Abstract
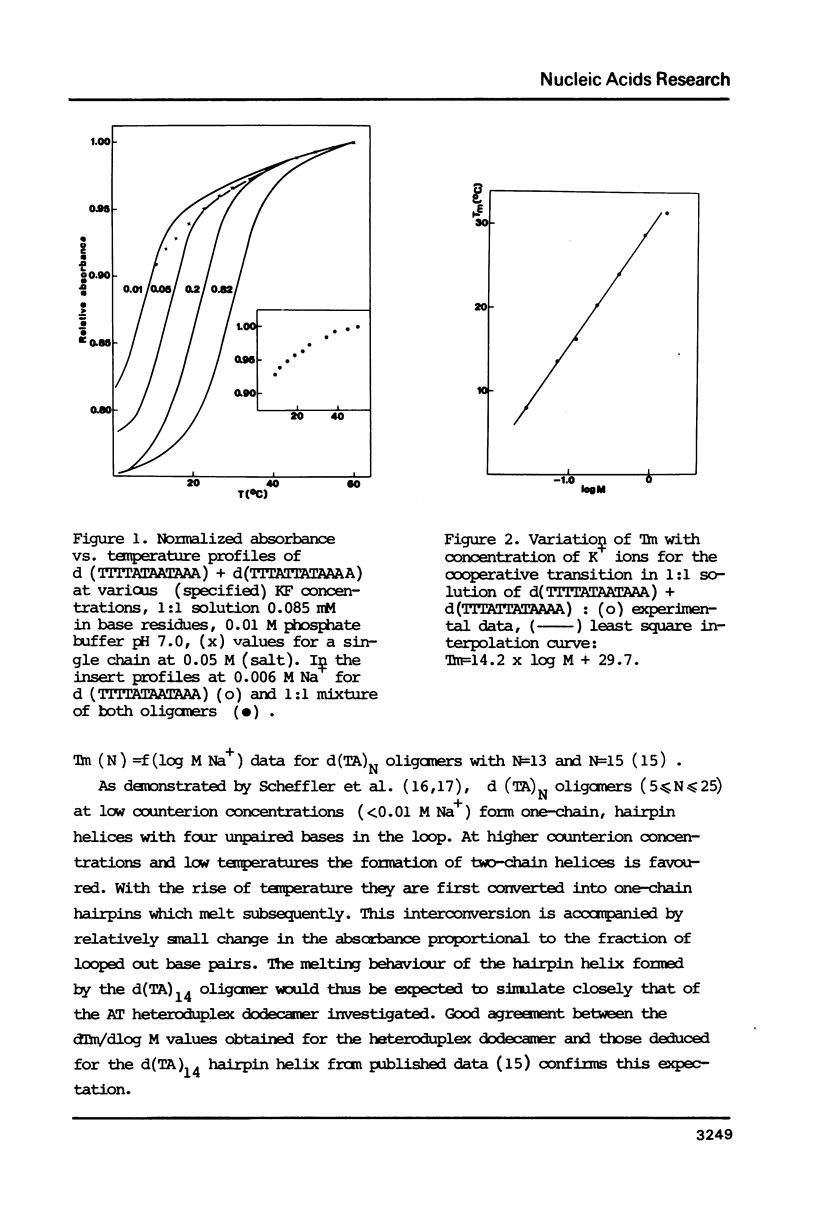
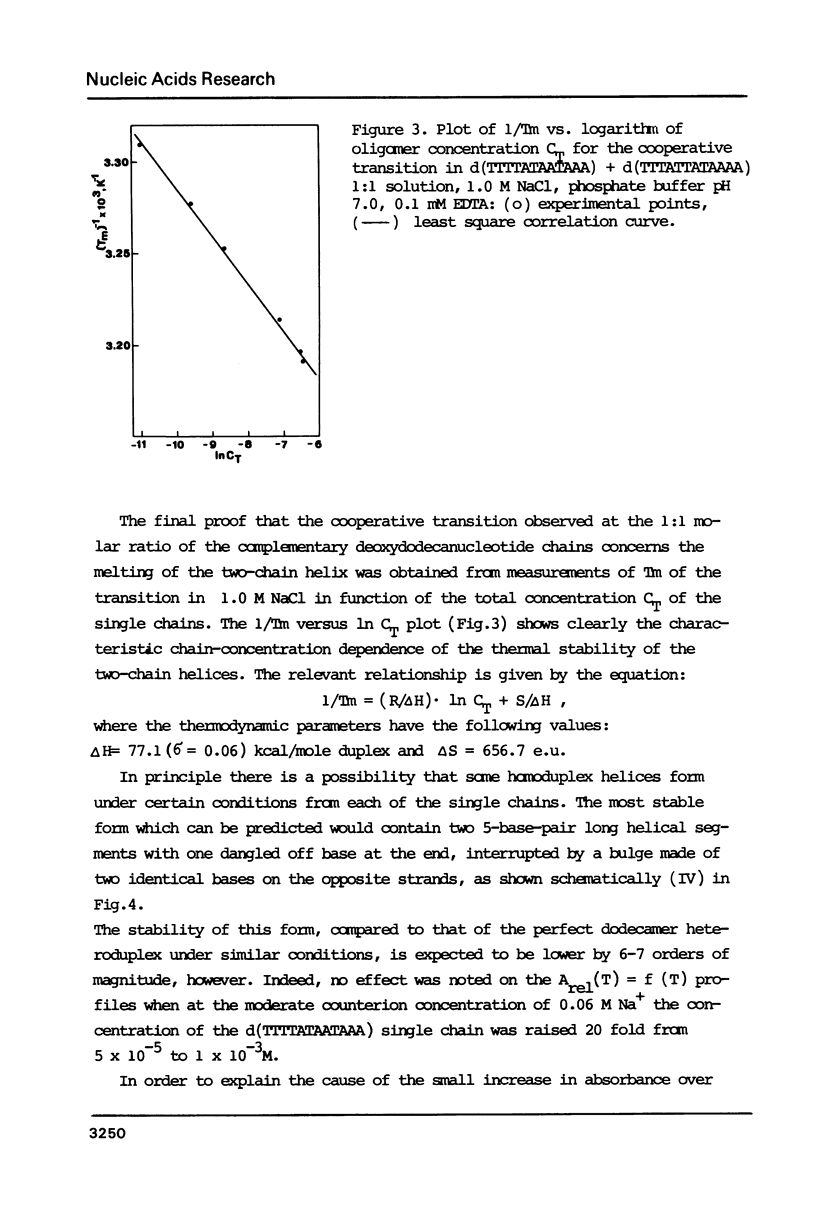
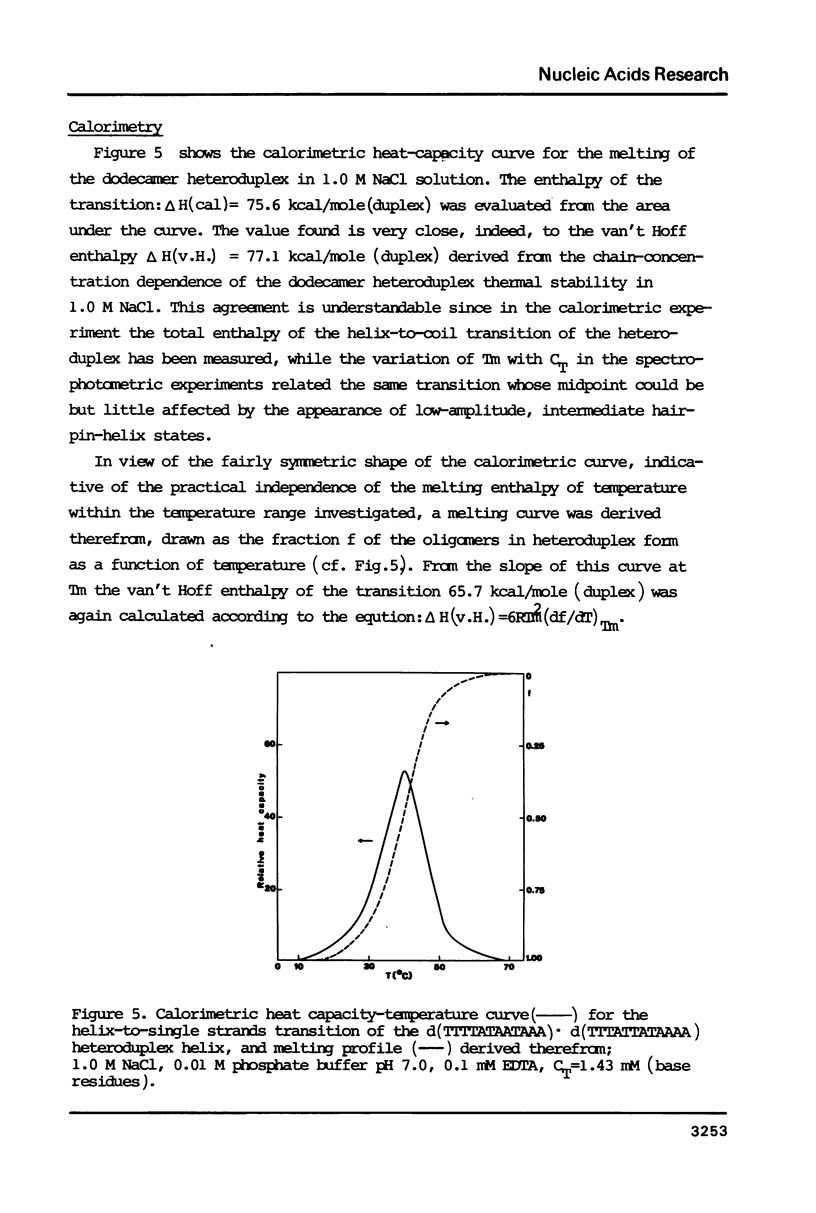
Thermally induced structural transition in the d(TTTTATAATAAA) d(TTTATTATAAAA) heteroduplex is characterized by UV-spectroscopy and differential scanning calorimetry. At low salt (less than 0.1 M) the occurrence of a cooperative transition in the lower temperature range, followed by a broad transition connected with small increase in absorbance is observed. At high salt (greater than or equal to 0.2 M) a single, monophasic transition appears. Linear dependence of the latter on log of salt concentration (dTm:dlogM = 14.2 degrees C) and of 1/Tm on log of oligomer concentration [derived therefrom delta H (v.H.) = 77.1 kcal/mole (duplex)] allows relating it to the melting of the heteroduplex helix. The non-cooperative transition, independent of oligomer concentration and similar to that of the single chain, was attributed to melting of short hairpin helices upon heteroduplex dissociation. Calorimetric enthalpy: 75.6 kcal/mole (duplex) proved significantly lower than predicted from known calorimetric data for poly[d(AT)] and poly d(A) X poly d(T).
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cantor C. R., Warshaw M. M., Shapiro H. Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9(9):1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- Elson E. L., Scheffler I. E., Baldwin R. L. Helix formation by d(TA) oligomers. 3. Electrostatic effects. J Mol Biol. 1970 Dec 28;54(3):401–415. doi: 10.1016/0022-2836(70)90118-x. [DOI] [PubMed] [Google Scholar]
- Gamper H. B., Hearst J. E. A topological model for transcription based on unwinding angle analysis of E. coli RNA polymerase binary, initiation and ternary complexes. Cell. 1982 May;29(1):81–90. doi: 10.1016/0092-8674(82)90092-7. [DOI] [PubMed] [Google Scholar]
- Hsiung H. M., Brousseau R., Michniewicz J., Narang S. A. Synthesis of human insulin gene. Part I. Development of reversed-phase chromatography in the modified triester method. Its application in the rapid and efficient synthesis of eight deoxyribooligonucleotides fragments constituting human insulin A DNA. Nucleic Acids Res. 1979 Apr;6(4):1371–1385. doi: 10.1093/nar/6.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- INMAN R. B., BALDWIN R. L. HELIX--RANDOM COIL TRANSITIONS IN DNA HOMOPOLYMER PAIRS. J Mol Biol. 1964 Apr;8:452–469. doi: 10.1016/s0022-2836(64)80003-6. [DOI] [PubMed] [Google Scholar]
- Itakura K., Katagiri N., Bahl C. P., Wightman R. H., Narang S. A. Improved triester approach for the synthesis of pentadecathymidylic acid. J Am Chem Soc. 1975 Dec 10;97(25):7327–7332. doi: 10.1021/ja00858a020. [DOI] [PubMed] [Google Scholar]
- Jay E., Bambara R., Padmanabhan R., Wu R. DNA sequence analysis: a general, simple and rapid method for sequencing large oligodeoxyribonucleotide fragments by mapping. Nucleic Acids Res. 1974 Mar;1(3):331–353. doi: 10.1093/nar/1.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri N., Itakura K., Narang S. A. The use of arylsulfonyltriazoles for the synthesis of oligonucleotides by the triester approach. J Am Chem Soc. 1975 Dec 10;97(25):7332–7337. doi: 10.1021/ja00858a021. [DOI] [PubMed] [Google Scholar]
- Marky L. A., Blumenfeld K. S., Kozlowski S., Breslauer K. J. Salt-dependent conformational transitions in the self-complementary deoxydodecanucleotide d(CGCAATTCGCG): evidence for hairpin formation. Biopolymers. 1983 Apr;22(4):1247–1257. doi: 10.1002/bip.360220416. [DOI] [PubMed] [Google Scholar]
- Marky L. A., Breslauer K. J. Calorimetric determination of base-stacking enthalpies in double-helical DNA molecules. Biopolymers. 1982 Nov;21(11):2185–2194. doi: 10.1002/bip.360211107. [DOI] [PubMed] [Google Scholar]
- Nelson J. W., Martin F. H., Tinoco I., Jr DNA and RNA oligomer thermodynamics: the effect of mismatched bases on double-helix stability. Biopolymers. 1981 Dec;20(12):2509–2531. doi: 10.1002/bip.1981.360201204. [DOI] [PubMed] [Google Scholar]
- Scheffler I. E., Elson E. L., Baldwin R. L. Helix formation by d(TA) oligomers. II. Analysis of the helix-coli transitions of linear and circular oligomers. J Mol Biol. 1970 Feb 28;48(1):145–171. doi: 10.1016/0022-2836(70)90225-1. [DOI] [PubMed] [Google Scholar]
- Scheffler I. E., Elson E. L., Baldwin R. L. Helix formation by dAT oligomers. I. Hairpin and straight-chain helices. J Mol Biol. 1968 Sep 28;36(3):291–304. doi: 10.1016/0022-2836(68)90156-3. [DOI] [PubMed] [Google Scholar]
- Siebenlist U. RNA polymerase unwinds an 11-base pair segment of a phage T7 promoter. Nature. 1979 Jun 14;279(5714):651–652. doi: 10.1038/279651a0. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Stawinski J., Hozumi T., Narang S. A., Bahl C. P., Wu R. Arylsulfonyltetrazoles, new coupling reagents and further improvements in the triester method for the synthesis of deoxyribooligonucleotides. Nucleic Acids Res. 1977 Feb;4(2):353–371. doi: 10.1093/nar/4.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswamitra M. A., Shakked Z., Jones P. G., Sheldrick G. M., Salisbury S. A., Kennard O. Structure of the deoxytetranucleotide d-pApTpApT and a sequence-dependent model for poly(dA-dT). Biopolymers. 1982 Mar;21(3):513–533. doi: 10.1002/bip.360210304. [DOI] [PubMed] [Google Scholar]


