Abstract
Irr and RirA, rather than Fur, serve as the major iron-responsive regulators in the alphaproteobacteria. With only a few exceptions, however, the relative contributions of these transcriptional regulators to the differential expression of specific iron metabolism genes in Brucella strains are unclear. The gene encoding the outer membrane heme transporter BhuA exhibits maximum expression in Brucella abortus 2308 during growth under iron-deprived conditions, and mutational studies indicate that this pattern of bhuA expression is mediated by the iron-responsive regulator Irr. Specifically, a bhuA-lacZ transcriptional fusion does not produce elevated levels of β-galactosidase in response to iron deprivation in the isogenic irr mutant BEA5, and, unlike the parental strain, B. abortus BEA5 cannot utilize heme as an iron source in vitro and is attenuated in mice. A derivative of the bhuA-lacZ transcriptional fusion lacking the predicted Irr binding site upstream of the bhuA promoter does not produce elevated levels of β-galactosidase in response to iron deprivation in the parental B. abortus 2308 strain, and a direct and specific interaction between a recombinant version of the Brucella Irr and the bhuA promoter region was observed in an electrophoretic mobility shift assay. Despite the fact that it lacks the heme regulatory element linked to the iron-responsive degradation of its counterpart in Bradyrhizobium japonicum, readily detectable levels of Irr were found only in B. abortus 2308 cells by Western blot analysis following growth under iron-deprived conditions.
INTRODUCTION
Brucella abortus is a Gram-negative intracellular pathogen that causes abortion and infertility in its natural bovine host. Incidental infection in humans results in a prolonged illness known as undulant fever. As with most bacteria, iron is an essential micronutrient for Brucella strains (5, 32). This requirement for iron presents a particular challenge for the brucellae, as in nature these bacteria are found exclusively in association with mammalian hosts (13, 23). Iron not incorporated into host tissues is sequestered by host iron-binding proteins such as transferrin and lactoferrin in extracellular spaces and by ferritin within host cells (6). This tight sequestration of iron in mammals serves to prevent iron toxicity in the host, as well as limiting the availability of this nutrient to invading microbes. Indeed, iron restriction plays an important role in both the innate and acquired immune responses of the host (3, 14, 33, 34).
Previous studies have shown that B. abortus 2308 can use heme as an iron source (17). The TonB-dependent outer membrane transporter BhuA is required for heme utilization by this strain in vitro, and an isogenic bhuA mutant cannot maintain a chronic spleen infection in experimentally infected mice. Like most other bacterial genes that encode heme transporters, bhuA expression in B. abortus 2308 is maximal during growth under iron-deprived conditions (17), but the regulator responsible for the iron-responsive nature of bhuA expression is unknown. A logical candidate for this regulatory role is the iron-responsive regulator (Irr), a member of the Fur family of transcriptional regulators. Irr was initially identified in Bradyrhizobium japonicum as a repressor of hemB, a gene involved in heme biosynthesis (7). The stability of the Bradyrhizobium japonicum Irr is modulated by cellular iron levels. When sufficient iron is available to allow heme biosynthesis, Irr is degraded and hemB repression is relieved. Iron-responsive degradation of Irr requires direct interaction with ferrochelatase (20), which allows Irr to coordinate the activity of the heme biosynthetic pathways with the cellular availability of iron (28). Further studies have linked Irr to the regulation of numerous iron metabolism genes (including those involved in heme acquisition) in Bradyrhizobium japonicum (25, 35), Rhizobium leguminosarum (31), and Bartonella quintana (16).
Experimental evidence indicates that Irr can serve as either a repressor or an activator (25, 35). Since Irr is functional when cellular iron levels are low, Irr often serves as an activator of genes involved in iron acquisition in the alphaproteobacteria (28). In contrast, Irr generally represses genes in these bacteria that encode proteins that require iron for their activity, proteins that participate in cellular processes that require iron (e.g., heme biosynthesis), or iron storage proteins (28). Recent studies have shown that Irr regulates siderophore and heme biosynthesis genes in B. abortus 2308 (10, 11), and the studies described in this report were designed to investigate the role of this regulator in the iron-responsive expression of the gene encoding the heme transporter BhuA in this strain.
MATERIALS AND METHODS
Culture media and growth conditions.
Routine cultivation of Escherichia coli strains was carried out in Luria-Bertani (LB) broth or on tryptic soy agar (TSA) plates with appropriate antibiotic supplementation as necessary. Brucella strains were routinely grown in brucella broth (Difco) at 37°C or on Schaedler agar (Difco) supplemented with 5% defibrinated bovine blood (SBA) incubated at 37°C under 5% CO2. Ampicillin, chloramphenicol, and kanamycin were added to these culture media as needed at final concentrations of 100, 5, and 45 μg/ml, respectively. Low-iron minimal medium was prepared as previously described (9), and 50 μM FeCl3 was added to this medium as a control for growth under iron-replete conditions.
Construction and genetic complementation of B. abortus irr mutants.
A 2,051-bp fragment containing the irr open reading frame (ORF) (BAB1_2175) and flanking sequences was amplified from B. abortus 2308 genomic DNA using Taq DNA polymerase (primers: Fwd, 5′-ACCGGCTTTCGGATCAAG-3′; Rev, 5′-GCGGCCGCATGAAAACTC-3′) and ligated into the SmaI site of pBlueScript KS+ (Stratagene). The resulting plasmid was linearized with NruI and ligated to a 987-bp SmaI/HincII fragment containing the chloramphenicol resistance gene (cat) gene from pBlue-Cm2 (22) or to a 1.4-kb SmaI fragment containing the kanamycin resistance gene (aph3a) from pKS-kan (8). These plasmids were then used to construct isogenic irr mutants from B. abortus 2308 via gene replacement using previously described procedures (4). The genotypes of the resulting B. abortus irr mutants (designated BEA2 [irr::cat] and KH3 [irr::aph3a]) were confirmed by PCR amplification of the mutated irr loci and restriction enzyme digestion and DNA sequence analysis of the amplified DNA fragments. To facilitate genetic complementation studies, the same 2,051-bp irr-containing PCR fragment described above was also cloned into pGEM-T Easy (Promega), excised from the resulting plasmid by EcoRI digestion, and ligated with EcoRI-digested pBBR1MCS-4 (8). This plasmid, pKHS2, was introduced into B. abortus BEA2 by electroporation (4).
Determination of the transcriptional start site for bhuA by primer extension.
The transcriptional start for the bhuA gene was determined by primer extension analysis performed on total RNA preparations obtained from B. abortus 2308 cultures grown for 120 h in low-iron minimal medium using the primer 5′-TGATTTGATTCCAAATGGTTCC-3′ and the methods described by Robertson et al. (22).
Construction of bhuA-lacZ transcriptional fusions and β-galactosidase assays.
The construction of pbhuA-lacZ, a pMR15-based plasmid containing a bhuA-lacZ transcriptional fusion, has been previously described (17). Derivatives of this plasmid carrying truncated versions of the bhuA-lacZ fusion with 29-bp and 120-bp 5′ deletions were constructed by amplifying B. abortus 2308 genomic DNA with the forward primers 6a.5aF (5′-CGCCGTGCCGTTTTCGCATAAA-3′) and 5aF (5′-TAGTCATCTTTATATCCCTCCC-3′) in conjunction with the same reverse primer (5′-GACGGTGGCAAGCAGAGA-3′) used to construct pbhuA-lacZ by PCR with Pfu polymerase and cloning the resulting PCR fragments into pMR15 (2). The resulting transcriptional reporters, designated pbhuA6.5A-lacZ and pbhuA5A-lacZ, respectively, were introduced into B. abortus 2308 by electroporation. Derivatives of B. abortus 2308 carrying pbhuA-lacZ, pbhuA6.5A-lacZ, pbhuA5A-lacZ, and pMR15 were grown in 50 ml low-iron minimal medium (9) or low-iron minimal medium supplemented with 50 μM FeCl3 in 250-ml Erlenmeyer flasks at 37°C with shaking at 175 rpm. β-Galactosidase production by these cultures was measured using the procedures described by Miller (12).
Capacity of the B. abortus strains to use heme as an iron source.
Free iron was removed from the hemin used to make stock solutions by treatment with 10 mM HCl (29). To test for the capacity of hemin to serve as an iron source, B. abortus strains were grown on SBA for 48 h at 37°C with 5% CO2. Bacterial cells were harvested into phosphate-buffered salinel (PBS) (pH 7.2) and adjusted to an optical density at 600 nm (OD600) of 0.15 (109 CFU/ml). Portions (100 μl) of these bacterial cell suspensions were then added to 250-ml flasks containing 50 ml low-iron minimal medium. These flasks were incubated at 37°C with shaking at 250 rpm. Following 96 h of growth, the bacterial cultures were adjusted to an OD600 of 0.15, and 100 μl of the bacterial cell suspensions was mixed with 3 ml tryptic soy broth (TSB) containing 0.7% agar and 300 μM EDDHA (ethylenediamine-di-o-hydroxyphenyl acetic acid). This mixture was overlaid onto 100-mm plates containing TSB with 1.5% agar and 300 μM EDDHA. Sterile filter paper disks (7-mm diameter, Whatman no. 3) were placed onto the plates, 10 μl of a 20 mM solution of hemin or a 50 mM solution of FeCl3 was added to the filter disks, and the plates were incubated at 37°C with 5% CO2. Following 96 h of incubation, the diameter of the zone of bacterial growth around each filter disk was measured and recorded in millimeters.
Production of recombinant Brucella Irr and electrophoretic mobility shift assays (EMSAs) with the bhuA promoter region.
A 444-bp fragment encompassing the irr coding region (BAB1_2175) was amplified from B. abortus 2308 genomic DNA by PCR (primers: F, 5′-CGCGCGGAATTCGTGAATATGCATTCTTCACATACCCACTC-3′; R, 5′-TCAGCGGGCCTGACGGCGCAGACGCACAATGATATCCACAT-3′) using Pfu polymerase and directionally cloned into the EcoRI and EcoRV sites of pASK-IBA6 (IBA). The resulting plasmid, pStrep-irr, produces a chimeric version of Irr carrying an OmpA secretion signal and Strep-Tactin affinity tag on its N terminus. pStrep-irr was introduced into E. coli strain DH5-α. Following the manufacturer's directions, the recombinant Irr protein was isolated from the periplasmic compartment of recombinant E. coli cultures and purified by passage over a Strep-Tactin Sepharose column.
For electrophoretic mobility shift assays, a 153-bp fragment encompassing the bhuA promoter region (primers: F, 5′-ATAAGGCGACCTTACCGAG-3′; R, 5′-GCGACCGATCCGGGAGGG-3′) was amplified from B. abortus 2308 genomic DNA by PCR with Pfu polymerase and radiolabeled with [32P]dATP using Taq polymerase as described previously (1). Radiolabeled probe was purified using a Qiaquick nucleotide removal kit (Qiagen) and resuspended in 30 μl double-distilled water (ddH2O). For a specific DNA competitor, an unlabeled version of the same fragment was used. A 175-bp NcoI/EcoRI restriction digestion fragment from pBlue-Cm2 was employed as a nonspecific competitor. Binding reaction mixtures contained 75 ng radiolabeled bhuA promoter DNA, 2 μg sheared herring sperm DNA, 10 μg bovine serum albumin (BSA), 12 μl of binding buffer (10 mM Tris-HCl, pH 8, 40 mM KCl, 1 mM MgCl2, 1 mM dithiothreitol [DTT], 5% glycerol), and purified Irr protein at 0.25, 0.5, 0.75, or 1 μg. Mixtures were incubated for 30 min at 37°C and then loaded onto a 5% nondenaturing acrylamide gel and subjected to electrophoresis at 100 V. The gel was physically transferred to a piece of Whatman 1 M paper using a Bio-Rad model 583 gel dryer and visualized by autoradiography.
Experimental infection of mice.
Previously described methods (21) were used to evaluate the spleen colonization profiles of B. abortus 2308 and BEA2 in C57BL/6 mice obtained from Harlan Laboratories. Briefly, mice were infected with 5 × 104 brucellae via the intraperitoneal route, and at each sampling point postinfection, the mice were euthanized, their spleens were removed, and spleen homogenates were serially diluted and plated on SBA to determine the number of viable brucellae present. These experiments were performed prior to receiving notification from the Centers for Disease Control and Prevention's Select Agent Program that the introduction of Brucella melitensis, Brucella suis, and B. abortus strains carrying chloramphenicol resistance genes into experimentally infected animals is no longer permitted.
Immunoblot analysis.
Whole-cell protein lysates were collected from Brucella cultures grown to the desired time points (i.e., 72, 96, or 120 h) in low-iron minimal medium and low-iron minimal medium supplemented with 50 μM FeCl3. Cells were collected by centrifugation (6,800 × g, 10 min, room temperature), and the pellet was suspended in 1 ml of protein sample buffer (0.3% SDS, 200 mM DTT, 22 mM Tris base, and 28 mM Tris-HCl). The cells were then boiled for 1 h, followed by 10 cycles (20 s per cycle) of disintegration in lysing matrix B (MP Biomedicals, Solon, OH) using a BIO101 FastPrep FP120 cell disruptor (Thermo Savant [Thermo Fisher Scientific Inc.], Waltham, MA). A Bradford assay with BSA standards was used to determine protein concentrations. Eighty micrograms of total protein per culture was separated by electrophoresis through a 13% SDS-PAGE gel. Proteins were transferred to a nitrocellulose membrane by electroblotting at 100 V for 2 h. Membranes were blocked at room temperature in 5% skim milk in PBST (155 mM NaCl, 8 mM Na2HPO4, 2 mM NaH2PO4, 0.05% Tween 20). Primary antibodies (anti-Irr [1:10,000]) were incubated with the membranes at room temperature in 5% skim milk. Secondary antibodies (anti-rabbit IgG horseradish peroxidase [HRP] conjugate) (Promega) were used at a 1:10,000 dilution. All washing steps were performed with PBST. Visualization of the HRP signal was performed using SuperSignal West Pico chemiluminescent substrate (Thermo Scientific). Anti-Irr serum was raised in New Zealand White rabbits using the recombinant Brucella Irr as the antigen and TiterMax Gold (Sigma) as an adjuvant for the primary immunization.
Statistical analysis.
All statistical analyses were performed using the Student two-tailed t test (24). P values of ≤0.05 were considered significant.
RESULTS AND DISCUSSION
Irr regulates the expression of the genes encoding the BhuA homologs, HmuR and HutA, in two close phylogenetic relatives of the brucellae, Bradyrhizobium japonicum and Bartonella quintana, respectively (16, 25). In Bradyrhizobium japonicum, Irr is required for the induction of hmuR in response to iron deprivation, and genetic and biochemical evidence suggest that Irr serves as a transcriptional activator of hmuR (25). In contrast, in Bartonella quintana, overproduction of Irr has a negative impact on hutA expression, and Irr has been shown to bind upstream of the hutA promoter in an electrophoretic mobility shift assay (16). As shown in Fig. 1, transcription of bhuA is induced during stationary phase following growth in low-iron minimal medium in B. abortus 2308 but not in the isogenic irr mutant BEA2. Moreover, introduction of a plasmid-borne copy of irr into BEA2 restores low-iron-responsive bhuA expression in this strain. These data indicate that Irr is required for the low-iron-responsive induction of bhuA transcription in B. abortus 2308. Such a role is reminiscent of the one that Irr plays in the iron-responsive expression of hmuR and several other genes encoding TonB-dependent outer membrane iron transporters in Bradyrhizobium japonicum, where Irr serves as a transcriptional activator (25, 27). Consistent with its inability to induce bhuA expression in response to iron deprivation, the B. abortus irr mutant BEA2 is also unable to use hemin as an iron source in an in vitro assay (Fig. 2A), while the parental 2308 strain and the complemented mutant (BEA2.C) grow equally well around disks containing hemin on plates containing the iron-specific chelator EDDHA. In contrast, BEA2 produces a zone of growth similar to that of the parental 2308 strain around disks containing FeCl3 (Fig. 2B).
Fig. 1.
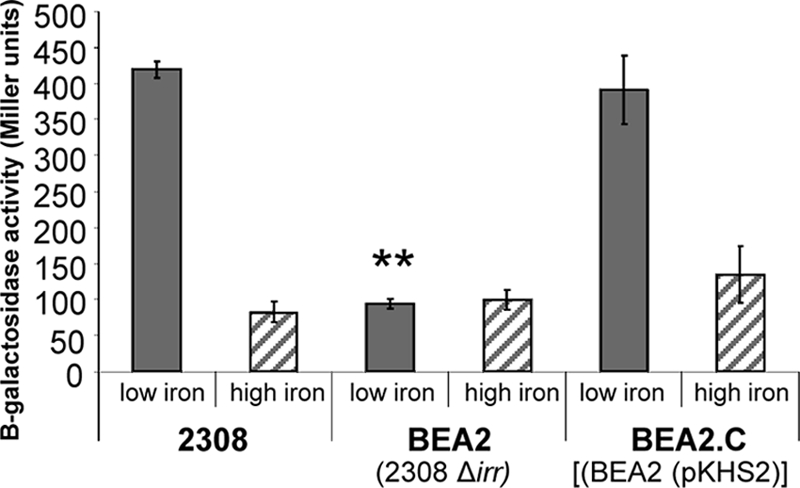
Transcriptional activity of a bhuA-lacZ reporter in B. abortus 2308, BEA2(2308 Δirr), and BEA2.C [BEA2(pKHS2)]. Gray bars show β-galactosidase production in Miller units (12) by B. abortus strains carrying pbhuA-lacZ following 96 h of growth in low-iron minimal medium (low iron). The crosshatched bars show β-galactosidase levels produced by these strains following 96 h of growth in low-iron medium supplemented with 50 μM FeCl3 (high iron). The results presented are means and standard deviations from a single experiment that is representative of multiple (≥3) experiments performed from which equivalent results were obtained. The average β-galactosidase level produced by the parental plasmid pMR15 in B. abortus 2308 during growth in low-iron minimal medium or this medium supplemented with 50 μM FeCl3 was subtracted from each data set to account for background activity. **, P ≤ 0.001.
Fig. 2.
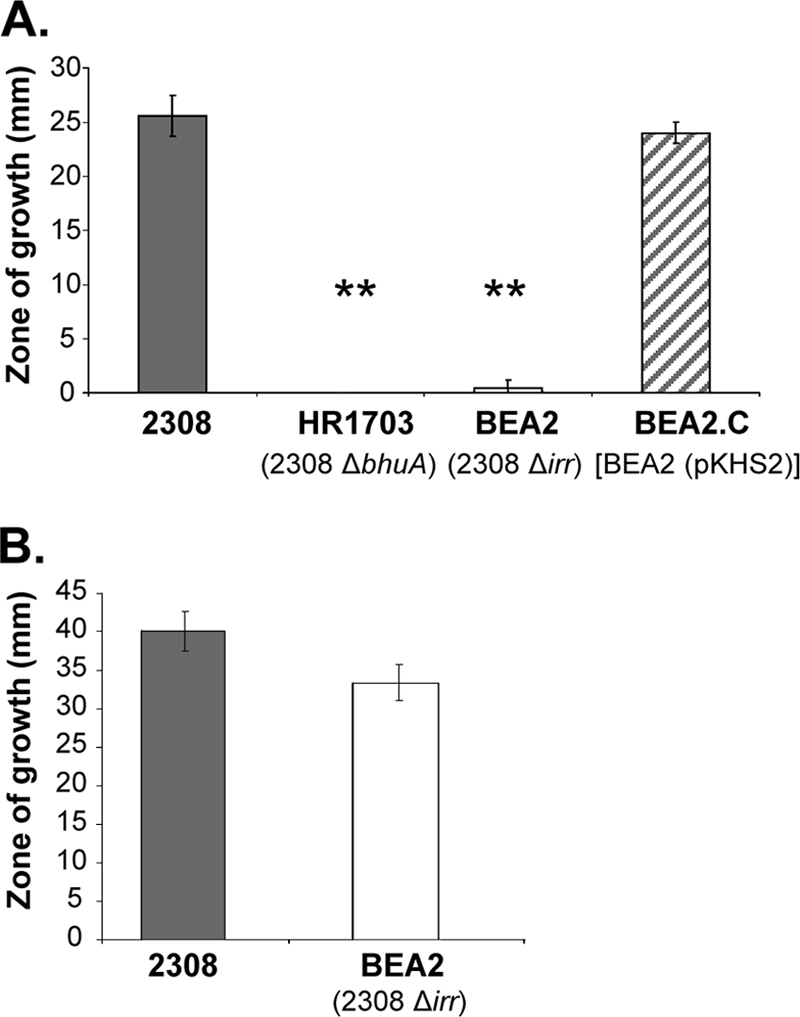
Capacity of B. abortus 2308, HR1703(2308 ΔbhuA), BEA2(2308 Δirr) and BEA2.C [BEA2(pKHS2)] to use hemin (A) or FeCl3 (B) as an iron source in a disk diffusion assay. The results presented are from a single experiment that is representative of multiple (≥3) experiments performed from which equivalent results were obtained. **, P ≤ 0.001.
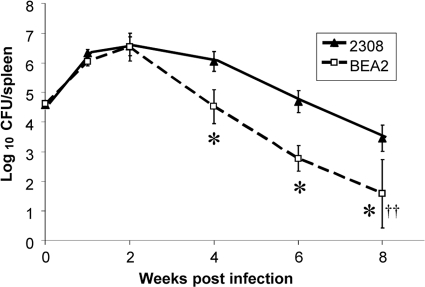
The capacity of B. abortus 2308 to use heme as an iron source has been linked to its ability to maintain chronic spleen infections in experimentally infected mice (17). Thus, the accelerated clearance of the B. abortus irr mutant from the spleens of infected mice compared to that of the parental strain (Fig. 3) is consistent with the inefficient bhuA expression exhibited by the irr mutant and its inability to use heme as an iron source in vitro. The attenuation of the B. abortus irr mutant in mice observed in these studies appears at first to be in contrast to the results reported by Martínez et al. (11), who reported that a B. abortus irr mutant is not attenuated at 1 and 3 weeks postinfection in BALB/c mice. However, it should be noted that no attenuation was observed for the irr mutant at 1 or 2 weeks postinfection in mice in the present study (Fig. 3), with attenuation being observed starting at week 4 postinfection. In addition, C57BL/6 mice were used in the study reported here, while BALB/c mice were employed in the study described by Martínez et al. (11). From a regulatory standpoint, it is important to point out that the mouse infections described in this report were performed prior to notification from the Centers for Disease Control and Prevention's Select Agent Program that the introduction of B. melitensis, B. suis, and B. abortus strains carrying chloramphenicol resistance genes into experimentally infected animals is no longer permitted.
Fig. 3.
Spleen colonization profiles of B. abortus 2308 and BEA2(2308 irr) in C57BL6 mice. The data are means and standard deviations for the number of brucellae detected in the spleens of five mice infected with each strain at each experimental time point in a single experiment. *, P ≤ 0.01 for comparisons of the data obtained with 2308 and BEA2. ††, no brucellae were detected in the spleens of 2 of the mice infected with the B. abortus irr mutant at 8 weeks postinfection.
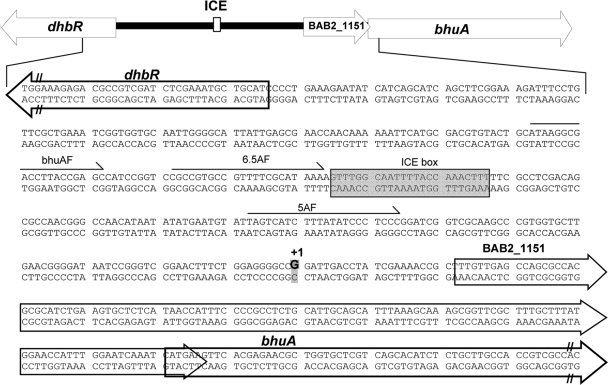
To begin to define the cis-acting regulatory elements that control the iron-responsive expression of bhuA in B. abortus 2308, primer extension was employed to determine the transcriptional start site for this gene. A guanine residue (G) 141 nucleotides (nt) upstream of the predicted bhuA start codon was identified as the +1 start site for the bhuA transcript (Fig. 4). This transcript would include the open reading frame (ORF) annotated as BAB2_1151 in the B. abortus 2308 genome sequence which is predicted to encode a hypothetical protein. Whether or not the BAB2_1151 ORF is translated is unknown and is currently under investigation. Notably, 156 nucleotides upstream of the transcriptional site for bhuA is a consensus Irr binding site (also known as an ICE box) (Fig. 4). The spacing of this putative Irr binding site relative to the bhuA transcriptional start site is similar to that found in the blr4504 gene (121 nt) in Bradyrhizobium japonicum, which requires Irr for its induction in response to iron deprivation (27).
Fig. 4.
Genetic organization of the dhbR-bhuA locus in B. abortus 2308. The transcriptional start site for the bhuA gene (which would also include the hypothetical gene designated BAB2_1151) is denoted by +1, and the corresponding guanine residue (G) is shown in boldface and highlighted. The nucleotides comprising the putative Irr binding site upstream of bhuA are enclosed in a box (ICE box). The oligonucleotide primers used to construct the truncated versions of the plasmid-borne bhuA-lacZ fusion are indicated by the arrows labeled 7AF, 6.5AF, and 5AF.
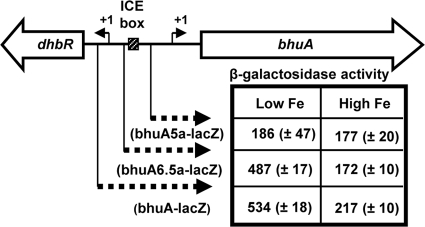
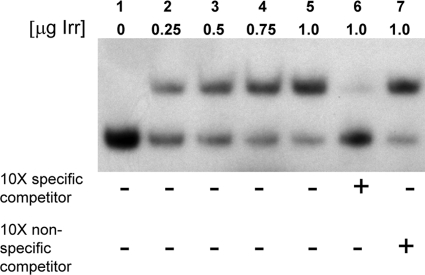
To evaluate the importance of the ICE box for iron-responsive expression of bhuA in B. abortus 2308, β-galactosidase production by derivatives of this strain carrying pbhuA-lacZ (a pMR15-based plasmid carrying a bhuA-lacZ transcriptional fusion [17]) and truncated versions of this plasmid was evaluated. Plasmid pbhuA6.5a-lacZ contains a lacZ fusion containing 179 bp upstream of the bhuA transcriptional start site that includes the putative Irr binding site (Fig. 4 and 5). Plasmid pbhu5a-lacZ, on the other hand, contains a lacZ fusion containing 88 bp upstream of the bhuA transcriptional start site lacking an Irr binding site. As shown in Fig. 4, both of the bhuA-lacZ transcriptional fusions (pbhuA-lacZ and pbhuA6.5a-lacZ) containing the putative Irr binding site exhibit enhanced expression in B. abortus 2308 following growth in low-iron minimal medium but basal expression when this strain is grown under iron-replete conditions. In contrast, the bhuA-lacZ transcriptional fusion lacking the Irr binding site (pbhuA5a-lacZ) displays basal levels of expression in both low-iron medium and low-iron medium supplemented with 50 μM FeCl3. These experimental findings verify that the putative Irr-binding site upstream of bhuA is required for the low-iron-responsive induction of this gene in B. abortus 2308 and, moreover, suggest that Irr is serving as a transcriptional activator. This proposition is further supported by the observation that Irr directly binds to the bhuA promoter region in a specific manner in an EMSA (Fig. 6).
Fig. 5.
Identification of the iron-responsive bhuA promoter in B. abortus 2308 by deletion analysis. β-Galactosidase production by B. abortus 2308 carrying the plasmid-borne bhuA-lacZ transcriptional fusions following 120 h of growth in low-iron minimal medium (low Fe) or low-iron minimal medium supplemented with 50 μM FeCl3 (high Fe) is shown. The results presented are from a single experiment that is representative of multiple (≥3) experiments performed from which equivalent results were obtained. The regions of the dhbR-bhuA locus included in the bhuA-lacZ fusions are shown as dashed arrows, and the location of the putative Irr binding motif is denoted as the ICE box.
Fig. 6.
Irr interacts directly with the bhuA promoter region in an EMSA. Lane 1, 75 ng 32P-labeled bhuA promoter-specific DNA fragment; lanes 2 through 5, 75 ng 32P-labeled bhuA promoter-specific DNA fragment plus increasing concentrations (0.25, 0.5, 0.75, and 1 μg, respectively) of the Irr protein; lane 6, 75 ng 32P-labeled bhuA promoter-specific DNA fragment plus 1 μg Irr plus 750 ng unlabeled bhuA promoter-specific DNA fragment (specific inhibitor); lane 7, 75 ng 32P-labeled bhuA promoter-specific DNA fragment plus 1 μg Irr plus 750 ng unlabeled pBlue-Cm2 specific DNA fragment (nonspecific inhibitor). The results presented are from a single experiment that is representative of multiple (≥3) experiments performed from which equivalent results were obtained.
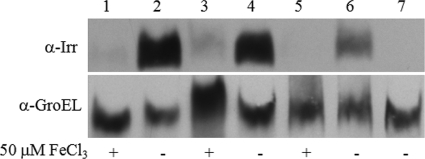
Considering the essential role that Irr plays in regulating the iron-responsive expression of bhuA in B. abortus 2308, an important question to address is how Irr activity is modulated in this strain. As noted previously, the stability of the Irr protein in Bradyrhizobium japonicum is controlled by cellular iron levels (20). When iron levels are sufficient to allow heme biosynthesis, Irr is degraded. The studies described by Martínez et al. (10) suggest that a similar relationship between cellular iron levels and Irr stability may also exist in B. abortus 2308. However, recent studies with another alphaproteobacterium, Rhizobium leguminosarum, suggest that Irr activity in this bacterium is modulated by heme, and Irr is inactivated, but not degraded, when cellular iron levels increase (26). Instead, heme binding to the R. leguminosarum Irr inhibits its DNA binding capacity. Distinguishing between these two models for regulating Irr function is important because the Brucella Irr, like the R. leguminsarum Irr, does not possess the heme regulatory element (GCPWAD) in its N terminus that is required for the iron-responsive degradation of Irr in Bradyrhizobium japonicum (19, 26). As shown in Fig. 7, Western blot analysis of cell lysates from B. abortus 2308 cultures grown in low-iron minimal medium and in low-iron minimal medium supplemented with FeCl3 suggests that Irr is degraded in this strain in response to increasing intracellular levels of iron. Thus, despite its lack of a conserved GCPWAD domain, it appears that the activity of the Brucella Irr is being modulated by iron-responsive degradation in a manner similar to that proposed for the Bradyrhizobium Irr (20, 28).
Fig. 7.
Analysis of Irr protein levels in B. abortus 2308 during growth under low- and high-iron conditions. Irr-specific antiserum was used in Western blot analysis to detect Irr levels in cell lysates from B. abortus 2308 cultures (lanes 1 thru 6) grown in low-iron minimal medium or low-iron minimal medium supplemented with 50 μM FeCl3 as indicated for 72 h (lanes 1 and 2), 96 h (lanes 3 and 4), or 120 h (lanes 5 and 6). Lane 7 shows a cell lysate from B. abortus KH3(2308 irr) grown for 96 h in low-iron minimal medium. Antiserum specific for the Brucella GroEL was used as a protein loading control.
The experimental findings presented here build upon those presented by Martínez et al. (10, 11) and demonstrate that Irr is a major iron-responsive regulator of gene expression in B. abortus 2308. Indeed, the presence of Irr appears to be essential for the wild-type expression of bhuA, which encodes a major virulence determinant (17). Because the brucellae must carefully balance cellular iron levels to avoid the enhancement of endogenous oxidative stress arising from their respiratory metabolism and exogenous oxidative stress derived from the NADPH and inducible nitric oxide synthase (iNOS) activity of host macrophages (23), it will be important to obtain further insight into the nature of the Irr regulon in Brucella. In addition to Irr, Brucella strains also possess a homolog of the rhizobial iron regulator RirA, another important regulator of iron metabolism genes in the alphaproteobatecteria (30), and preliminary studies indicate that RirA also plays an important role in controlling the expression of iron metabolism genes in B. abortus 2308, including bhuA (18). Thus, it will also be important to determine how Irr and RirA work together to regulate iron metabolism in this strain.
ACKNOWLEDGMENT
This work was supported by a grant from NIAID (AI-63516) to R.M.R.
Footnotes
Published ahead of print on 29 July 2011.
REFERENCES
- 1. Anderson E. S., Paulley J. T., Roop R. M., II 2007. The AraC-like transcriptional regulator DhbR is required for maximum expression of the 2,3-dihydroxybenzoic acid biosynthesis genes in Brucella abortus 2308 in response to iron deprivation. J. Bacteriol. 190:1838–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bellaire B. H., Elzer P. H., Baldwin C. L., Roop R. M., II 2003. Production of the siderophore 2,3-dihydroxybenzoic acid is required for the wild-type growth of Brucella abortus in the presence of erythritol under low-iron conditions in vitro. Infect. Immun. 71:2927–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cellier M. F., Courville P., Campion C. 2007. Nramp1 phagocyte intracellular metal withdrawal defense. Microbes Infect. 9:1662–1670 [DOI] [PubMed] [Google Scholar]
- 4. Elzer P. H., Phillips R. W., Kovach M. E., Peterson K. M., Roop R. M., II 1994. Characterization and genetic complementation of a Brucella abortus high-temperature-requirement A (htrA) deletion mutant. Infect. Immun. 62:4135–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Evenson M. A., Gerhardt P. 1955. Nutrition of brucellae: utilization of iron, magnesium and manganese for growth. Proc. Soc. Exp. Biol. Med. 89:678–680 [DOI] [PubMed] [Google Scholar]
- 6. Griffiths E. 1999. Iron in biological systems, p. 1–25 In Bullen J. J., Griffiths E. (ed.), Iron and infection. Molecular, physiological and clinical aspects, 2nd ed John Wiley & Sons, New York, NY [Google Scholar]
- 7. Hamza I., Chauhan S., Hassett R., O'Brian M. R. 1998. The bacterial Irr protein is required for coordination of heme biosynthesis through iron availability. J. Biochem. 273:21669–21674 [DOI] [PubMed] [Google Scholar]
- 8. Kovach M. E., et al. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
- 9. López-Goñi I., Moriyón I., Neilands J. B. 1992. Identification of 2,3-dihydroxybenzoic acid as a Brucella abortus siderophore. Infect. Immun. 60:4496–4503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martínez M., Ugalde R. A., Almirón M. 2005. Dimeric Brucella abortus Irr protein controls its own expression and binds haem. Microbiology 151:3427–3433 [DOI] [PubMed] [Google Scholar]
- 11. Martínez M., Ugalde R. A., Almirón M. 2006. Irr regulates brucebactin and 2,3-dihydroxybenzoic acid biosynthesis, and is implicated in the oxidative stress resistance and intracellular survival of Brucella abortus. Microbiology 152:2591–2598 [DOI] [PubMed] [Google Scholar]
- 12. Miller J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 13. Moreno E., Moriyón I. 2002. Brucella melitensis: a nasty bug with hidden credentials for virulence. Proc. Natl. Acad. Sci. U. S. A. 99:1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nemeth E., et al. 2004. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 306:2090–2093 [DOI] [PubMed] [Google Scholar]
- 15. Reference deleted.
- 16. Parrow N. L., Abbott J., Lockwood A. R., Battisti J. M., Minnick M. F. 2009. Function, regulation, and transcriptional organization of the hemin utilization locus of Bartonella quintana. Infect. Immun. 77:307–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paulley J. T., Anderson E. S., Roop R. M., II 2007. Brucella abortus requires the heme transporter BhuA for maintenance of chronic infection in BALB/c mice. Infect. Immun. 75:5248–5254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Paulley J. T. 2007. Ph.D. thesis East Carolina University, Greenville, NC [Google Scholar]
- 19. Qi Z., Hamza I., O'Brian M. R. 1999. Heme is an effector molecule for iron-dependent degradation of the bacterial iron response regulator (Irr) protein. Proc. Natl. Acad. Sci. U. S. A. 96:13056–13061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qi Z., O'Brian M. R. 2002. Interaction between the bacterial iron response regulator and ferrochelatase mediates genetic control of heme biosynthesis. Mol. Cell 9:155–162 [DOI] [PubMed] [Google Scholar]
- 21. Robertson G. T., Roop R. M., II 1999. The Brucella abortus host factor I (HF-I) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol. Microbiol. 34:690–700 [DOI] [PubMed] [Google Scholar]
- 22. Robertson G. T., et al. 2000. The Brucella abortus CcrM DNA methyltransferase is essential for viability, and its overexpression attenuates intracellular replication in murine macrophages. J. Bacteriol. 182:3482–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roop R. M., II, Gaines J. M., Anderson E. S., Caswell C. C., Martin D. W. 2009. Survival of the fittest: how Brucella strains adapt to their intracellular niche in the host. Med. Microbiol. Immunol. 198:221–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rosner B. 2000. Fundamentals of biostatistics, 5th ed Duxbury, Pacific Grove, CA [Google Scholar]
- 25. Rudolph G., et al. 2006. The iron control element, acting in positive and negative control of iron-regulated Bradyrhizobium japonicum genes, is a target for the Irr protein. J. Bacteriol. 188:733–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Singleton C., et al. 2010. Heme-responsive DNA binding by the global iron regulator Irr from Bradyrhizobium leguminosarum. J. Biol. Chem. 285:16023–16031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Small S. K., Puri S., Sangwan I., O'Brian M. R. 2009. Positive control of ferric siderophore receptor gene expression by the Irr protein in Bradyrhizobium japonicum. J. Bacteriol. 191:1361–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Small S. K., Puri S., O'Brian M. R. 2009. Heme-dependent metalloregulation by the iron response regulator (Irr) protein in Rhizobium and other alpha-proteobacteria. Biometals 22:89–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Staggs T. M., Perry R. D. 1991. Identification and cloning of a fur regulatory gene in Yersinia pestis. J. Bacteriol. 173:417–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Todd J. D., et al. 2002. RirA, an iron-responsive regulator in the symbiotic bacterium Rhizobium leguminosarum. Microbiology 148:4059–4071 [DOI] [PubMed] [Google Scholar]
- 31. Todd J. D., Sawers G., Rodionov D. A., Johnston A. W. B. 2006. The Rhizobium leguminosarum regulator IrrA affects the transcription of a wide range of genes in response to Fe availability. Mol. Genet. Genomics 275:564–577 [DOI] [PubMed] [Google Scholar]
- 32. Waring W. S., Elberg S. S., Schneider P., Green W. 1953. The role of iron in the biology of Brucella suis. J. Bacteriol. 66:82–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Weinberg E. D. 1995. Acquisition of iron and other nutrients in vivo, p. 79–93 In Roth J. A., Bolin C. A., Brogden K. A., Minion F. C., Wannemuehler M. J. (ed.), Virulence mechanisms of bacterial pathogens. ASM Press, Washington, DC [Google Scholar]
- 34. Weiss G. 2005. Modification of iron regulation by the inflammatory response. Best Pract. Res. Clin. Haematol. 18:183–201 [DOI] [PubMed] [Google Scholar]
- 35. Yang J., et al. 2006. Bradyrhizobium japonicum senses iron through the status of haem to regulate iron homeostasis and metabolism. Mol. Microbiol. 60:427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]