorophore BODIPY FL-GTP (10 nM) were studied by recording the emission spectrum from 490 nm to 600 nm after excitation at 488 nm. A buffer containing BODIPY FL-GTP served as a control. (B) Spectrum recorded from 490 to 600 nm by adding increasing concentrations of MtbEf-Tuunphos (1 to 10 μM) to a buffer containing BODIPY FL-GTP (10 nM). (C) Saturation curve for the MtbEf-Tuunphos–BODIPY FL-GTP interaction. (D) Spectrum recorded from 490 to 600 nm by adding increasing concentrations of MtbEf-Tuphos (1 to 10 μM) to a buffer containing BODIPY FL-GTP (10 nM). (E) Saturation curve for the MtbEf-Tuphos–BODIPY FL-GTP interaction.
Table 3.
Comparison of MtbEf-Tu and E. coli Ef-Tu
| Enzymatic property | E. coli Ef-Tu |
M. tuberculosis Ef-Tu |
|
|---|---|---|---|
| MtbEf-Tuphos | MtbEf-Tuunphos | ||
| Interaction with GTP (Kd) | 5 × 105 M−1 s−1 (±1 × 105) (time dependent)a | 4.8 × 10−6 M (±0.698 × 10−6) (steady state) | 1.6 × 10−6 M (±0.386 × 10−6) (steady state) |
| Interaction with GDP (rate of dissociation) | 2 × 10−3 s−1 (±1 × 10−3)a | 5.5 × 10−3 s−1 (±1.76 × 10−4) | 2.07 × 10−3 s−1 (±0.86 × 10−4) |
| Effect of Ef-Ts | |||
| Dissociation of GDP | 125 s−1 (±25)a | 0.64 s−1 (±14.9 × 10−3) | 0.53 s−1 (±0.707 × 10−3) |
| Phosphorylation of Ef-Tu | Enhances Ef-Tu phosphorylationb | No effect | |
| Effect of kirromycin | |||
| Nucleotide binding | Stalls Ef-Tu in activated state; does not bind to phosphorylated Ef-Tub | Does not interact | Enhances interaction |
| Phosphorylation of Ef-Tu | Inhibits Ef-Tu phosphorylationb | No effect | |
| Phosphorylated residue(s) | Thr382 (domain III)c | Thr9, Thr17, Thr29, Thr46, Thr64, Thr74, Thr118 (domain I), Thr227, Thr256, Thr259 (domain II), Thr337, Ser362 (domain III) | |
| Effect of phosphorylation | Facilitates its release from the ribosome and prevents ternary complex formationb | Decreases affinity for guanine nucleotides (GTP and GDP) | |
Importance of MtbEf-Tu Thr118 for interaction with GTP.
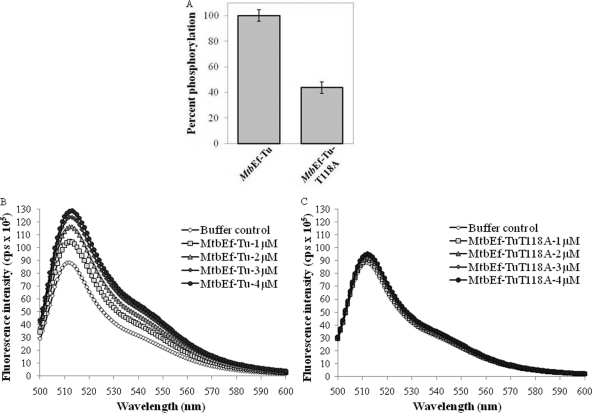
Within Ef-Tu, domain I retains the GTPase activity. Thus, the sites of phosphorylation in domain I may affect its interaction with the nucleotide. In a study of the M. tuberculosis phosphoproteome, MtbEf-Tu was found to be phosphorylated on Thr118 (37). To investigate the specific role of Thr118, MtbEf-TuT118A was generated. A phosphorylation assay with PknBc showed a ∼50% loss of the phosphorylation signal of the mutant protein (Fig. 4A). This partial—not complete—loss of phosphorylation also confirmed earlier results showing that in addition to Thr118, other residues are also phosphorylated in MtbEf-Tu. We next assessed the interaction of GTP with MtbEf-Tu and MtbEf-TuT118A (Fig. 4B and C). MtbEf-TuT118A displayed a significant loss of interaction with GTP, suggesting that Thr118 is important for the interaction with GTP, in addition to its role in phosphorylation-mediated regulation.
Fig. 4.
Phosphorylation and interaction of MtbEf-TuT118A with BODIPY FL-GTP. (A) Bar graph showing partial loss of the phosphorylation signal on the MtbEf-TuT118A mutant compared to the native protein when phosphorylated by PknBc. (B and C) Interaction of increasing concentrations of MtbEf-Tu (B) or MtbEf-TuT118A (C) (1 μM to 4 μM) with a constant concentration of BODIPY FL-GTP (200 nM).
Effect of Ef-Ts on the interaction of guanine nucleotides with MtbEf-Tu.
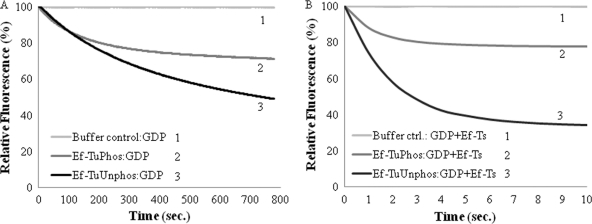
Another elongation factor that works in close association with Ef-Tu is Ef-Ts, which helps Ef-Tu to recycle from the GDP-bound inactive state to the GTP-bound active state. The dissociation of BODIPY FL-GDP from MtbEf-Tuunphos and MtbEf-Tuphos was studied in the presence or absence of Ef-Ts. The complex of MtbEf-Tuunphos or MtbEf-Tuphos with BODIPY FL-GDP was titrated an with excess of unlabeled GDP (500 nM), and the dissociation of labeled GDP was studied in the absence of Ef-Ts (Fig. 5A). The spontaneous dissociation of BODIPY FL-GDP was very slow. However, the rate of dissociation of BODIPY FL-GDP from MtbEf-Tuphos (5.5 × 10−3 s−1 ± 1.76 × 10−4) was ∼2.5-fold higher than the dissociation rate obtained for MtbEf-Tuunphos (2.07 × 10−3 s−1 ± 0.86 × 10−4), suggesting reduced affinity for the guanine nucleotide after phosphorylation (Table 4). As expected, Ef-Ts catalyzed the dissociation of BODIPY FL-GDP at much higher rates (Fig. 5B), and the dissociation of BODIPY FL-GDP was achieved in few seconds. However, the rates of GDP dissociation from MtbEf-Tuunphos and MtbEf-Tuphos, 0.53 s−1 (±0.71 × 10−3) and 0.64 s−1 (±14.9 × 10−3), respectively, were very similar. Thus, Ef-Ts mediated the dissociation of GDP from Ef-Tu regardless of its phosphorylation status. Additionally, Ef-Ts was not phosphorylated by PknBc, and its presence did not affect the levels of phosphorylation of MtbEf-Tu by PknBc (see Fig. S3 in the supplemental material).
Fig. 5.
Dissociation of BODIPY FL-GDP from MtbEf-Tu and effect of Ef-Ts. Spontaneous dissociation of BODIPY FL-GDP from MtbEf-Tu (1 μM) was measured by titrating the MtbEf-Tu–BODIPY FL-GDP complex with an excess of unlabeled GDP (500 nM) at 25°C. BODIPY FL-GDP was excited at 488 nm, and emission was measured at 512 nm. (A) The dissociation of BODIPY FL-GDP from MtbEf-Tuphos (curve 2), MtbEf-Tuunphos (curve 3), and buffer F, used as a control (curve 1), was measured up to 800 s. (B) The dissociation of BODIPY FL-GDP from MtbEf-Tuphos (curve 2) and MtbEf-Tuunphos (curve 3) was measured in the presence of Ef-Ts (200 nM).
Table 4.
Effect of Ef-Ts on interaction of GDP with MtbEf-Tu
| Ef-Ts | GDP off-rate |
Difference | |
|---|---|---|---|
| MtbEf-Tuunphos | MtbEf-Tuphos | ||
| − | 2.07 × 10−3 s−1 (±0.86 × 10−4) | 5.5 × 10−3 s−1 (±1.76 × 10−4) | ∼2.5-fold faster |
| + | 0.53 s−1 (±0.71 × 10−3) | 0.64 s−1 (±14.9 × 10−3) | Negligible |
Role of kirromycin in PknB-mediated phosphorylation of MtbEf-Tu.
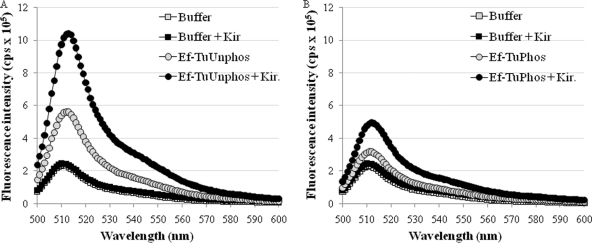
Kirromycin, a well-established inhibitor of translation, specifically arrests Ef-Tu in a nucleotide-bound form, enhances its GTPase activity, and prevents its rearrangement for another round of protein elongation (53, 54). The interactions of BODIPY FL-GTP with MtbEf-Tuunphos and MtbEf-Tuphos were examined in the presence of kirromycin. In agreement with its effect on E. coli Ef-Tu, kirromycin maintained the nucleotide-bound state of MtbEf-Tu, as indicated by an increase in fluorescence counts on the addition of kirromycin (Fig. 6). The effect of kirromycin on unphosphorylated MtbEf-Tu (Fig. 6A) was more prominent than that on the phosphorylated protein (Fig. 6B). Further, kirromycin had no effect on the phosphorylation status of MtbEf-Tu (see Fig. S4 in the supplemental material). Thus, unphosphorylated MtbEf-Tu appears to be more prone to inhibition by kirromycin than is the phosphorylated protein. These results suggest that phosphorylated MtbEf-Tu is not effectively inhibited by kirromycin.
Fig. 6.
Effect of kirromycin on the interaction of MtbEf-Tu with GTP. The interactions of MtbEf-Tuunphos (1 μM) (A) and MtbEf-Tuphos (1 μM) (B) with BODIPY FL-GTP (10 nM) were studied in the presence of kirromycin (10 μM) by recording the emission spectrum from 500 to 600 nm.
Kirromycin, an effective inhibitor of protein synthesis, inhibited the growth of M. smegmatis (data not shown). To understand the inhibition of phosphorylated Ef-Tu by kirromycin, an M. smegmatis strain overexpressing PknB was grown in the presence of kirromycin, and growth was measured. Cells transformed with a vector carrying PknBK40M served as a control. Addition of kirromycin to the cells expressing PknB caused the doubling time to increase by 2 h, whereas this increase was ∼5 h when PknBK40M was expressed in M. smegmatis and the cells were grown in the presence of kirromycin (Table 5). The M. smegmatis strain overexpressing PknB is likely to have more phosphorylated MtbEf-Tu than the strain expressing PknBK40M, whereas the doubling time was significantly less in the strain expressing PknB on the addition of kirromycin. These results suggest that the phosphorylation of Ef-Tu reduced its sensitivity to kirromycin, as was also observed by fluorimetric analysis.
Table 5.
Effect of kirromycin on the generation time of M. smegmatis expressing PknB or PknBK40M
| Gene overexpressed | Generation time (h): |
Difference in generation time (h) | |
|---|---|---|---|
| In the absence of kirromycin | In the presence of kirromycin (10 μM) | ||
| PknB | 10.94 | 12.94 | 2 |
| PknBK40M | 8.5 | 13.8 | 5.3 |
DISCUSSION
The protein translation process comprises three main steps: initiation, peptide elongation, and termination. The elongation phase is the most dynamic process among the three steps and involves several elongation factors together with many ribosomal components. Ef-Tu is a conserved protein that is involved in the elongation phase of translation and is known to be regulated by posttranslational modifications. Phosphorylation of Ef-Tu has been suggested to facilitate its release from the ribosome in E. coli (3), while for L. monocytogenes, Ef-Tu phosphorylation is implicated in acclimation to the stress conditions encountered during the course of infection (6). In a recent report on the M. tuberculosis phosphoproteome, MtbEf-Tu was identified as one of the phosphorylated proteins (37). However, the effect of phosphorylation on Ef-Tu activity and its role in cellular protein synthesis or growth are not known.
PknB is an essential kinase that phosphorylates a large number of substrates regulating various pathways. Ef-Tu has been found to be phosphorylated in several bacteria that contain homologs of PknB (see Fig. S1 in the supplemental material). In this study, we show that MtbEf-Tu is phosphorylated by several kinases, including PknB (Fig. 1D and 2; see also Fig. S2 in the supplemental material). Overexpression of PknB has been reported to reduce the growth and alter the cell morphology of M. smegmatis and Mycobacterium bovis (23). We found that PknB overexpression led to reduced protein synthesis, as observed by reduced incorporation of radiolabeled methionine (Fig. 1A). Therefore, an effort was made to understand the effect of PknB-mediated phosphorylation on MtbEf-Tu activity. Purified MtbEf-Tu was observed to be phosphorylated when incubated in the presence of PknB (Fig. 1D). Phosphorylation of MtbEf-Tu was also confirmed by coexpression of MtbEf-Tu with PknB or PknBK40M in E. coli and probing of Ef-Tu with an anti-pThr antibody (Fig. 2A). Phosphoamino acid analysis showed that MtbEf-Tu was phosphorylated mainly on Thr residues (Fig. 2C). Most of the substrates of protein kinases in M. tuberculosis are known to be phosphorylated mainly on Thr residues (37). Differential site phosphorylation of substrates by Ser/Thr kinases under different conditions have been reported in both eukaryotes and prokaryotes (21, 44, 47). Mass spectrometric analysis of phosphorylated MtbEf-Tu showed that 11 Thr residues and 1 Ser residue were phosphorylated (Fig. 2D). Due to phosphorylation on multiple residues, several species of MtbEf-Tu were observed by 2D-PAGE analysis of MtbEf-Tuphos purified from E. coli and coexpressed with PknB (Fig. 2B).
The structure of MtbEf-Tu was predicted by homology modeling using Swiss-Model (7, 45), by taking E. coli Ef-Tu as the template (48) (Protein Data Bank [PDB] identification code 1OB2-A). The homology modeling broadly revealed the tridomain structure of MtbEf-Tu, oriented in a manner similar to that of its E. coli homolog (see Fig. S5 in the supplemental material). Domain I of Ef-Tu is known to interact with GTP (26), and this domain had the maximum phosphorylation sites (Fig. 2D). Binding of GTP is an essential step for translation elongation; therefore, the impact of phosphorylation on GTP binding was studied. The results showed that phosphorylation reduced the affinity of MtbEf-Tu for GTP (Fig. 3). The reduced affinity of phosphorylated MtbEf-Tu for GTP is possibly due to the negative charge on the protein gained by phosphorylation or the structural perturbations due to phosphorylation. The reduced affinity was reflected in the lower dissociation rate constant (Kd) of unphosphorylated MtbEf-Tu than of the phosphorylated protein for GTP. Since Ef-Tu is an unstable protein with low intrinsic GTPase activity (36), only freshly purified protein was used for all the studies. Also, MtbEf-Tu was kept essentially free of nucleotides during purification and storage and was also treated with EDTA to remove intrinsically bound nucleotides, as shown previously (19).
Since mass spectrometry data showed that Thr118 is phosphorylated, which was also reported in a recent study on the phosphoproteome of M. tuberculosis (37), we tried to understand the role of this residue. MtbEf-TuT118A showed reduced phosphorylation and was less competent in interaction with GTP (Fig. 4). The significant loss in the phosphorylation of the mutant indicated that Thr118 is a major site for phosphorylation. The experiments on GTP binding also suggested that in addition to its role in phosphorylation, Thr118 is also required for optimal activity of Ef-Tu.
Ef-Ts mediates the exchange of GDP with GTP bound to Ef-Tu, thus accelerating the overall rate of protein translation (1, 24). In E. coli, Ef-Ts is known to interact principally with domains I and III of Ef-Tu and to disrupt the Mg2+ binding site, bringing about the release of bound GDP (24). Subsequently, binding of GTP with Ef-Tu causes the release of Ef-Ts from its complex with Ef-Tu. In order to assess the role of phosphorylation in the overall cycle, MtbEf-Tu was allowed to bind with Ef-Ts prior to phosphorylation by PknBc. No changes were observed in the phosphorylation status of MtbEf-Tu (see Fig. S3 in the supplemental material), indicating that binding of Ef-Ts had no impact on the phosphorylation of MtbEf-Tu. This could possibly be due to the different sites of phosphorylation and Ef-Ts interaction. These results differ from those of the report on E. coli, where enhanced phosphorylation of Ef-Tu was observed in the presence of Ef-Ts (3). A possible reason for this could be the difference in the sites and extent of phosphorylation between M. tuberculosis and E. coli. Also, in the presence of Ef-Ts, the rates of GDP dissociation from MtbEf-Tuunphos and MtbEf-Tuphos were enhanced as much as >250-fold and >100-fold, respectively, over the rates of GDP release without Ef-Ts, thus equalizing the rates of GDP dissociation.
The protein synthesis machinery has been used as a target of antibiotics such as streptomycin for the control of bacterial growth, including that of M. tuberculosis (17). Subsequently, a number of antibiotics were developed that manipulate the translation machinery of the pathogen. Kirromycin is an antibiotic that inhibits protein synthesis by irreversibly blocking the rearrangement of Ef-Tu from the GTP- to the GDP-bound conformation even after GTP hydrolysis, thus stabilizing the GTPase activated state (53–55). The resulting Ef-Tu–GDP complex remains associated with the ribosome, causing interruption of protein synthesis (55). In solution, kirromycin blocks Ef-Tu in the GTPase activated state and increases its GTPase activity (53). In E. coli, kirromycin blocks Ef-Tu phosphorylation, and phosphorylated Ef-Tu does not bind kirromycin (3). In the present study, we observed that phosphorylation of MtbEf-Tu by PknBc is not altered in the presence of kirromycin, even at high concentrations (see Fig. S4 in the supplemental material). Thus, kirromycin binding and MtbEf-Tu phosphorylation appear to be independent events. The amount of MtbEf-Tu bound to GTP was enhanced in the presence of kirromycin, possibly due to stabilization of the complex by kirromycin. Since MtbEf-Tu will be continuously in a GTPase activated state, it may tend to interact more with GTP without changing to the inactive conformation after GTP hydrolysis. A more pronounced effect of kirromycin was observed with MtbEf-Tuunphos than with the phosphorylated protein (Fig. 6), indicating that kirromycin does not seem to bind phosphorylated MtbEf-Tu effectively, as was also shown for E. coli Ef-Tu (3). In view of the fact that phosphorylated Ef-Tu is less susceptible to inhibition by kirromycin, M. smegmatis cells expressing PknB were less sensitive to kirromycin than cells expressing inactive PknB (Table 5).
In summary, we show that MtbEf-Tu is regulated through PknB-mediated phosphorylation and that the phosphorylated protein has less affinity for GTP, which possibly leads to reduced protein synthesis and decreased growth of M. smegmatis. We also show that phosphorylated MtbEf-Tu is poorly targeted by kirromycin. Thus, in the development of inhibitors, the targeting of M. tuberculosis proteins that are phosphorylated requires additional consideration.
Supplementary Material
ACKNOWLEDGMENTS
We thank Zachary Waldon (Proteomics Core Facility, Children's Hospital, Boston, MA) for mass spectrometric analysis. The pVV16 vector was kindly provided by the Tuberculosis Vaccine Testing and Research Materials contract, Colorado State University.
Financial support was provided by the Council of Scientific and Industrial Research (NWP-0038). A. Sajid, G. Arora, and M. Gupta are Senior Research Fellows, and A. Singhal is a Junior Research Fellow, of the Council of Scientific and Industrial Research, India.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 29 July 2011.
REFERENCES
- 1. Abel K., Jurnak F. 1996. A complex profile of protein elongation: translating chemical energy into molecular movement. Structure 4:229–238 [DOI] [PubMed] [Google Scholar]
- 2. Agirrezabala X., Frank J. 2009. Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu. Q. Rev. Biophys. 42:159–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alexander C., et al. 1995. Phosphorylation of elongation factor Tu prevents ternary complex formation. J. Biol. Chem. 270:14541–14547 [DOI] [PubMed] [Google Scholar]
- 4. Ames G. F., Niakido K. 1979. In vivo methylation of prokaryotic elongation factor Tu. J. Biol. Chem. 254:9947–9950 [PubMed] [Google Scholar]
- 5. Arai K., et al. 1980. Primary structure of elongation factor Tu from Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 77:1326–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Archambaud C., Gouin E., Pizarro-Cerda J., Cossart P., Dussurget O. 2005. Translation elongation factor EF-Tu is a target for Stp, a serine-threonine phosphatase involved in virulence of Listeria monocytogenes. Mol. Microbiol. 56:383–396 [DOI] [PubMed] [Google Scholar]
- 7. Arnold K., Bordoli L., Kopp J., Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201 [DOI] [PubMed] [Google Scholar]
- 8. Arora G., et al. 2010. Understanding the role of PknJ in Mycobacterium tuberculosis: biochemical characterization and identification of novel substrate pyruvate kinase A. PLoS One 5:e10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bach H., Wong D., Av-Gay Y. 2009. Mycobacterium tuberculosis PtkA is a novel protein tyrosine kinase whose substrate is PtpA. Biochem. J. 420:155–160 [DOI] [PubMed] [Google Scholar]
- 10. Barik S., Sureka K., Mukherjee P., Basu J., Kundu M. 2010. RseA, the SigE specific anti-sigma factor of Mycobacterium tuberculosis, is inactivated by phosphorylation-dependent ClpC1P2 proteolysis. Mol. Microbiol. 75:592–606 [DOI] [PubMed] [Google Scholar]
- 11. Bendt A. K., et al. 2003. Towards a phosphoproteome map of Corynebacterium glutamicum. Proteomics 3:1637–1646 [DOI] [PubMed] [Google Scholar]
- 12. Bourne H. R., Sanders D. A., McCormick F. 1991. The GTPase superfamily: conserved structure and molecular mechanism. Nature 349:117–127 [DOI] [PubMed] [Google Scholar]
- 13. Boyle W. J., van der Geer P., Hunter T. 1991. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 201:110–149 [DOI] [PubMed] [Google Scholar]
- 14. Bullard J. M., Cai Y. C., Zhang Y., Spremulli L. L. 1999. Effects of domain exchanges between Escherichia coli and mammalian mitochondrial EF-Tu on interactions with guanine nucleotides, aminoacyl-tRNA and ribosomes. Biochim. Biophys. Acta 1446:102–114 [DOI] [PubMed] [Google Scholar]
- 15. Canova M. J., Kremer L., Molle V. 2009. The Mycobacterium tuberculosis GroEL1 chaperone is a substrate of Ser/Thr protein kinases. J. Bacteriol. 191:2876–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cole S. T., et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544 [DOI] [PubMed] [Google Scholar]
- 17. Corper H. J., Cohn M. L. 1947. The remote sustained threshold therapeutic action of streptomycin in tuberculosis. Science 106:446–447 [DOI] [PubMed] [Google Scholar]
- 18. Dasgupta A., Datta P., Kundu M., Basu J. 2006. The serine/threonine kinase PknB of Mycobacterium tuberculosis phosphorylates PBPA, a penicillin-binding protein required for cell division. Microbiology 152:493–504 [DOI] [PubMed] [Google Scholar]
- 19. Gromadski K. B., Wieden H. J., Rodnina M. V. 2002. Kinetic mechanism of elongation factor Ts-catalyzed nucleotide exchange in elongation factor Tu. Biochemistry 41:162–169 [DOI] [PubMed] [Google Scholar]
- 20. Gupta M., Sajid A., Arora G., Tandon V., Singh Y. 2009. Forkhead-associated domain-containing protein Rv0019c and polyketide-associated protein PapA5, from substrates of serine/threonine protein kinase PknB to interacting proteins of Mycobacterium tuberculosis. J. Biol. Chem. 284:34723–34734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huttlin E. L., et al. 2010. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143:1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jones M. D., et al. 1980. The complete amino-acid sequence of elongation factor Tu from Escherichia coli. Eur. J. Biochem. 108:507–526 [DOI] [PubMed] [Google Scholar]
- 23. Kang C. M., et al. 2005. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 19:1692–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kawashima T., Berthet-Colominas C., Wulff M., Cusack S., Leberman R. 1996. The structure of the Escherichia coli EF-Tu · EF-Ts complex at 2.5 Å resolution. Nature 379:511–518 [DOI] [PubMed] [Google Scholar]
- 25. Khan S., et al. 2010. Phosphorylation of enoyl-acyl carrier protein reductase InhA impacts mycobacterial growth and survival. J. Biol. Chem. 285:37860–37871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Krab I. M., Parmeggiani A. 1998. EF-Tu, a GTPase odyssey. Biochim. Biophys. Acta 1443:1–22 [DOI] [PubMed] [Google Scholar]
- 27. Levine A., et al. 2006. Analysis of the dynamic Bacillus subtilis Ser/Thr/Tyr phosphoproteome implicated in a wide variety of cellular processes. Proteomics 6:2157–2173 [DOI] [PubMed] [Google Scholar]
- 28. Lippmann C., et al. 1993. Prokaryotic elongation factor Tu is phosphorylated in vivo. J. Biol. Chem. 268:601–607 [PubMed] [Google Scholar]
- 29. McEwen D. P., Gee K. R., Kang H. C., Neubig R. R. 2001. Fluorescent BODIPY-GTP analogs: real-time measurement of nucleotide binding to G proteins. Anal. Biochem. 291:109–117 [DOI] [PubMed] [Google Scholar]
- 30. Mikulik K., Zhulanova E. 1995. Sequencing of the tuf1 gene and the phosphorylation pattern of EF-Tu1 during development and differentiation in Streptomyces collinus producing kirromycin. Biochem. Biophys. Res. Commun. 213:454–461 [DOI] [PubMed] [Google Scholar]
- 31. Molle V., Kremer L. 2010. Division and cell envelope regulation by Ser/Thr phosphorylation: Mycobacterium shows the way. Mol. Microbiol. 75:1064–1077 [DOI] [PubMed] [Google Scholar]
- 32. O'Hare H. M., et al. 2008. Regulation of glutamate metabolism by protein kinases in mycobacteria. Mol. Microbiol. 70:1408–1423 [DOI] [PubMed] [Google Scholar]
- 33. Parikh A., Verma S. K., Khan S., Prakash B., Nandicoori V. K. 2009. PknB-mediated phosphorylation of a novel substrate, N-acetylglucosamine-1-phosphate uridyltransferase, modulates its acetyltransferase activity. J. Mol. Biol. 386:451–464 [DOI] [PubMed] [Google Scholar]
- 34. Parish T., Stoker N. G. 1998. Electroporation of mycobacteria. Methods Mol. Biol. 101:129–144 [DOI] [PubMed] [Google Scholar]
- 35. Park S. T., Kang C. M., Husson R. N. 2008. Regulation of the SigH stress response regulon by an essential protein kinase in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 105:13105–13110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Parmeggiani A., Sander G. 1981. Properties and regulation of the GTPase activities of elongation factors Tu and G, and of initiation factor 2. Mol. Cell. Biochem. 35:129–158 [DOI] [PubMed] [Google Scholar]
- 37. Prisic S., et al. 2010. Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. Proc. Natl. Acad. Sci. U. S. A. 107:7521–7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ramakrishnan V. 2002. Ribosome structure and the mechanism of translation. Cell 108:557–572 [DOI] [PubMed] [Google Scholar]
- 39. Rodnina M. V., Gromadski K. B., Kothe U., Wieden H. J. 2005. Recognition and selection of tRNA in translation. FEBS Lett. 579:938–942 [DOI] [PubMed] [Google Scholar]
- 40. Rosenkrands I., et al. 2000. Towards the proteome of Mycobacterium tuberculosis. Electrophoresis 21:3740–3756 [DOI] [PubMed] [Google Scholar]
- 41. Sajid A., et al. 2011. Phosphorylation of Mycobacterium tuberculosis Ser/Thr phosphatase by PknA and PknB. PLoS One 6:e17871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sassetti C. M., Boyd D. H., Rubin E. J. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77–84 [DOI] [PubMed] [Google Scholar]
- 43. Schmeing T. M., et al. 2009. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326:688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schmidl S. R., et al. 2010. The phosphoproteome of the minimal bacterium Mycoplasma pneumoniae: analysis of the complete known Ser/Thr kinome suggests the existence of novel kinases. Mol. Cell. Proteomics 9:1228–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schwede T., Kopp J., Guex N., Peitsch M. C. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sharma K., et al. 2006. Transcriptional control of the mycobacterial embCAB operon by PknH through a regulatory protein, EmbR, in vivo. J. Bacteriol. 188:2936–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sheu G. T., Traugh J. A. 1997. Recombinant subunits of mammalian elongation factor 1 expressed in Escherichia coli. Subunit interactions, elongation activity, and phosphorylation by protein kinase CKII. J. Biol. Chem. 272:33290–33297 [DOI] [PubMed] [Google Scholar]
- 48. Song H., Parsons M. R., Rowsell S., Leonard G., Phillips S. E. 1999. Crystal structure of intact elongation factor EF-Tu from Escherichia coli in GDP conformation at 2.05 Å resolution. J. Mol. Biol. 285:1245–1256 [DOI] [PubMed] [Google Scholar]
- 49. Starck J., Kallenius G., Marklund B. I., Andersson D. I., Akerlund T. 2004. Comparative proteome analysis of Mycobacterium tuberculosis grown under aerobic and anaerobic conditions. Microbiology 150:3821–3829 [DOI] [PubMed] [Google Scholar]
- 50. Sun X., et al. 2010. Phosphoproteomic analysis reveals the multiple roles of phosphorylation in pathogenic bacterium Streptococcus pneumoniae. J. Proteome Res. 9:275–282 [DOI] [PubMed] [Google Scholar]
- 51. Veyron-Churlet R., et al. 2009. The Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III activity is inhibited by phosphorylation on a single threonine residue. J. Biol. Chem. 284:6414–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Veyron-Churlet R., Zanella-Cleon I., Cohen-Gonsaud M., Molle V., Kremer L. 2010. Phosphorylation of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein reductase MabA regulates mycolic acid biosynthesis. J. Biol. Chem. 285:12714–12725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wilson D. N. 2009. The A-Z of bacterial translation inhibitors. Crit. Rev. Biochem. Mol. Biol. 44:393–433 [DOI] [PubMed] [Google Scholar]
- 54. Wolf H., Chinali G., Parmeggiani A. 1974. Kirromycin, an inhibitor of protein biosynthesis that acts on elongation factor Tu. Proc. Natl. Acad. Sci. U. S. A. 71:4910–4914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wolf H., Chinali G., Parmeggiani A. 1977. Mechanism of the inhibition of protein synthesis by kirromycin. Role of elongation factor Tu and ribosomes. Eur. J. Biochem. 75:67–75 [DOI] [PubMed] [Google Scholar]
- 56. Wong D. K., Lee B. Y., Horwitz M. A., Gibson B. W. 1999. Identification of Fur, aconitase, and other proteins expressed by Mycobacterium tuberculosis under conditions of low and high concentrations of iron by combined two-dimensional gel electrophoresis and mass spectrometry. Infect. Immun. 67:327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xolalpa W., et al. 2007. Identification of novel bacterial plasminogen-binding proteins in the human pathogen Mycobacterium tuberculosis. Proteomics 7:3332–3341 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.