Abstract
We characterized Orf5 and SepQ, two type III secretion (T3S) system proteins in enteropathogenic Escherichia coli, and showed that they are essential for T3S, associated with the bacterial membrane, and interact with EscN. Our findings suggest that Orf5 and SepQ are homologs of YscL and YscQ from Yersinia, respectively.
TEXT
Enteropathogenic Escherichia coli (EPEC) belongs to a family of bacterial pathogens that colonize the gut epithelium via formation of distinct histopathological attaching and effacing (A/E) lesions (15). The A/E phenotype is linked to the locus of enterocyte effacement (LEE), the pathogenicity island of EPEC that encodes components of a type III secretion system (T3SS), transcriptional regulators, chaperones, and effector proteins (4, 12).
The T3SS is a multiprotein complex that transports effector proteins into target cells. The overall structure of the type III secretion (T3S) apparatus is conserved among different pathogens and resembles the flagellar apparatus. The T3S apparatus consists of three ring structures that transverse the bacterial cytoplasm (C ring), inner membrane (inner ring), and outer membrane (outer ring), joined together by a cylinder (inner rod) and linked to a needle and a filament that reach the host cell membrane to form a pore (translocators) (23, 28–31). The cytoplasmic ring is assumed to consist of an oligomer of a protein from the YscQ/FliN family. This assumption is based on the crystal structures of FliN and HrcQB, which were previously shown to form doughnut-shaped tetramers that conform in size and shape to electron microscopy (EM) images of the C ring (5, 26). Although PSI-BLAST searches failed to find any YscQ homolog among the EPEC LEE-encoded proteins, a search of the NCBI conserved-domain database (18) identified an FliN domain in the C terminus of the LEE-encoded SepQ protein (25). A recent study by Lara-Tejero et al. suggested that SpaO, the Salmonella YscQ homolog, has a critical function that ensures the hierarchy in T3S, as it serves as a sorting platform for effectors and translocators (17). This finding reinforces the importance of identifying the YscQ/SpaO homolog in EPEC.
The Yersinia YscQ protein was previously shown to interact with YscL, the soluble cytoplasmic T3SS component (2, 11). Moreover, YscL was shown to bind YscN, the T3SS ATPase, and inhibit its activity (2). A similar protein organization was found in the flagellar export apparatus, where FliH, the flagellar homolog of YscL, was shown to bind both FliI (YscN homolog) and FliN (YscQ homolog) as well as to inhibit FliI ATPase activity (19, 21).
Pallen et al. proposed that, based on the results of PSI-BLAST searches restricted to bacterial proteins, the missing YscL/FliH component in the EPEC T3SS is Orf5 and suggested renaming it EscL (25). In this study, we biochemically characterized Orf5 and SepQ. Based on our results, we support the suggestion of Pallen et al. to change the names of Orf5 and SepQ to EscL and EscQ, respectively (25); thus, we use that terminology throughout this paper.
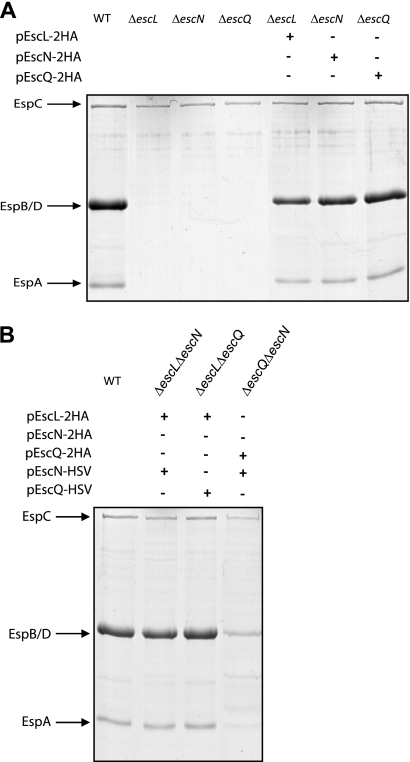
To determine whether EPEC EscL, EscN, and EscQ are essential for T3S, nonpolar deletion mutants were constructed for escL and escQ by the use of the sacB-based allelic exchange method (14). The respective primer pairs used are summarized in Table 1. The EPEC ΔescN mutant used here was previously described (7). We analyzed the secretion profiles of the translocators EspA, EspB, and EspD in EPEC ΔescL, ΔescN, and ΔescQ strains, after induction of T3S (8). The null strains showed defective secretion, indicating that EscL, EscN, and EscQ are essential for T3S (Fig. 1A), in agreement with the observations previously reported for Citrobacter rodentium and EHEC (3, 16). Complementation of the null strains with double-hemagglutinin (HA)-tagged EscL, EscN, or EscQ in trans (cloned into pTOPO-2HA [3] by the use of HindIII and XhoI sites with the primer pairs shown in Table 1) restored secretion of the translocators, thus confirming that the deletion mutations in escL, escN, and escQ were nonpolar and that the tagged versions of these proteins were functional (Fig. 1A) (the plasmids used are listed in Table 2). Double-deletion strains missing two of the three genes (ΔescLΔescN, ΔescLΔescQ, and ΔescNΔescQ) were also complemented for T3S when both missing proteins were expressed from plasmids (pTOPO-2HA and pACYC-HSV [7]) (Fig. 1B). The ΔescNΔescQ strain complemented with pEscN-HSV and pEscQ-2HA showed a reduced ability to secrete translocators compared to the wild type (WT), possibly due to interference from the tags.
Table 1.
Primers used in this study
| Construct and primer | Sequence (5′–3′) |
|---|---|
| escL deletion mutant | |
| F1-escL | TAAAGGTACCTACAATGAAGCAAGGAACTG |
| R1-escL | TATGCTAGCTTCCGTAAGCACTAAATCAAT |
| F2-escL | TATGCTAGCATTATTCAGGAATGATGATATG |
| R2-escL | AATGAGCTCCCCAGCCGCATCTATTGCC |
| escQ deletion mutant | |
| F1-escQ | TCGGTACCTTTGCGTGCGGCG |
| R1-escQ | TAGCTAGCAACAACGGCAAATAAAAATCG |
| F2-escQ | CAGCTAGCACGGAATAATACCACAATCG |
| R2-escQ | AAAGACTCACAAATAAATTACCTATGAACACATCA |
| lysA/argH deletion mutant | |
| F1-argH | CGGGGTACCCGGGTTTGCCGTTCAGGGCGA |
| R1-argH | CTAGCTAGCACGCCCGCCCCAAAGTGCCAT |
| F2-argH | CTAGCTAGCGCAAGGGCGCGGTTGGGGTAA |
| R2-argH | ATCGAGCTCACCTGCCGCCACCATGTTGCG |
| F1-lysA | CGGGGTACCATAATTCACCCAAAGCCAGCG |
| R1- lysA | CTAGCTAGCCTGGCGCTGGAATTGCTTTAA |
| F2- lysA | CTAGCTAGCGCTGAACAGTGAATGTGGCAT |
| R2- lysA | ATCGAGCTCAAATGCTCACTAAACGCCTGC |
| pEscN-2HA | |
| F-escN-HA | CCAAGCTTTCTTTTCAGGAGGTTGGGAAT |
| R-escN-HA | CACTCGAGGGCAACCACTTTGAATAGG |
| pEscL-2HA | |
| F-escL-HA | AAAAGCTTTTACCGAAAAGTGGAGAAAT |
| R-escL-HA | TACTCGAGTTCCTGAATAATGCTAAGATAATTATC |
| pEscQ-2HA | |
| F-escQ-HA | TAAAGCTTAAAAAAGACTATGTTTTCGATAAG |
| R-escQ-HA | CCCTCGAGATCACATACTACGCTAATAGTA |
| pEmpty-2HA | |
| F-empty-HA | AGCTTAGTGGAGAAATAAAACCAACTCATATTAATGATTTATTTCTTAC |
| R-empty-HA | TCGAGTAAGAAATAAATCATTAATATGAGTTGGTTTTATTTCTCCACTA |
| pEscQ-HSV | |
| F-escQ-HSV | ATGGATCCTGATGAAGCCATTGAGTT |
| R-escQ-HSV | TGGTCGACCACATACTACGCTAATAGTAAAATAAA |
Fig. 1.
EscL, EscN, and EscQ are essential for type III secretion in EPEC. (A) Protein secretion profiles of the EPEC WT, ΔescL, ΔescN, and ΔescQ strains as well as the null mutants complemented with a double-HA-tagged version of the missing protein (pEscL-2HA, pEscN-2HA, or pEscQ-2HA). Secreted proteins were concentrated from supernatants of bacterial cultures grown in DMEM and analyzed by SDS–12.5% PAGE and Coomassie staining. The locations of the translocators EspA, EspB, and EspD are indicated at the left of the gel. The location of EPEC EspC, which is not secreted via the LEE-encoded T3SS, is also indicated. (B) Protein secretion profiles of EPEC double-deletion strains complemented with the two missing proteins. Secreted proteins were processed as described for panel A.
Table 2.
Strains and plasmids used in this study
| Strain | Description | Source or reference |
|---|---|---|
| ΔescL | EPEC E2348/E2369 escL deletion mutant | This study |
| ΔescN | EPEC E2348/E2369 escN deletion mutant | 7 |
| ΔescQ | EPEC E2348/E2369 escQ deletion mutant | This study |
| ΔescLΔescN | EPEC E2348/E2369 escL and escN deletion mutant | This study |
| ΔescLΔescQ | EPEC E2348/E2369 escL and escQ deletion mutant | This study |
| ΔescQΔescN | EPEC E2348/E2369 escQ and escN deletion mutant | This study |
| ΔlysAΔargH | EPEC E2348/E2369 lysA and argH deletion mutant | This study |
| pEscL-2HA | pCR2.1-TOPO carrying EscL-2HA | This study |
| pEscN-2HA | pCR2.1-TOPO carrying EscN-2HA | This study |
| pEscQ-2HA | pCR2.1-TOPO carrying EscQ-2HA | This study |
| pEmpty-2HA | pCR2.1-TOPO expressing double-HA tag | This study |
| pEscN-HSV | pACYC184 carrying EscN-HSV | 7 |
| pEscQ-HSV | pACYC184 carrying EscQ-HSV | This study |
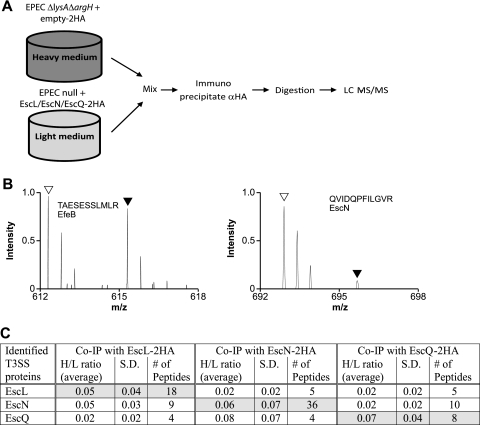
To identify the interaction partners of EscL, EscN, and EscQ of the EPEC T3SS, we employed a mass spectrometry (MS)-based stable isotope labeling of amino acids in cell culture (SILAC) technique. SILAC is a quantitative proteomic method for identification of protein-protein interactions with high confidence and without a priori knowledge of interaction partners (24). The parental strain, EPECΔlysAΔargH expressing the double-HA tag without a protein attached (empty-2HA), was grown in media supplemented with the stable (heavy) isotopes 13C6 l-arginine (L-Arg6) and 4,4,5,5-D4 l-lysine (L-Lys4) for 3 h under T3S conditions. EPECΔlysAΔargH was completely auxotrophic for arginine and lysine; therefore, the proteome of this strain would be preferentially labeled by heavy lysine and arginine derivatives. The null strains EPECΔescL, EPECΔescN, and EPECΔescQ, expressing their corresponding double HA-tagged proteins, were grown under similar conditions but supplemented with normal (light) l-arginine and l-lysine. Lysates of heavy and light bacterial populations were mixed at a 1:1 ratio and immunoprecipitated (IP) using α-HA antibodies. Eluted proteins were digested into peptides, separated by high-performance liquid chromatography (LC), and analyzed by tandem mass spectrometry (LC-MS/MS) on an LTQ Orbitrap XL system (27) (Fig. 2A). The relative abundances of proteins present in two different samples can thus be quantified based on the ratios of the peak intensities of arginine- and lysine-containing peptide pairs for calculation of a heavy/light (H/L) ratio. Peptides with H/L ratios below 0.3 are considered significant, and the corresponding protein is considered an interaction partner candidate for the tagged protein.
Fig. 2.
SILAC screen for interacting proteins EscL, EscN, and EscQ. (A) Schematic representation of the SILAC method. The EPECΔlysAΔargH strain expressing the double-HA tag and an EPEC null mutant complemented with the HA-tagged protein were differentially labeled, and lysates were mixed in a 1:1 ratio. Proteins were immunoprecipitated with the α-HA antibody and then trypsinized and analyzed by LC-MS/MS. (B) Representative mass spectra for peptides recovered from the SILAC experiment performed with EscL-2HA. Right panel: open and filled triangles indicate the expected mass charge (m/z) of light and heavy forms, respectively, of a peptide (QVIDQPFILGVR) from the specifically bound EscN from pulldowns with EscL-2HA and 2HA, respectively. The light peptide of EscN is present in a ratio >3.0 higher than the heavy peptide, indicating a specific interaction with EscL. Left panel: open and filled triangles indicate the expected m/z of light and heavy forms, respectively, of a peptide (TAESESSLMLR) from the nonspecifically bound EfeB protein from pulldowns with EscL-2HA and 2HA, respectively. (C) Summary of LEE-encoded proteins identified by LC-MS/MS as specifically immunoprecipitated with EscL-2HA, EscN-2HA, or EscQ-2HA in the SILAC experiments. H/L ratios represent the average values for the heavy/light isotope ratio that were determined among several peptides from either two or three independent immunoprecipitation experiments. S. D., standard deviations.
Immunoprecipitation of EscL-2HA showed significant interaction with the LEE-encoded proteins EscN and EscQ. Nine unique peptides from EscN and four unique peptides from EscQ were identified. All peptides showed a very low H/L ratio (Fig. 2B, right panel), indicating specific binding to EscL. Peptides from nonspecific binding proteins, such as the iron transporter EfeB, showed an H/L ratio near 1.0 (Fig. 2B, left panel). Furthermore, immunoprecipitation of EscN-2HA identified interactions with EscL and EscQ whereas immunoprecipitation of EscQ-2HA identified peptides of EscL and EscN (summarized in Fig. 2C). Overall, the SILAC results suggested formation of a ternary protein complex, EscL-EscN-EscQ. The H/L ratios of these interactions were very low (<0.1), suggesting that the interactions were specific and probably strong. Interestingly, no other component of the T3SS was identified in the SILAC screen. This result is in agreement with a recent report that failed to identify chaperones or proteins associated with either YscN or YscL (2) but is inconsistent with previous reports that identified chaperones of the ATPase (6, 32, 33). These conflicting results can be explained by the different experimental setups: interactions with chaperones and associated proteins were observed using purified proteins. In contrast, interactions between T3SS substrates and chaperones are highly regulated and probably transient in vivo and therefore would not be detected by our analysis. Moreover, the C ring must interact with inner-membrane components of the T3SS, but these were not identified in this study. Recently, Johnson et al. showed interactions between CdsD, CdsQ, and CdsL of Chlamydophila pneumoniae, suggesting that the C ring is targeted to the T3S apparatus through a YscD homolog (13).
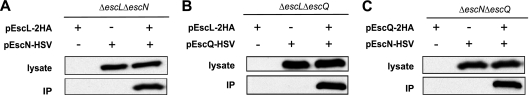
Confirmation of the EscL-EscN-EscQ complex was obtained by coimmunoprecipitation. EPECΔescLΔescN was complemented with pEscL-2HA and pEscN-HSV, EPECΔescLΔescQ with pEscL-2HA and pEscQ-HSV, and EPECΔescQΔescN with pEscQ-2HA and pEscN-HSV. All strains were subcultured at a 1:40 dilution in Dulbecco's modified Eagle's medium (DMEM) and grown for 3 h under T3S-inducing conditions, including centrifugation at a relative centrifugation force (RCF) of 5,000 for 10 min, and resuspended in 1 ml of lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 3 mM MgCl2, 1 mM CaCl2, 2 mM β-mercaptoethanol, protease inhibitor cocktail). The cells were lysed using a probe sonicator, and then 0.1% NP-40 was added. Unlysed bacteria were removed by centrifugation for 10 min at an RCF of 8,000, and the supernatant representing the cleared lysate was collected. Cell lysates were precleaned on protein A-Sepharose beads (GE Healthcare) by incubation on a rotary wheel for 30 min at 4°C. The beads were removed by centrifugation for 4 min at an RCF of 1,000, and the precleared lysates were transferred to a clean tube. For the immunoprecipitation, 5 μg of anti-HA antibody and 20 μl of washed protein A-Sepharose beads were added simultaneously to the lysates. Samples were incubated with rotating for 2 h at 4°C, and then the beads were pelleted by centrifugation for 4 min at an RCF of 1,000, followed by three washes in lysis buffer (plus 0.1% NP-40). The washed beads were resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, boiled for 10 min, and subjected to Western blot analysis.
EscN and EscQ, tagged with a herpes simplex virus (HSV) epitope, coeluted with EscL-2HA (Fig. 3A and B), and HSV-tagged EscN coeluted with EscQ-2HA (Fig. 3C). HSV-tagged EscN and EscQ did not show nonspecific binding to Sepharose beads (when expressed in the absence of HA-tagged bait proteins), indicating that the interactions between EscL-EscN, EscL-EscQ, and EscQ-EscN are specific. These results confirm the data obtained by the SILAC method showing that EscL, EscQ, and EscN are all part of the same complex and are in agreement with the idea that the ATPase–C-ring complex of the flagellar system is composed of FliI, FliH, and FliN (19) and that the C ring of the T3SS of Yersina is composed of YscN, YscL, YscK, and YscQ (11). To date, no homolog of YscK has been identified in EPEC.
Fig. 3.
EscL, EscN, and EscQ interact with each other. (A) EscL-2HA and EscN-HSV were expressed alone or together in an EPECΔescLΔescN mutant. Lysates were immunoprecipitated with α-HA antibody. Lysates and eluted fractions (IP) were blotted against HSV. EscN-HSV was immunoprecipitated only in the presence of EscL-2HA. (B) EscL-2HA immunoprecipitated EscQ-HSV by the use of a procedure similar to that described for panel A. (C) EscQ-2HA immunoprecipitated EscN-HSV by the use of a procedure similar to that described for panel A.
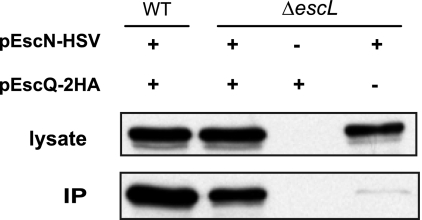
To examine whether EscL targets the EscN ATPase to EscQ, the basal secretion component, we performed coimmunoprecipitation using the EPECΔescL strain expressing both EscN-HSV and EscQ-2HA. Interestingly, we observed that EscN interacts with EscQ in the absence of EscL (Fig. 4). It should be noted that, although we observed similar expression levels of EscN in both EPEC WT and the ΔescL strains, less EscN was detected in the immunoprecipitated fraction in the absence of EscL. This suggests that, although EscL is not essential for EscN-EscQ interaction, it may stabilize the EscN-EscQ complex. Similar results were reported for the flagellar system in Salmonella, where FliI can interact with the export apparatus in the absence of FliH (20), but are contradictory to recent reports that FliH is responsible for efficient localization of the FliI to the C-ring complex (9, 19, 22).
Fig. 4.
EscN-EscQ interaction is independent of EscL. EscN-HSV and EscQ-2HA were individually expressed or coexpressed in WT EPEC and in EPECΔescL strains. Lysates were immunoprecipitated with an α-HA antibody. Lysates and eluted fractions (IP) were immunoblotted against the HSV epitope.
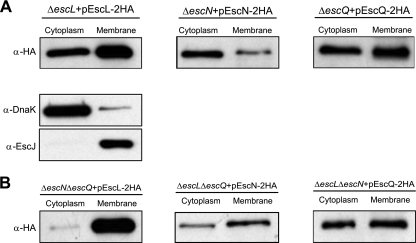
The interaction between EscL, EscN, and EscQ suggests similar localizations of these proteins in the T3SS. Therefore, we expressed EscL, EscN, and EscQ tagged with the C-terminal double-HA tag and examined their subcellular localization. Bacterial cell fractionation was done as previously described (7, 10). We found EscN in both the soluble and membrane fractions (Fig. 5A) as previously reported (7). EscL and EscQ are 27-kDa and 35-kDa proteins, respectively, and are predicted to be cytoplasmic. EscL and EscQ were both found in the soluble fraction as well as associated with the membrane (Fig. 5A). This is in agreement with the predicted homology of EscL to the YscL/FliH family and the homology of EscQ to YscQ/FliN (1, 25). Collectively, EscN, EscL, and EscQ demonstrated similar localizations. Western blots were probed with markers for the soluble protein, DnaK, and the membrane protein, EscJ, to confirm proper fractionation. To determine whether the localization of EscL, EscN, and EscQ to the membrane fraction is intrinsic or requires binding to specific components of the basal body, cell fractionation was performed with strains that expressed EscL-EscN-EscQ in a double-deletion mutant for the other two proteins. As seen in Fig. 5B, the localization profile of EscQ-2HA was not affected in EPECΔescLΔescN compared to the WT. EscL-2HA and EscN-2HA, on the other hand, showed higher membrane association in EPECΔescNΔescQ and in EPECΔescLΔescQ, respectively (Fig. 5B). These results indicate that EscL, EscN, and EscQ are capable of localizing to the membrane regardless of the other complex components. These data support observations reported by Auvray et al. that FliI and FliH, the EscN and EscL homologs, respectively, are intrinsically targeted to the membrane before contacting the secretion machinery (1).
Fig. 5.
Subcellular localization of EscL, EscN, and EscQ. Bacteria expressing tagged versions of EscL, EscN, or EscQ were fractionated into soluble and membrane fractions. Samples (2 μg of each fraction) were resolved by SDS–12.5% PAGE, transferred to a polyvinylidene fluoride membrane, and probed with α-HA antisera. DnaK and EscJ were used as markers for the cytoplasmic and membrane fractions, respectively. (A) Localization was examined in the complemented strains. (B) Localization of the proteins was examined in double-mutant strains.
In this study, we demonstrated that EscL and EscQ are essential for secretion, suggesting that they are part of the secretion apparatus. Localization of EscL and EscQ indicated that these proteins fractionate to both the cytosol and the membrane. These data suggest that these proteins are part of the C-ring complex that assembles in a location adjacent to the inner membrane. Moreover, using the SILAC technique and coimmunoprecipitation experiments, we confirmed formation of a protein complex between EscL, EscN, and EscQ that is similar to the flagellar system and the T3SS in Yersinia. Overall, our results validate the bioinformatics prediction by Pallen et al. whereby a YscQ/FliN domain was identified in SepQ and Orf5 had homology to YscL-FliH (25). Based on our results, we support the suggestion of Pallen et al. to rename SepQ and Orf5 as EscQ and EscL, respectively (25). Interestingly, we found that EscL is not essential for the EscN and EscQ interaction but most likely stabilizes the interaction.
Acknowledgments
We thank members of the Finlay laboratory for critical reading of the manuscript.
Work in B.B.F.'s laboratory is funded by the Canadian Institutes of Health Research (CIHR). The work done in L.J.F.'s laboratory was funded by CIHR (MOP-77688). B.B.F. is the University of British Columbia Peter Wall Distinguished Professor. L.J.F. is the Canada Research Chair in Quantitative Proteomics. E.B.-O. and N.S.-M. are supported by postdoctoral fellowships from the Michael Smith Foundation for Health Research (MSFHR). N.S.-M. is also supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Rothschild Foundation.
Footnotes
Published ahead of print on 29 July 2011.
REFERENCES
- 1. Auvray F., Ozin A. J., Claret L., Hughes C. 2002. Intrinsic membrane targeting of the flagellar export ATPase FliI: interaction with acidic phospholipids and FliH. J. Mol. Biol. 318:941–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blaylock B., Riordan K. E., Missiakas D. M., Schneewind O. 2006. Characterization of the Yersinia enterocolitica type III secretion ATPase YscN and its regulator, YscL. J. Bacteriol. 188:3525–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Deng W., et al. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. U. S. A. 101:3597–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Elliott S. J., et al. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1–4 [DOI] [PubMed] [Google Scholar]
- 5. Fadouloglou V. E., et al. 2004. Structure of HrcQB-C, a conserved component of the bacterial type III secretion systems. Proc. Natl. Acad. Sci. U. S. A. 101:70–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gauthier A., Finlay B. B. 2003. Translocated intimin receptor and its chaperone interact with ATPase of the type III secretion apparatus of enteropathogenic Escherichia coli. J. Bacteriol. 185:6747–6755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gauthier A., Puente J. L., Finlay B. B. 2003. Secretin of the enteropathogenic Escherichia coli type III secretion system requires components of the type III apparatus for assembly and localization. Infect. Immun. 71:3310–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gauthier A., et al. 2005. Transcriptional inhibitor of virulence factors in enteropathogenic Escherichia coli. Antimicrob. Agents Chemother. 49:4101–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. González-Pedrajo B., Minamino T., Kihara M., Namba K. 2006. Interactions between C ring proteins and export apparatus components: a possible mechanism for facilitating type III protein export. Mol. Microbiol. 60:984–998 [DOI] [PubMed] [Google Scholar]
- 10. Ilan O., et al. 1999. Protein tyrosine kinases in bacterial pathogens are associated with virulence and production of exopolysaccharide. EMBO J. 18:3241–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jackson M. W., Plano G. V. 2000. Interactions between type III secretion apparatus components from Yersinia pestis detected using the yeast two-hybrid system. FEMS Microbiol. Lett. 186:85–90 [DOI] [PubMed] [Google Scholar]
- 12. Jerse A. E., Yu J., Tall B. D., Kaper J. B. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. U. S. A. 87:7839–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johnson D. L., Stone C. B., Mahony J. B. 2008. Interactions between CdsD, CdsQ, and CdsL, three putative Chlamydophila pneumoniae type III secretion proteins. J. Bacteriol. 190:2972–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kaniga K., Delor I., Cornelis G. R. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137–141 [DOI] [PubMed] [Google Scholar]
- 15. Knutton S., Lloyd D. R., McNeish A. S. 1987. Adhesion of enteropathogenic Escherichia coli to human intestinal enterocytes and cultured human intestinal mucosa. Infect. Immun. 55:69–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ku C. P., Lio J. C., Wang S. H., Lin C. N., Syu W. J. 2009. Identification of a third EspA-binding protein that forms part of the type III secretion system of enterohemorrhagic Escherichia coli. J. Biol. Chem. 284:1686–1693 [DOI] [PubMed] [Google Scholar]
- 17. Lara-Tejero M., Kato J., Wagner S., Liu X., Galan J. E. 2011. A sorting platform determines the order of protein secretion in bacterial type III systems. Science 331:1188–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marchler-Bauer A., et al. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33:D192–D196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McMurry J. L., Murphy J. W., Gonzalez-Pedrajo B. 2006. The FliN-FliH interaction mediates localization of flagellar export ATPase FliI to the C ring complex. Biochemistry 45:11790–11798 [DOI] [PubMed] [Google Scholar]
- 20. Minamino T., Gonzalez-Pedrajo B., Kihara M., Namba K., Macnab R. M. 2003. The ATPase FliI can interact with the type III flagellar protein export apparatus in the absence of its regulator, FliH. J. Bacteriol. 185:3983–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Minamino T., MacNab R. M. 2000. FliH, a soluble component of the type III flagellar export apparatus of Salmonella, forms a complex with FliI and inhibits its ATPase activity. Mol. Microbiol. 37:1494–1503 [DOI] [PubMed] [Google Scholar]
- 22. Minamino T., et al. 2009. Roles of the extreme N-terminal region of FliH for efficient localization of the FliH-FliI complex to the bacterial flagellar type III export apparatus. Mol. Microbiol. 74:1471–1483 [DOI] [PubMed] [Google Scholar]
- 23. Morita-Ishihara T., et al. 2006. Shigella Spa33 is an essential C-ring component of type III secretion machinery. J. Biol. Chem. 281:599–607 [DOI] [PubMed] [Google Scholar]
- 24. Ong S. E., et al. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1:376–386 [DOI] [PubMed] [Google Scholar]
- 25. Pallen M. J., Beatson S. A., Bailey C. M. 2005. Bioinformatics analysis of the locus for enterocyte effacement provides novel insights into type-III secretion. BMC Microbiol. 5:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paul K., Blair D. F. 2006. Organization of FliN subunits in the flagellar motor of Escherichia coli. J. Bacteriol. 188:2502–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rogers L. D., et al. 2008. Identification of cognate host targets and specific ubiquitylation sites on the Salmonella SPI-1 effector SopB/SigD. J. Proteomics 71:97–108 [DOI] [PubMed] [Google Scholar]
- 28. Sani M., et al. 2007. Structural organization of the needle complex of the type III secretion apparatus of Shigella flexneri. Micron 38:291–301 [DOI] [PubMed] [Google Scholar]
- 29. Schraidt O., et al. 2010. Topology and organization of the Salmonella typhimurium type III secretion needle complex components. PLoS Pathog. 6:e1000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sekiya K., et al. 2001. Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc. Natl. Acad. Sci. U. S. A. 98:11638–11643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Spreter T., et al. 2009. A conserved structural motif mediates formation of the periplasmic rings in the type III secretion system. Nat. Struct. Mol. Biol. 16:468–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thomas J., Stafford G. P., Hughes C. 2004. Docking of cytosolic chaperone-substrate complexes at the membrane ATPase during flagellar type III protein export. Proc. Natl. Acad. Sci. U. S. A. 101:3945–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thomas N. A., et al. 2005. CesT is a multi-effector chaperone and recruitment factor required for the efficient type III secretion of both LEE- and non-LEE-encoded effectors of enteropathogenic Escherichia coli. Mol. Microbiol. 57:1762–1779 [DOI] [PubMed] [Google Scholar]