Abstract
Bacillus subtilis is known to accumulate large amounts of the compatible solute proline via de novo synthesis as a stress protectant when it faces high-salinity environments. We elucidated the genetic determinants required for the osmoadaptive proline production from the precursor glutamate. This proline biosynthesis route relies on the proJ-encoded γ-glutamyl kinase, the proA-encoded γ-glutamyl phosphate reductase, and the proH-encoded Δ1-pyrroline-5-caboxylate reductase. Disruption of the proHJ operon abolished osmoadaptive proline production and strongly impaired the ability of B. subtilis to cope with high-osmolarity growth conditions. Disruption of the proA gene also abolished osmoadaptive proline biosynthesis but caused, in contrast to the disruption of proHJ, proline auxotrophy. Northern blot analysis demonstrated that the transcription of the proHJ operon is osmotically inducible, whereas that of the proBA operon is not. Reporter gene fusion studies showed that proHJ expression is rapidly induced upon an osmotic upshift. Increased expression is maintained as long as the osmotic stimulus persists and is sensitively linked to the prevalent osmolarity of the growth medium. Primer extension analysis revealed the osmotically controlled proHJ promoter, a promoter that resembles typical SigA-type promoters of B. subtilis. Deletion analysis of the proHJ promoter region identified a 126-bp DNA segment carrying all sequences required in cis for osmoregulated transcription. Our data disclose the presence of ProA-interlinked anabolic and osmoadaptive proline biosynthetic routes in B. subtilis and demonstrate that the synthesis of the compatible solute proline is a central facet of the cellular defense to high-osmolarity surroundings for this soil bacterium.
INTRODUCTION
The soil-dwelling bacterium Bacillus subtilis (20) is frequently exposed to osmotic fluctuations in its environment as a consequence of wetting and drying cycles of the upper layers of the soil (8). As a result of these changes in the osmolarity and salinity of the habitat, the B. subtilis cell has to cope with osmotically instigated water fluxes across the cytoplasmic membrane. Consequently, the integrity of the cell is threatened under hypo-osmotic conditions, or its growth is impaired under hyperosmotic conditions (8). No microorganism can actively pump water in or out of the cell to compensate for water fluxes caused by changes in the external osmotic condition. Instead, microorganisms determine the direction and scale of water permeation across the cytoplasmic membrane indirectly by actively controlling the osmotic potential of their cytoplasm (9, 66). This is often accomplished under high-osmolarity conditions by first importing large amounts of potassium as an emergency stress reaction (9, 41, 66). Subsequently, the cell replaces part of the accumulated potassium by a distinct class of highly water-soluble organic compounds, the compatible solutes (11). These osmolytes have been specifically selected in the course of evolution as effective osmo- and cytoprotectants in all three kingdoms of life and are highly congruous with metabolism and other physiological functions of the cell (12, 15, 30, 32, 41, 66, 67).
Turgor is considered essential for cell expansion and growth (42), and Whatmore and Reed (65) have determined the turgor pressure of B. subtilis to a value of ∼1.9 MPa. Whatmore et al. (64) found that turgor was strongly reduced in B. subtilis cells immediately after an osmotic up-shock due to rapid water outflow from the cell. Its subsequent recovery by water influx was dependent on the prior accumulation of potassium ions (64), and the KtrAB and KtrCD potassium uptake systems are critically involved in this process (31). Prolonged high levels of potassium ions are detrimental to cellular physiology and are therefore not a sustainable way for the cell to adjust to high-osmolarity surroundings. B. subtilis therefore reduces the initially increased cellular potassium pool by accumulating compatible solutes (e.g., proline and glycine betaine) and concomitantly exporting potassium (64). In this way, the ionic strength of the cytoplasm is reduced without compromising turgor. One of the potassium extrusion systems operating in B. subtilis is the cation/proton antiporter YhaTU (22), whose structural genes (yhaSTU) are upregulated in response to a salt shock (25, 59).
B. subtilis makes intensive use of compatible solutes to adjust its physiology to sustained high-osmolarity environments (8). It is equipped with a set of five high-affinity uptake systems, the Opu family of transporters, for the scavenging of a broad spectrum of compatible solutes from scarce environmental sources (30, 32, 36–40, 47, 62). The trimethylammonium compound glycine betaine and the amino acid proline are particularly important osmostress protectants for B. subtilis since they can be synthesized (5, 6, 39, 64), as well as taken up by osmotically stressed cells (38–40, 62).
In the absence of an exogenous supply of preformed compatible solutes (8) or the precursor (choline) for glycine betaine synthesis (5, 6, 39), B. subtilis is left to its own devices for cellular adjustment to high-osmolarity surroundings. Under such growth conditions, large amounts of the compatible solute proline are produced by B. subtilis (46, 64). Detailed physiological studies conducted by Whatmore et al. (64) revealed that the cellular proline pool rose from a basal level of 16 mM to about 500 to 700 mM via de novo synthesis within 7 h of growth subsequent to a moderate osmotic upshift with 0.4 M NaCl. Proline is a compatible solute that is widely used in nature by both pro- and eukaryotic cells to fend off the detrimental effects of high osmolarity on cell physiology (15, 41, 66). It also exhibits protein-stabilizing properties and promotes protein folding under unfavorable conditions (21, 35, 60).
In many microorganisms, proline biosynthesis proceeds from the precursor glutamate and involves three enzyme-catalyzed steps (Fig. 1D) (16). Synthesis of proline begins with the ATP-dependent phosphorylation of glutamate by the γ-glutamyl kinase (ProB). The resulting γ-glutamyl phosphate is then reduced to glutamate-5-semialdehyde by the proA-encoded γ-glutamyl phosphate reductase (ProA). The ProB and ProA enzymes likely form a complex, thereby preventing the release of the unstable intermediate γ-glutamyl-phosphate into the cytoplasm (57). Glutamate-5-semialdehyde cyclizes spontaneously to Δ1-pyrroline-5-carboxylate, and this compound is then reduced by the Δ1-pyrroline-5-carboxylate reductase (ProC) to the end product, proline (Fig. 1D). The anabolic proline biosynthesis pathway in microorganisms is frequently regulated biochemically through allosteric feedback inhibition of the activity of the first proline-biosynthetic enzyme (ProB) by the end product proline (16, 23, 45, 49–51).
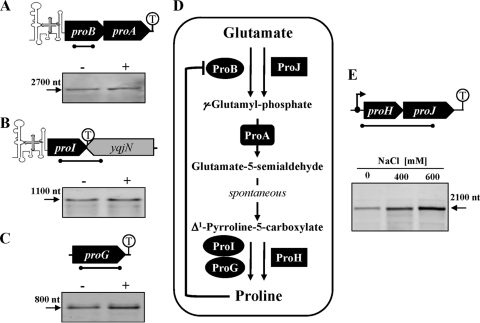
Fig. 1.
Anabolic and osmoadaptive pathways for the synthesis of proline from the precursor glutamate in B. subtilis. The transcriptional organization of the proBA operon (A), the proI (B) and proG (C) genes, and the proHJ operon (E) are shown and the T-box element present in the proBA and proI loci is indicated. The transcription of the proline biosynthetic genes was analyzed by Northern blot analysis. Total RNA was isolated from cultures of B. subtilis wild-type JH642 that were grown in SMM either in the absence (−) or the presence of 0.4 M NaCl (+). Specific RNA transcripts were identified by hybridization to DIG-labeled single-stranded antisense RNA probes. (E) Northern blot analysis of the transcriptional induction of the proHJ operon in response to increasing osmolarity. Cultures of wild-type JH642 were grown in SMM without NaCl or in the presence of NaCl (400 or 600 mM), and the proHJ mRNA was detected as described above. Localization of the single-stranded antisense RNAs used as probes in the Northern blot analysis are indicated as black bars below the gene symbols. (D) Anabolic and osmoadaptive proline biosynthetic routes of B. subtilis. The ProG protein has Δ1-pyrroline-5-carboxylate reductase activity, but its physiological role in connection with proline biosynthesis in B. subtilis is unclear (4).
Inspection of the B. subtilis genome sequence (3, 44) and mutant studies conducted by Belitsky et al. (4) revealed the presence of two proline biosynthetic routes that are interconnected via the γ-glutamyl phosphate reductase (ProA) (Fig. 1D). Paralogous enzymes for the first (the γ-glutamyl kinases ProB and ProJ) and last (Δ1-pyrroline-5-carboxylate reductases ProI, ProG, and ProH) steps of proline biosynthesis (Fig. 1D) are present in B. subtilis, but no additional ProA-related protein can be found (4). The expression of the proBA and proI loci encoding the enzymes for the anabolic proline biosynthesis route (ProB-ProA-ProI) is genetically controlled through a T-box system, an RNA-based regulatory device that keeps proBA and proI expression low until cellular starvation for proline sets (described in reference 10). In addition, the activity of the ProB enzyme from B. subtilis is sensitively regulated through feedback inhibition (14). Hence, both transcriptional and posttranscriptional control systems tie the anabolic proline biosynthesis route to the protein biosynthetic activities of the B. subtilis cell. As a consequence, a high-level buildup of the cellular proline pool is prevented, and a wasteful overproduction for this energetically costly to synthesize amino acid (1) is avoided.
It is apparent from the above-described genetic (10) and biochemical (14) regulatory mechanisms that the anabolic ProB-ProA-ProI proline biosynthetic route (Fig. 1D) is unsuited to providing the very large amounts of proline that are found in osmotically stressed B. subtilis cells (64). Therefore, the following two questions arise: (i) how does B. subtilis synthesize proline when it is osmotically challenged, and (ii) does the production of this compatible solute actually confer osmostress protection to the B. subtilis cell, as one would predict? The data presented here unravel the molecular details of the osmoadaptive proline biosynthesis route and demonstrate that proline production is a physiologically very important facet of the cellular adjustment process of B. subtilis to high-osmolarity surroundings.
MATERIALS AND METHODS
Chemicals.
Glycine betaine, proline, and the chromogenic substrate for the TreA enzyme, para-nitrophenyl-α-d-glucopyranoside (α-PNPG), were purchased from Sigma-Aldrich (Steinheim, Germany). The antibiotics kanamycin, erythromycin, spectinomycin, tetracycline, ampicillin, and chloramphenicol were purchased from Sigma-Aldrich (Steinheim, Germany). Zeocin (used for the selection of B. subtilis strains carrying a ble resistance cassette) was purchased from Invitrogen (Carlsbad, CA). [γ-32P]ATP was purchased from Amersham Pharmacia Biotech (Freiburg, Germany), and high-pressure liquid chromatography (HPLC)-grade solvents were purchased from Carl Roth (Karlsruhe, Germany). FMOC (9-Fluorenylmethoxycarbonyl-chloride) and 1-adamanatanamine (ADAM) for amino acid derivatization were obtained from Grom Analytic (Rottenburg-Hailfingen, Germany).
Media and growth conditions.
Escherichia coli and B. subtilis strains were routinely maintained and propagated on Luria-Bertani (LB) agar plates. Spizizen minimal medium (SMM) with 0.5% (wt/vol) glucose as the carbon source, l-tryptophan (20 mg liter−1), l-phenylalanine (18 mg liter−1), and a solution of trace elements (26) was used as defined medium for the growth of B. subtilis strains derived from the wild-type strain JH642 (Table 1). Bacterial growth was monitored spectrophotometrically as the optical density at 578 nm (OD578). The osmotic strength of SMM was increased by the addition of NaCl, KCl, sucrose, lactose, and glycerol from concentrated stock solutions. The osmolarity values of these media were determined with a vapor pressure osmometer (Vapor Pressure 5500; Wescor, Inc., Utah). The osmolarity of SMM is 340 mosmol kg of water−1. For experiments in which we continuously monitored the growth of B. subtilis cultures, we inoculated 75 ml of prewarmed minimal medium in 500-ml Erlenmeyer flasks to an OD578 of 0.1 and grew these cultures aerobically at 37°C in a shaking water bath set at 220 rpm. For experiments where we wanted to determine the growth yield of cultures propagated at various osmolarities, we inoculated 75 ml of prewarmed medium in 500-ml Erlenmeyer flasks to an OD578 of 0.05 and grew these cultures for 17 h. For osmotic up-shock experiments, 75-ml cultures of B. subtilis proH-treA fusion strains were grown to mid-log phase (OD578 = 0.4), and the osmolarity of the cultures was then suddenly increased by the addition of 0.4 M NaCl from a stock of NaCl dissolved in SMM. For long-term osmotic stress experiments, cells of proH-treA fusion strains were pregrown overnight in SMM at various osmolarities; aliquots were then used to inoculate 75-ml SMM cultures (in 500-ml Erlenmeyer flasks) with the appropriate osmolarity and were grown to mid-log phase (OD578 = 0.5 to 0.8) on an orbital shaker (220 rpm) at 37°C. The antibiotics chloramphenicol, kanamycin, tetracycline, erythromycin, spectinomycin, and zeocin were used with B. subtilis strains at final concentrations of 5, 10, 15, 1, 100, and 35 μg ml−1, respectively. Ampicillin was used at a final concentration of 100 μg ml−1 for E. coli cultures. The production of the extracellular α-amylase AmyE by B. subtilis strains was tested by flooding colonies grown on LB plates containing 1% starch with Gram's iodine stain (0.5% [wt/vol] iodine, 1% potassium iodide) for 1 min and scoring for zones of clearing around the colonies after the strain was decanted (17).
Table 1.
B. subtilis strains used in this study
| Straina | Relevant genotype | Source or referenceb |
|---|---|---|
| JH642 | trpC2 pheA1 | J. Hoch (BGSC 1A96) |
| BB1951 | SMY Δ(proG::ble)1 | 4 |
| FSB1 | Δ(treA::neo)1 | 58 |
| JSB7 | amyE::[Φ(proH474bp′-treA)1 cat] Δ(treA::neo)1 | This study |
| JSB8 | Δ(proHJ::tet)1 | 10 |
| JSB9 | Δ(proI::spc)1 | This study |
| JSB11 | Δ(proBA::cat)1 | This study |
| JSB13 | Δ(proHJ::tet)1 Δ(proI::spc)1 | This study |
| JSB15 | Δ(proJ::neo)1 | This study |
| JSB17 | proA × pEPV1T (tet) | This study |
| JSB19 | proH × pJS23 (erm) | This study |
| JSB36 | amyE::[Φ(proH153bp′-treA)3 cat] Δ(treA::neo)1 | This study |
| JSB1951 | Δ(proG::ble)1 | This study |
| MDB47 | amyE::[Φ(proH126bp′-treA)2 cat] Δ(treA::neo)1 | This study |
| TRB0 | Δ(treA::neo)1 amyE::[Φ(treA) cat] | 58 |
All strains, except BB1951, are derivatives of the B. subtilis wild-type strain JH642 and therefore also carry the trpC2 pheA1 mutations.
BGSC, Bacillus Genetic Stock Center.
Bacterial strains and construction of B. subtilis mutants.
The E. coli strain DH5α (Invitrogen, Carlsbad, CA) was used for the routine propagation of plasmids. The B. subtilis strains used in the present study are all derivatives of the wild-type strain JH642 (trpC2 pheA1; BGSC 1A96), kindly provided by J. Hoch, Scripps Research Institute (Table 1). The genetic construction of these strains is described in the supplemental material. The genotypes for all of the B. subtilis strains used in the present study are given in Table 1.
Methods used with nucleic acids and construction of plasmids.
Routine manipulations of plasmid DNA, PCR, the construction of recombinant DNA plasmids, the isolation of chromosomal DNA from B. subtilis, and the detection of homologous sequences by Southern hybridization using digoxigenin (DIG)-labeled DNA probes were all carried out according to standard procedures (54). The nucleotide sequence of cloned PCR proH′ fragments was verified by DNA sequence analysis using the chain termination method and a Thermo Sequenase fluorescence-labeled primer cycle sequencing kit (Amersham Pharmacia Biotech, Freiburg, Germany). The DNA sequencing reactions were primed with synthetic oligonucleotides labeled at their 5′ ends with the infrared dye IRD-800 (Eurofins MWG Operon, Ebersberg, Germany) and analyzed on a LI-COR DNA sequencer (model 4000; Eurofins MWG Operon). The construction of the plasmids used here is described in the supplemental material.
Northern blot analysis.
Total RNA was isolated from log-phase (OD578 = 0.5 to 0.8) cultures (1 ml) of the B. subtilis wild-type strain JH642 grown in SMM or SMM with NaCl (0.4 or 0.6 M) using a Total RNA minikit (Qiagen, Hilden, Germany). Then, 4 to 10 μg of total RNA was electrophoretically separated on a 1.4% agarose gel, transferred to a Schleicher & Schuell NY13N membrane, and hybridized with DIG-labeled single-strand RNA probes specific for proHJ, proBA, proI, or proG. Single-stranded antisense RNA probes were prepared from plasmids pJS22 (proHJ), pFSB66 (proBA), and pJS18 (proI) using a DIG RNA labeling kit (SP6/T7; Roche Diagnostics GmbH, Mannheim, Germany) with either T7 RNA polymerase (for plasmids pJS22 and pFSB66) or T3 RNA polymerase (for plasmid pJS18). For the preparation of a proG-specific antisense RNA probe, a 618-bp PCR-fragment covering part of the proG coding region was generated with synthetic oligonucleotides that carried an artificially added T7 promoter sequence to the 5′ end of the reverse primer (32). T7 RNA polymerase and a DIG-RNA labeling kit (Roche Diagnostics) were then used to prepare the DIG-labeled proG-specific antisense RNA probe. RNA-RNA hybridization and the detection of specific transcripts were performed according to the DIG hybridization and detection procedure described by the manufacturer (Roche Diagnostics).
Primer extension analysis.
For primer extension analysis of the proHJ genes, total RNA was isolated using the acid phenol extraction method from the B. subtilis JH642 wild-type strain (20-ml cultures) grown either in SMM or in SMM with 0.4 M NaCl to log phase (OD578 = 0.8). A reverse transcriptase reaction was carried out with 15 μg of total RNA and ∼2 pmol of a 32P-5′-end-labeled specific oligonucleotide (5′-GTAACGCAGATGTTTTGTTTCGGG-3′; bp 96 to 119 in the proH coding region). The RNA and the oligonucleotide were resuspended in 5 μl of reverse transcriptase buffer (50 mM Tris-HCl [pH 8.3], 10 mM MgCl2, 80 mM KCl), heated to 70°C for 4 min and then placed in ethanol-dry ice. After thawing on ice, 5 μl of a mixture containing 2 mM dATP, 2 mM dCTP, 2 mM dGTP, 2 mM dTTP, and 8 mM dithiothreitol in reverse transcriptase buffer and 4 U of avian myeloblastosis virus reverse transcriptase (Eurogentec, Cologne, Germany) were added. The samples were incubated for 30 min at 48°C, precipitated, washed, dried, resuspended in 12 μl of formamide loading buffer–Tris-EDTA (1:2), and run on a denaturing 5% polyacrylamide gel. A DNA sequencing ladder produced by using the same primer with plasmid pJS13 as a template was run in parallel to determine the exact position of the 5′ ends of the proHJ mRNA.
TreA activity assays.
An aliquot (0.6 ml) from cultures of proH-treA B. subtilis fusion strains was harvested by centrifugation for 5 min in an Eppendorf microcentrifuge and resuspended in 0.25 ml of Z-buffer (24) adjusted to pH 6.0 and containing 1 mg of lysozyme ml−1. After incubation for 5 min at 37°C in an Eppendorf Thermomixer, cellular debris was removed by centrifugation, and the supernatant was then used for TreA activity assays with para-nitrophenyl-α-d-glucopyranoside as the substrate (24, 58). TreA [phospho-α-(1,1)-glucosidase] specific activity is expressed in units per mg of protein. Protein concentrations were determined using the Bio-Rad protein assay with acetylated bovine serum albumin as the standard.
HPLC analysis of proline and glutamate.
Cells for quantitative HPLC analysis of the amino acids proline and glutamate were grown in SMM of different osmolarities. The cells were harvested by centrifugation when they had reached an OD578 between one and two and were subsequently lyophilized. The dry weight (DW) of the cell material was determined, and the cells were extracted as described by Kuhlmann and Bremer (43). The extracts containing the cytoplasmic amino acids were reacted with FMOC, and HPLC analysis of the derivatization mixtures was performed as described previously (43) using the HPLC system from SYKAM (Gilching, Germany). The amounts of fluorescently labeled amino acids were monitored by using a fluorescence detector (SYKAM, model S3400) set at an excitation wavelength of 245 nm and an emission wavelength of 316 nm.
Natural abundance 13C-NMR spectroscopy.
The presence of dominant organic osmolytes in the cytoplasm of the B. subtilis wild-type strain JH642 and its proHJ mutant JSB8 was monitored by using natural-abundance 13C-nuclear magnetic resonance (13C-NMR) spectroscopy. For these experiments, the cells (300-ml cultures) were propagated in SMM or SMM with 0.8 NaCl until they reached an OD578 of 1.5. The cells were then harvested by centrifugation, the cell pellet was extracted with 10 ml of 80% (vol/vol) ethanol, cellular debris was removed by centrifugation, and the supernatant was evaporated to dryness. Ethanolic cell extracts from three 300-ml cultures were then dissolved in 1 ml of 2H2O supplemented with 1.2 mg of D4-3-(trimethylsilyl)propionate as a standard. 13C-NMR spectra were recorded with a Bruker AC300 spectrometer operating at 75 MHz (47).
Database searches and alignments of amino acid sequences.
Proteins that are homologous to the proline biosynthetic enzymes of B. subtilis were searched for via the web server of the DOE Joint Genome Institute (JGI; http://www.jgi.doe.goc/) or that of the National Center for Biotechnology Information Institute (http://www.ncbi.nlm.nih.gov/) using the BLAST algorithm (2). Protein sequences were aligned and analyzed by using CLUSTAL W (61).
RESULTS
Osmotically induced proline biosynthesis.
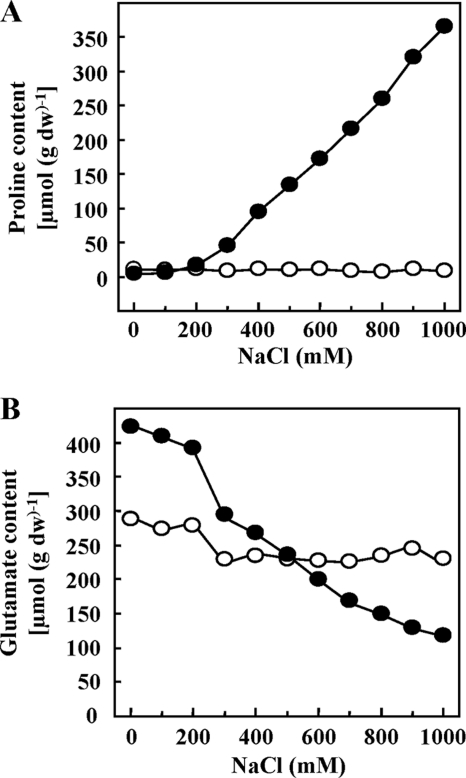
To investigate the correlation between the osmolarity of the growth medium and the buildup of the intracellular proline pool in B. subtilis via de novo synthesis under osmotic stress conditions (64), we grew cultures of the wild-type strain JH642 in a minimal medium (SMM) with increasing NaCl concentrations to an OD578 of 2 and then determined the proline content of the cells by HPLC analysis. This experiment revealed that the intracellular proline concentration is proportionally linked to the osmolarity of the growth medium over a considerable range of salt concentrations (Fig. 2A). The proline content increased from a basal level of 8 μmol per g (dry weight) in cells that were cultivated in SMM to a proline content of 362 μmol per g (dry weight) in cells that were cultivated in SMM with 1 M NaCl, a 45-fold rise in proline content in osmotically stressed cells (Fig. 2A). We also monitored the glutamate level in these cells by HPLC analysis and found that it fell as the osmolarity of the growth medium was raised (Fig. 2B). Apparently, the strongly increased proline production observed under hypertonic growth conditions progressively drains the cell of the proline precursor glutamate (Fig. 1D). However, one needs also to consider that glutamate is not an effective compatible solute for bacilli (19, 43), and therefore it is possible that the B. subtilis cell prefers proline over glutamate as an osmolyte and begins to convert glutamate into proline as soon as the osmotic stress sets in (Fig. 2B).
Fig. 2.
Proline and glutamate content of osmotically stressed B. subtilis cells. Cultures of the B. subtilis wild-type strain JH642 (●) and its Δ(proHJ::tet)1 mutant derivative JSB8 (○) were grown in SMM with different salinities to an optical density (OD578) of ∼2. The proline (A) and glutamate (B) content of cell extracts was determined by HPLC analysis. The values presented are the averages of three independently grown cultures. For each sample analyzed, the proline and glutamate content was determined twice. Standard deviations were typically ca. 10%.
Glutamate dominates the amino acid pool of B. subtilis, and Whatmore et al. (64) reported a value of ∼100 mM in osmotically nonstressed cells that were cultivated in a minimal medium at 25°C. These authors also reported a moderate increase in the glutamate pool (from 100 to 160 mM) within 7 h of growth of B. subtilis cells subjected to an osmotic upshift with 0.4 M NaCl (64). We did not observe such an increase (Fig. 2B), and these differences might be explained either by the bacterial strains used or by the fact that we analyzed cells that were fully adapted to growth at 37°C at high salinity, whereas Whatmore et al. (64) studied cells cultivated at 25°C that were subjected to an osmotic up-shock.
The proHJ operon and the proA gene are essential for osmoadaptive proline biosynthesis.
Our finding that the proline content of osmotically stressed B. subtilis cells is strongly increased raised the question regarding what enzymes were responsible for the increased proline production under these growth conditions. As outlined in Fig. 1D, paralogous enzymes for the first (ProB and ProJ) and third (ProI, ProG, and ProH) steps of proline biosynthesis are present in B. subtilis, whereas only one enzyme (ProA) is present that catalyzes the second step in the proline biosynthetic pathway from the precursor glutamate (4). The ProB and ProJ γ-glutamyl kinases exhibit a degree of amino acid sequence identity of 43%; the ProI and ProH Δ1-pyrroline-5-carboxylate reductases have a similar level of amino acid identity (42%), but the amino acid identities of ProH and ProI to ProG are only 24 and 21%, respectively. The genes that might be involved in the osmoadaptive proline biosynthesis route were suggested by genome-wide transcriptional profiling studies of salt-stressed B. subtilis cells that showed a strong increase in the level of proHJ expression and unchanged transcription of the other pro genes (25, 59; see below and Fig. 1).
To identify the genes required for the osmoregulatory synthesis of proline, we carried out gene disruption experiments in the various pro loci, determined the Pro growth phenotype of the resulting mutants on minimal agar plates, and tested their ability to accumulate proline via de novo synthesis at elevated osmolarity (SMM with 0.4 M NaCl) through HPLC analysis (Table 2). Deletion of the proHJ locus (strain JSB8) or the individual disruption of the proJ (strain JSB15) or the proH (strain JSB19) genes resulted in B. subtilis mutants that were prototrophic for growth in the absence of proline, but none of these strains was able to accumulate proline via de novo synthesis under hyperosmotic growth conditions (Table 2). Hence, the proHJ locus is instrumental for the osmoadaptive de novo production of proline, but it is not required for the anabolic synthesis of this amino acid.
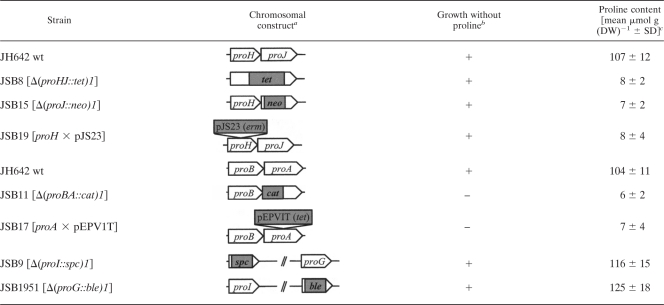
Table 2.
Influence of mutations in the proHJ, proBA, proI, and proG loci on cell growth in the absence of proline and their effects on intracellular proline accumulation under osmotic stress
Genetic organization of the proHJ, proBA, proI, and proG loci of the B. subtilis wild-type strain JH642 and its mutant derivatives is shown schematically. The cat, neo, tet, spc, and ble antibiotic resistance cassettes and the plasmids pJS23 and pEPV1T inserted into the various pro genes are not drawn to scale. In strain JSB19, plasmid pJS23 was inserted via a single Campbell-type recombination event into the proH gene.
The growth of the various mutant strains was determined on minimal agar plates (SMM) without proline after 2 days of incubation at 37°C (+, growth; –, no growth). Disruption of the proBA operon or of the proA gene does not yield a tight proline auxotrophic growth phenotype since such mutant strains form microcolonies on proline-free minimal agar plates with glucose as the carbon source.
The proline content of cells grown in SMM containing 0.4 M NaCl was measured by HPLC analysis. Strains JSB11 and JSB17 are proline auxotrophs and were grown in SMM with 0.4 M NaCl containing 0.4 mM proline; at this concentration of proline, there is no significant osmoprotective effect on cell growth (data not shown). The data presented are the means of two independently grown cultures. The proline concentration of each sample was determined twice. DW, dry weight.
Disruption of the proBA genes by insertion of a chloramphenicol resistance cassette (strain JSB11) or the destruction of the proA gene via a Campbell-type single-crossover event with plasmid pEPV1T (strain JSB17) yielded a proline auxotrophic growth phenotype of the resulting mutant strains (Table 2). When we partially satisfied the proline auxotrophy of strains JSB11 and JSB17 by the addition of 0.4 mM proline to the minimal medium (10) and then grew these strains in SMM with 0.4 M NaCl, we found that both mutants were unable to accumulate proline via de novo synthesis. Since the proBA locus is organized as an operon (10) (Fig. 1A), we conclude the proA-encoded γ-glutamyl phosphate reductase is required for both the anabolic and the osmoregulatory production of proline (Table 2). This is consistent with the previous finding that no protein functionally related to the ProA enzyme is present in B. subtilis (4).
The disruption of the proI or proG genes did not impair the ability of B. subtilis to produce large quantities of proline in high-salinity media (Table 2). Such mutants also did not cause a proline-auxotrophic growth phenotype (Table 2), since we expected from the report of Belitsky et al. (4) that multiple enzymes (ProI, ProG, and ProH) could catalyze the last step of proline biosynthesis through the conversion of Δ1-pyrroline-5-carboxylate to proline (Fig. 1D).
Taken together, these gene disruption experiments demonstrate that the ProJ-ProA-ProH enzymes mediate the synthesis of the compatible solute proline in osmotically stressed B. subtilis cells (Fig. 1D). The iso-enzymes of ProJ (ProB) and ProH (ProI and ProG) are not involved in this osmostress responsive process (Fig. 1B to D), and only the disruption of the proA-encoded γ-glutamyl phosphate reductase both impairs the osmoadaptive proline biosynthesis and simultaneously causes a proline auxotrophic growth phenotype (see above) (Table 2).
Phenotypic characterization of a proHJ mutant strain.
To study the role of the proHJ genes in the osmostress response of B. subtilis in greater detail, we performed growth experiments with both the wild-type strain JH642 and its Δ(proHJ::tet)1 derivative, strain JSB8. Both strains grow equally well in SMM (Fig. 3A), demonstrating that the proHJ-encoded γ-glutamyl kinase (ProJ) and Δ1-pyrroline-5-carboxylate reductase (ProH) are not required by B. subtilis cells that are not placed under osmotic stress conditions. However, this picture changed completely when the Δ(proHJ::tet)1 mutant strain JSB8 was grown in a high-osmolarity medium (SMM with 1.2 M NaCl). As expected from previous studies (5), such a strong increase in the osmolarity negatively affected the growth of the wild-type strain JH642 (Fig. 3A); however, this strain was eventually able to reach the OD of the osmotically unstressed culture (data not shown) (5). In contrast, the proHJ mutant strain JSB8 exhibited a strong growth disadvantage under hyperosmotic conditions (Fig. 3A), implying that the ability to synthesize large quantities of the compatible solute proline is an important facet in cellular defense of B. subtilis against high-osmolarity surroundings. The osmosensitive growth phenotype of the Δ(proHJ::tet)1 mutant strain JSB8 could be rescued by the addition of the potent osmoprotectant glycine betaine (1 mM) to the growth medium (Fig. 3A). This latter result demonstrates that the genetic disruption of the proHJ locus does not cause a nonspecific sensitivity to high-salinity environments.
Fig. 3.
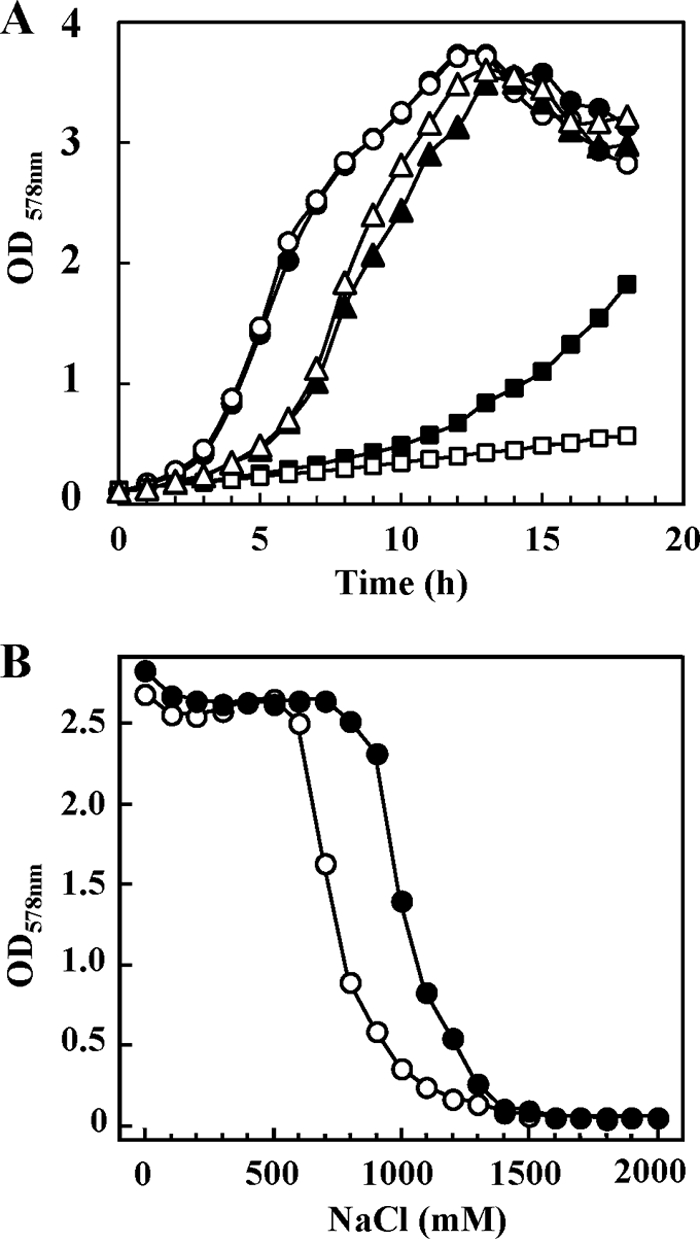
The proHJ mutant strain JSB8 exhibits a strong growth disadvantage at high osmolarity. (A) Cultures of the proHJ wild-type strain JH642 (solid symbols) and its Δ(proHJ::tet)1 derivative, strain JSB8 (open symbols), were inoculated to an optical density (OD578) of 0.1 from overnight cultures pregrown in SMM without NaCl. The cultures were grown in SMM (circles) and SMM with 1.2 M NaCl in the absence (squares) or presence (triangles) of 1 mM glycine betaine. (B) Strains JH642 (proHJ) (●) and JSB8 [Δ(proHJ::tet)1] (○) were grown in SMM containing various concentrations of NaCl. The cultures were inoculated to an optical density (OD578) of 0.05 from overnight cultures pregrown in SMM, and the growth yield of the cultures was determined after 17 h of incubation.
We also compared the growth yield of the wild-type strain JH642 and its isogenic proHJ mutant derivative strain JSB8 when they were propagated for prolonged periods (17 h) in minimal media of different osmolarities. Strain JSB8 was unable to reach the ODs of the wild-type culture once the salinity of the growth medium exceeded 0.6 M NaCl (Fig. 3B).
To ascertain whether the growth defect of JSB8 [Δ(proHJ::tet)1] under hyperosmotic growth conditions was due to a lack of proline buildup, we monitored the proline content of JSB8 cultures grown in the presence of various concentrations of NaCl by HPLC analysis. We found that the disruption of the proHJ locus in strain JSB8 entirely prevented the accumulation of proline under osmotic stress conditions (Fig. 2A). Hence, there is a correlation between the growth defect of strain JSB8 at high salinities (Fig. 3A and B) and its inability to synthesize proline in response to osmotic stress (Fig. 2A).
We also monitored the glutamate content of the osmotically stressed JSB8 cells and found a pattern that differed from that observed for the wild-type strain JH642. For reasons that are not clear to us, the level of glutamate in osmotically nonstressed cultures of JSB8 was somewhat reduced compared to the proHJ+ parent strain JH642 (Fig. 2B). This glutamate level was practically constant over the entire range of medium osmolarities tested, whereas there was steady decline in glutamate content of the wild-type strain JH642 under the identical growth conditions (Fig. 2B). The internal glutamate pool in the Δ(proHJ::tet)1 mutant JSB8 is therefore higher than that found in JH642 once the osmolarity has been raised by the addition of more than 0.5 M NaCl. It thus seems that the proHJ mutant partially compensates for its loss of proline biosynthetic capacity by maintaining an increased glutamate pool.
To test whether strain JSB8 compensates for the loss of proline biosynthetic capacity under hyperosmotic conditions by synthesizing different organic osmolytes, we used natural-abundance 13C-NMR spectroscopy that allows the detection of the dominant organic solutes within the B. subtilis cell (43, 47, 64). 13C-NMR spectra of the wild-type and the proHJ mutant strain JSB8 grown in SMM with 0.8 M NaCl were recorded. As observed previously (43, 47, 64), glutamate and proline were the dominant organic osmolytes detectable in salt-stressed cells of the B. subtilis wild-type strain JH642. However, in salt-stressed cells of the proHJ mutants strain JSB8, only the resonances of glutamate were detected by 13C-NMR spectroscopy (data not shown). This latter finding demonstrates that the proHJ mutant does not produce substantial quantities of a novel organic osmolyte to compensate for the lacking compatible solute proline.
Osmotic induction of proHJ transcription.
The physiological data presented above demonstrate that proline synthesis for osmoadaptive purposes in B. subtilis is under osmotic control (Fig. 2A) and involves the ProH-ProA-ProJ enzymes (Tables 2 and 3). To define whether the observed increase in the proline pool of osmotically stressed cells involved an increase in the level of the transcription the corresponding genes, we performed Northern blot analysis. Total RNA was isolated from cultures of strain JH642 that had been grown to mid-log phase in minimal medium (SMM) or SMM with 0.4 or 0.6 M NaCl. This RNA was then hybridized with a DIG-labeled proHJ-specific single-stranded RNA probe; a single proHJ transcript with the size of ∼2.1-kb was detected (Fig. 1E). The amount of the proHJ mRNA increased as the salt concentration of the growth medium was increased, demonstrating that the transcription of the proHJ operon was osmotically inducible. The size of this mRNA species closely corresponds to the calculated distance (2.07 kb) between the proHJ promoter (see below) and a putative factor-independent transcription terminator positioned downstream of the proHJ coding region (Fig. 1E).
Table 3.
Response of the proH-treA reporter fusion strain JSB7 in response to ionic and nonionic osmolytes
| Medium | TreA activitya (mean U mg of protein−1 ± SD) |
|---|---|
| SMM | 38 ± 2 |
| SMM + 0.4 M NaCl | 70 ± 4 |
| SMM + 0.4 M KCl | 74 ± 3 |
| SMM + 0.62 M sucrose | 208 ± 9 |
| SMM + 0.61 M lactose | 264 ± 9 |
| SMM + 0.68 M glycerol | 32 ± 2 |
The cultures were grown to early exponential growth phase (OD578 = 0.5 to 0.8), and their TreA enzyme activities were then assayed. The values given are from two independently grown cultures, and each TreA assay was performed twice. The osmolarity of the growth medium was raised in each culture to 1,100 mosmol kg of water−1 through the addition of salts, sugars, or glycerol.
We also carried out a Northern blot analysis of the proBA (Fig. 1A), proI (Fig. 1B), and proG (Fig. 1C) loci in salt-stressed and nonstressed cultures of the wild-type strain JH642 to determine whether any of these genes was under osmotic control. Transcripts of the size expected from the genetic organization of these loci were readily detected, but transcription of the proBA operon or the proI genes was not induced under hyperosmotic growth conditions (Fig. 1A to C), although a minor osmotic stimulation of proG transcription might exist (Fig. 1C).
Reporter gene fusion analysis of proHJ expression in response to hypertonicity.
To further characterize the pattern of proHJ expression in response to increases in medium osmolarity, we performed a reporter gene fusion analysis. A transcriptional fusion was constructed between the proH regulatory region and a promoterless treA reporter gene, which encodes a highly salt tolerant phospho-α(1,1)-glucosidase (TreA) from B. subtilis (24, 58). For this purpose, a 474-bp DNA fragment carrying the 3′ end of the rtp gene, the intergenic region between rtp and proH, and 51 bp of the proH coding region (Fig. 4A) was fused to the treA reporter gene. This transcriptional fusion construct was then integrated via homologous recombination as a single copy into the amyE locus of the chromosome of strain FSB1 [(treA::neo)1], yielding the Φ(proH-treA)1 fusion strain JSB7.
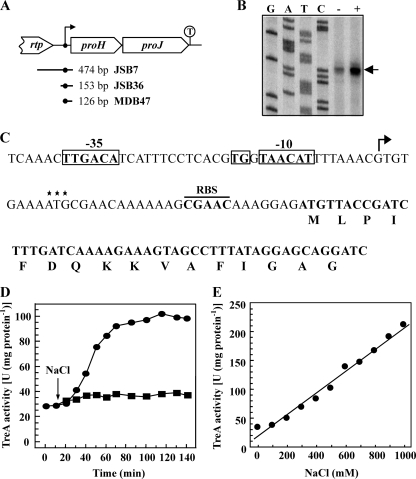
Fig. 4.
Osmotic control of proHJ transcription. (A) The genetic organization of the proHJ region is shown. The position of the proHJ promoter and a putative factor-independent transcription terminator (T) are indicated. The size and localization of the DNA fragments used to construct the various Φ(proH-treA)1 operon fusions present in strains JSB7, JSB36, and MDB47 are shown. (B) Total RNA was prepared from cells of the B. subtilis strain JH642 grown in SMM (−) and SMM with 0.4 M NaCl (+) and used for a reverse transcriptase reaction primed with a proH-specific radiolabeled oligonucleotide. The same oligonucleotide was used for sequencing reactions with plasmid pJS13 as a template to size the proH transcript. (C) Nucleotide sequence of the proHJ promoter region that is present in the Φ(proH-treA)2 operon reporter fusion of strain MDB47. The transcription initiation site is indicated by an arrow, and the −10 and −35 sequences and the TG motif at position −16 of the proHJ promoter are marked; the putative ribosome-binding site (RBS) for the proH gene is also indicated. The start of the proH-coding region deduced by us from alignments of the amino acid sequences of different ProB proteins is shown in bold letters. The translation initiation codon for the proJ gene annotated by Kunst et al. (44) and Barbe et al. (3) in the genome sequence of the B. subtilis 168 wild-type strain is marked by three asterisks; we note that there is no appropriate ribosome-binding site present upstream of the predicted start codon and therefore assume that the start codon was incorrectly predicted. (D and E) Operon fusion analysis of proHJ expression in response to increases in the osmolarity of the growth medium. Expression of the chromosomally encoded Φ(proH-treA474)1 fusion present in strain JSB7 (● and ■) (D) and the Φ(proH-treA153)3 fusion present in strain JSB36 (●) (E) was monitored. (D) TreA activities were determined in strain JSB7 cells cultivated in SMM that were subjected to a sudden osmotic up-shock (arrow) with 0.4 M NaCl (●). A culture of strain JSB7 was grown in parallel in SMM that was not subjected to an osmotic upshift (■). (E) Cultures of the Φ(proH-treA153)3 reporter fusion strain JSB36 (●) were pregrown in SMM of different osmolarities, freshly inoculated into minimal medium of the same osmolarity as the preculture, and then assayed for TreA activity at the mid-exponential growth phase (OD578 = 0.8). Strain TRB0 [(treA::neo)1] carrying the promoterless treA gene inserted into the amyE locus (amyE::treA) was used as a control for these experiments and yielded less than three units of TreA activity, which was not influenced by increased salinity. The values given, represent the averages of two independently grown cultures. For each sample analyzed, the TreA activity was determined twice. Standard deviations were typically ca. 10 to 15%.
We first tested whether the expression of Φ(proH-treA)1 reporter gene fusion construct was increased in response to an increase in high salinity and whether this was due to an osmotic or a salt-specific effect. We therefore grew the cells in SMM or in SMM whose osmolarity had been increased to equal values (1,100 mosmol per kg of water) by the addition of salts (NaCl or KCl) or by the addition of sugars (sucrose or lactose). Both ionic and nonionic osmolytes triggered the expression of the Φ(proH-treA)1 reporter gene fusion (Table 3), demonstrating that enhanced transcription of the proHJ operon observed in salt-stressed cells (Fig. 1E) is a true osmotic effect. Surprisingly, we found that an increase in the osmolarity of the growth medium caused by the addition of the sugars sucrose and lactose triggered a 3- to 4-fold-higher level of Φ(proH-treA)1 induction compared to cultures whose osmolarity was raised by the additions of either NaCl or KCl (Table 3). In contrast to what was observed by the addition of either salts or sugars to the growth medium of the reporter fusion strain JSB7, the addition of an equi-osmotic concentration of glycerol to the growth medium did not trigger enhanced Φ(proH-treA)1 transcription (Table 3). Taken together, these data strongly suggest that the induction of the expression of the proHJ operon requires the establishment of an osmotically effective gradient since solutes that cannot effectively permeate the cytoplasmic membrane (NaCl, KCl, sucrose or lactose) trigger proHJ transcription, whereas high concentrations of the membrane-permeable solute glycerol do not elicit enhanced proHJ expression (Table 3).
We then determined the expression of the Φ(proH-treA)1 reporter gene fusion in response to a sudden osmotic upshift. TreA activity was low in cells of strain JSB7 grown in SMM and rapidly increased following an osmotic up-shock with 0.4 M NaCl (Fig. 4D). When we monitored TreA activity in cells of strain JSB7 grown over longer time periods in media of various osmolarities, we found that the expression of the Φ(proH-treA)1 operon fusion was linearly correlated with the osmotic strength of the growth medium (Fig. 4E). It follows from these data that B. subtilis not only can detect a sudden increase in the external osmolarity (Fig. 4D) but also can monitor and respond with fine-tuned gene expression to incremental increases in osmolarity on a sustained basis (Fig. 4E).
Primer extension analysis of the proHJ transcript.
To identify the promoter of the proHJ operon, we mapped the transcription initiation site by primer extension analysis. Total RNA was prepared from the wild-type JH642 grown to mid-exponential phase in SMM or SMM with 0.4 M NaCl. A proH-specific primer radiolabeled at its 5′ end was annealed to the isolated total RNA and extended with reverse transcriptase. We detected a single mRNA species whose production was under osmotic control (Fig. 4B). The transcription initiation site corresponds to a G residue positioned 35 bp upstream of the proH start codon (Fig. 4C). Inspection of the DNA sequence upstream of the proHJ transcription initiation site revealed the presence of putative −10 and −35 sequences that closely resemble the consensus sequences of promoters recognized by the main vegetative sigma factor (σA) of B. subtilis (28). The −10 and −35 sequences of the proHJ promoter region are separated by 16 bp and the spacer region contains a TG motif at position −16 (Fig. 4C) that is frequently found in σA-dependent B. subtilis promoters and is often crucial for promoter activity (28).
A 126-bp proHJ promoter fragment is sufficient to confer osmotically controlled transcription.
The Φ(proH-treA)1 operon fusion used for the above-described transcriptional studies carries a 474-bp DNA fragment with 353 bp upstream of the −35 region of the proHJ promoter and a fusion junction in codon 17 of the proH reading frame (Fig. 4C). To define more closely the DNA segment that is required in cis for osmoregulated proHJ transcription, we shortened the 5′ end of the DNA segment used for the construction of the Φ(proH-treA)1 operon fusion. In this way we constructed proH-treA reporter fusions with (i) a 153-bp DNA segment that carries the proHJ promoter with a 32-bp region upstream of the −35 region (strain JSB36) and (ii) a 126-bp segment that carries the proHJ promoter with a 6-bp region upstream of the −35 region (strain MDB47). Both of these proH-treA fusion constructs carry the same 3′-fusion junction to the proH gene that is also present in the reporter fusion strain JSB7 (Fig. 4A and C). The expression of the proH-treA reporter fusions present in strains JSB36 and MDB47 were osmotically inducible (Fig. 5A) to the same extent as the fusion in strain JSB7 (Fig. 4E), which has the longest 5′-upstream region (353-bp) of proH tested in this study (Fig. 4A). The reporter strain MDB47 carrying the 126-bp proH-treA fusion construct was further tested for its response to incremental increases in the salinity of the growth medium. The expression of the proH-treA126bp fusion increased linearly in response to increases in the external salinity (Fig. 6) as observed for strain JSB7 carrying the proH-treA474bp fusion construct (Fig. 4E). We conclude from this set of experiments that a 126-bp DNA fragment carrying the proHJ promoter, a 6-bp segment upstream of the −35 region, the SigA-type promoter, a 42-bp nontranslated region including the ribosome binding site, and 17 codons of the proH reading frame carry all DNA sequences required in cis for the osmoregulated transcription of the proHJ operon of B. subtilis (Fig. 1E). The DNA sequence of this regulatory region is shown in Fig. 4C. We note that a palindrome (5′-CACGT-n13-ACGTG-3′) overlapping the transcriptional initiation site and part of the promoter spacer region is present in the proHJ promoter region; the role of this palindrome in controlling proHJ expression is currently unknown.
Fig. 5.
TreA-reporter fusion analysis of the 126-bp proHJ promoter fragment. (A) The B. subtilis strains JSB36 carrying the Φ(proH-treA153)2 reporter fusion and strain MDB47 carrying the Φ(proH-treA126)2 fusion (see Fig. 4A) were grown in SMM (▩) or in SMM with 0.4 M NaCl (■) to mid-exponential phase (OD578 = 0.8). The TreA activity was determined from three biological replicates. (B) Cultures of the Φ(proH-treA126)2 reporter fusion strain MDB47 (see Fig. 4A) were grown in SMM containing various NaCl concentrations to the mid-exponential growth phase (OD578 = 0.8), harvested by centrifugation, and assayed for TreA activity. The values given present the averages of two independently grown cultures. For each sample analyzed, the TreA activity was determined twice. Standard deviations were typically ca. 10 to 15%.
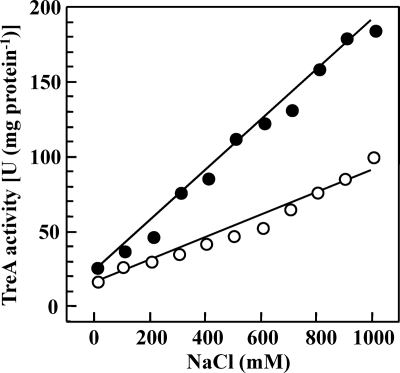
Fig. 6.
Repression of proHJ-treA expression by an exogenous supply of osmoprotectants. (A) The Φ(proH-treA474)1 reporter fusion strain JSB7 was grown in SMM or in SMM with increasing salinities in the absence (●) or the presence (○) of 1 mM glycine betaine. Cells were harvested in the mid-exponential phase (OD578 = 0.8), and the TreA activities were measured. The values given present the averages of two independently grown cultures. For each sample analyzed, the TreA activity was determined twice. Standard deviations were typically ca. 10 to 15%.
An exogenous supply of glycine betaine partially suppresses proHJ expression.
The uptake of compatible solutes by microorganisms is typically preferred, possibly for energetic reasons (48), over their de novo synthesis. It is thus often observed that the uptake of compatible solutes suppresses the expression of biosynthetic genes for these compounds (53). To test whether this was also the case for the proHJ genes, we grew the Φ(proH-treA)1 reporter gene fusion strain JSB7 in SMM with increasing concentrations of NaCl in either the absence or presence of glycine betaine (1 mM). The addition of glycine betaine to the high-salinity growth media repressed the expression of the Φ(proH-treA)1 reporter gene fusion to a considerable degree but still allowed a transcriptional response pattern to osmotic stress similar to that exhibited by the fusion strain grown in the absence of glycine betaine (Fig. 6). Hence, B. subtilis prefers physiologically the uptake of the osmoprotectant glycine betaine to the de novo synthesis of proline. We also observed that an exogenous supply of proline (1 mM) to high-salinity-stressed cells also repressed the expression of the Φ(proH-treA)1 reporter gene fusion, albeit to a lesser extent than that observed for glycine betaine (data not shown).
DISCUSSION
The cellular response of B. subtilis to salt stress is certainly complex (25, 33, 34, 59), but effective water management is the cornerstone for responses to fluctuations in the environmental osmolarity (8). This applies both to an osmotic downshift that requires the rapid expulsion of water-attracting solutes from the cell through the transient opening of mechanosensitive channels to avert cell lysis (29, 63) and to an increase in the osmolarity that requires the accumulation of water-attracting solutes to prevent dehydration of the cytoplasm and growth arrest (8, 9, 64). We address here the latter aspect of the B. subtilis osmostress response by focusing on the osmoadaptive synthesis of proline, a compatible solute that is widely used by microorganisms as an osmostress protectant (15, 21, 41, 66).
It has been known through the pioneering study by Measures (46) and subsequent detailed physiological investigations by Whatmore et al. (64) that B. subtilis synthesizes very large quantities of proline in response to high salinity. Proline is the only compatible solute that can be synthesized de novo by B. subtilis (43, 64). The data presented here now unravel the molecular details of the osmoadaptive proline biosynthesis route and demonstrate that (i) cellular proline content increases linearly over a considerable span of salt concentrations in the medium (Fig. 2A), (ii) proline production is required for the effective adjustment of the B. subtilis cell to sustained high-salinity growth conditions (Fig. 3A), (iii) osmoadaptive proline synthesis centrally relies on the osmotic control of proHJ transcription (Fig. 4D and E), and (iv) the osmoadaptive and anabolic proline biosynthetic routes of B. subtilis are interlinked via the γ-glutamyl phosphate reductase (ProA) (Fig. 1D).
Whatmore et al. (64) reported that a sudden osmotic upshift triggers a rapid onset of proline production. Our data now show that the B. subtilis cell can also adjust its proline pool via de novo synthesis in accord with the prevailing salinity of its growth medium when the cells are cultivated on a sustained basis under high osmolarity conditions (Fig. 2A). These two biosynthetic responses allow the B. subtilis cell to adjust sensitively to both sudden and prolonged increases in environmental osmolarity by producing levels of proline that are in tune with the degree of the imposed osmotic stress. The genetic disruption of the osmoadaptive proline biosynthesis route (Fig. 2A) greatly impairs growth of B. subtilis at high salinity (Fig. 3A), thereby underscoring the physiological role of the production of this compatible solute for the cellular acclimatization process to high-salinity/osmolarity surroundings. However, the growth of a mutant with a defect in the osmoadaptive proline biosynthesis (Fig. 2A) is not completely prevented in high-salinity media (Fig. 3B). Perhaps the somewhat increased glutamate pool in the proHJ mutant relative to the wild-type strain (Fig. 2B) contributes to osmotic adjustment. One needs to consider, however, that glutamate is not a very effective compatible solute for bacilli since Bacillus species that rely solely on glutamate production for their osmotic adjustment (e.g., Bacillus cereus) (19, 43) tend to be rather salt sensitive compared to those that synthesize proline or ectoine as their dominant compatible solute (13, 43). It might also be that the proHJ mutant of B. subtilis partially compensates for the loss of its osmoadaptive proline biosynthetic capacity by increasing the pool of ions (e.g., potassium) (55).
B. subtilis possesses paralogous enzymes for the first and last steps of proline biosynthesis from the precursor glutamate, but only a single γ-glutamyl phosphate reductase (ProA) is encoded by the B. subtilis genome (3, 4, 44) (Fig. 1D). From a physiological point of view, such an interconnection of the two pathways is perplexing, since the amounts of proline needed for anabolic purposes differ greatly from those required for osmoadaptation (10, 64). The expression of the operon (proBA) encoding the ProA enzyme is under T-box control (Fig. 1A), an RNA-based genetic regulatory device that allows a strong upregulation of proBA transcription only in cells that are starving for proline (10). In contrast to the transcription of the proBA gene cluster, the transcription of the operon (proHJ) that encodes the enzymes for the first (ProJ) and last (ProH) steps of the osmoadaptive proline biosynthetic route is osmotically inducible (Fig. 1A and E). The ProB and ProA enzymes typically form a complex, probably to prevent the release of the labile intermediate γ-glutamyl phosphate into the cytoplasm (16, 57). Since the amount of ProJ will increase in osmotically stressed cells as a result of enhanced proHJ transcription, the question arises as to how the B. subtilis cell ensures an adequate supply of ProA in order not to limit osmoadaptive proline biosynthesis. Our data clearly rule out a transcriptional control mechanism to boost ProA production under osmotic stress conditions, but we have not yet experimentally addressed the possibility that the amount of ProA is increased relative to that of ProB to match the amount of ProJ through a translational control mechanism or that ProA is a particularly stable protein.
Another aspect of the osmoadaptive proline biosynthetic pathway of B. subtilis that needs to be considered is the biochemical properties of the γ-glutamyl kinases ProB and ProJ (Fig. 1D). The enzyme activity of γ-glutamyl kinase is frequently regulated by feedback inhibition by the end product of the biosynthesis pathway, proline, in order to prevent a wasteful overproduction of this amino acid. Feedback inhibition of the ProB-catalyzed enzymatic reaction typically sets in at very low concentrations of proline (16), and this is true also for the ProB enzyme of B. subtilis (Fig. 1D), where proline concentrations of 7.5 × 10−6 M inhibit ProB activity by 50% (14). Conceptually, it is apparent that the γ-glutamyl kinase activity of the B. subtilis ProJ enzyme cannot be feedback inhibited by proline to a significant degree because it could otherwise not appropriately function in a cytoplasm that contains very large proline pools as found in osmotically stressed B. subtilis cells (64).
Single amino acid substitutions that abrogate the allosteric regulation by proline can be found at many positions in the γ-glutamyl kinase protein sequence of both prokaryotes and eukaryotes (for an overview, see the study by Perez-Arellano et al. [50]). The crystal structure of the E. coli γ-glutamyl kinase (49, 51) revealed that the glutamate substrate binding site partially overlaps with that of the feedback inhibitor proline. The interactions of both molecules with the γ-glutamyl kinases protein is modulated by a 16-amino-acid long flexible loop that links β-sheet 4 with α-helix E (49). Glu-153 is part of this loop and a change of the corresponding residue to a positively charged Lys residue in the γ-glutamyl kinase from tomato (tomPRO1) result in a 3,500-fold drop in the apparent Ki for proline compared to the wild-type enzyme and a concomitant 900-fold increase in cellular proline content (23). Substitutions at the corresponding site of the ProB enzyme of Listeria monocytogenes or E. coli also lead to a drop in feedback inhibition, resulting in a strong proline overproduction and increased osmotic tolerance (18, 56). Hence, this Glu residue is key in setting the degree of proline feedback-mediated inhibition of both bacterial and plant γ-glutamyl kinases. We note with interest that the feedback inhibited ProB enzyme from B. subtilis (14) possesses a Glu residue (Glu-142) at the corresponding region, whereas the ProJ enzyme possesses a positively charged Arg residue (Arg-146) at this position.
The pattern of transcription of the proHJ operon that encodes the enzymes for the first and last steps of the osmoadaptive proline biosynthesis route (Fig. 1D) is well suited to allow B. subtilis to physiologically respond to either suddenly imposed or prolonged osmotic challenges. Both a sudden osmotic upshift and the sustained imposition of salt stress trigger enhanced transcription of the proHJ gene cluster (Fig. 4D and E). It follows from our proH-treA reporter gene studies that B. subtilis must possess perception and signal transduction mechanisms that allow the cell to detect not only sudden increases (Fig. 4D) but also long-lasting and incremental rises in the salinity/osmolarity of the growth medium (Fig. 4E). Elevated levels of proHJ expression are maintained as long as the osmotic stimulus persists. This pattern of gene transcription is different from that of salt-inducible promoters recognized by SigB, the master regulator of the general stress response system of B. subtilis (27, 52). Transcription initiating from SigB-controlled promoters is typically rapidly induced in response to a suddenly imposed salt stress but then levels off again (7, 25, 58). Furthermore, these types of promoters are not active under sustained high-salinity growth conditions as exemplified through the analysis of the SigB-controlled opuE-P2 promoter for the osmotically inducible OpuE proline import system of B. subtilis (58).
It is unclear how the B. subtilis cell measures increases in the osmolarity of its environment to turn on the transcription of the proHJ operon. One wonders whether the cell monitors fluctuations in the external osmolarity directly or whether it derives information about the osmotic conditions in its habitat indirectly by recording changes in its intercellular ionic and nonionic solute pool. The signal that triggers proHJ expression requires the establishment of an osmotically effective gradient across the cytoplasmic membrane since the addition of high concentrations of the membrane-permeable glycerol did not elicit the induction of proHJ transcription (Table 3). It is noteworthy that in terms of proHJ regulation, the B. subtilis cell perceives an osmotic signal rather than a signal solely elicited by the addition of NaCl (Table 3). However, ionic and nonionic solutes have different effects on the level of proHJ transcription despite the fact that the overall osmolarity of the growth medium was raised to an equal degree (Table 3). At present, we have no adequate physiological or genetic explanation for this surprising finding, but it seems that either B. subtilis does not perceive osmotic stress elicited by ionic or nonionic solutes in the same way, or it responds to these environmental cues with a different strength of gene expression. The data reported by Shabala et al. (55) for osmotically stressed E. coli cultures suggest specificity of the cellular osmotic acclimatization process to ionic and nonionic osmotica.
We have narrowed the DNA sequences required in cis for the full osmotic control of proHJ transcription to a 126-bp DNA segment (Fig. 5). Visual inspection of this DNA segment (Fig. 4C) has thus far not yielded any clues as to why the activity of the proHJ promoter is turned on in response to high osmolarity in the environment since its −10 and −35 regions and the spacing between these sequences resemble typical SigA-type promoters of B subtilis (28). Genome-wide transcriptional profiling studies of cells continuously cultivated at high salinity (SMM with 1.2 M NaCl) have shown that the proHJ operon exhibits one of the strongest levels of induction among the ∼100 osmotically induced genes of B. subtilis (59). The proHJ regulatory region (Fig. 4C) might therefore serve as a useful tool to search for mutants that alter the ability of the B. subtilis cell to perceive and respond to osmotic stress.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to J. Gade for her expert technical assistance. We thank B. Belitsky, M. Jebbar, B. Kempf, M. Itaya, F. Spiegelhalter, and T. Tanaka for generously providing bacterial strains and plasmids. We are very grateful to V. Koogle for her kind help in editing the manuscript.
Financial support for this study was provided by the Deutsche Forschungsgemeinschaft, by the Fonds der Chemischen Industrie, and by grants from the BMBF via the Bacell-SysMo2 consortium and the LOEWE program of the State of Hessen (via the Centre for Synthetic Microbiology, SynMicro, Marburg, Germany).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 22 July 2011.
REFERENCES
- 1. Akashi H., Gojobori T. 2002. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 99:3695–3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barbe V., et al. 2009. From a consortium sequence to a unified sequence: the Bacillus subtilis 168 reference genome a decade later. Microbiology 155:1758–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Belitsky B. R., Brill J., Bremer E., Sonenshein A. L. 2001. Multiple genes for the last step of proline biosynthesis in Bacillus subtilis. J. Bacteriol. 183:4389–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boch J., Kempf B., Bremer E. 1994. Osmoregulation in Bacillus subtilis: synthesis of the osmoprotectant glycine betaine from exogenously provided choline. J. Bacteriol. 176:5364–5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boch J., Kempf B., Schmid R., Bremer E. 1996. Synthesis of the osmoprotectant glycine betaine in Bacillus subtilis: characterization of the gbsAB genes. J. Bacteriol. 178:5121–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boylan S. A., Redfield A. R., Brody M. S., Price C. W. 1993. Stress-induced activation of the σB transcription factor of Bacillus subtilis. J. Bacteriol. 175:7931–7937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bremer E. 2002. Adaptation to changing osmolarity, p. 385–391 In Sonenshein A. L., Hoch J. A., Losick R. (ed.), Bacillus subtilis and its closest relatives. ASM Press, Washington, DC [Google Scholar]
- 9. Bremer E., Krämer R. 2000. Coping with osmotic challenges: osmoregulation through accumulation and release of compatible solutes in bacteria, p. 79–97 In Storz G., Hengge-Aronis R. (ed.), Bacterial stress responses. ASM, Washington, DC [Google Scholar]
- 10. Brill J., Hoffmann T., Putzer H., Bremer E. 2011. T-box-mediated control of the anabolic proline biosynthetic genes of Bacillus subtilis. Microbiology 157:977–987 [DOI] [PubMed] [Google Scholar]
- 11. Brown A. D. 1976. Microbial water stress. Bacteriol. Rev. 40:803–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Burg M. B., Ferraris J. D. 2008. Intracellular organic osmolytes: function and regulation. J. Biol. Chem. 283:7309–7313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bursy J., Pierik A. J., Pica N., Bremer E. 2007. Osmotically induced synthesis of the compatible solute hydroxyectoine is mediated by an evolutionarily conserved ectoine hydroxylase. J. Biol. Chem. 282:31147–31155 [DOI] [PubMed] [Google Scholar]
- 14. Chen M., et al. 2007. Expression of Bacillus subtilis proBA genes and reduction of feedback inhibition of proline synthesis increases proline production and confers osmotolerance in transgenic Arabidopsis. J. Biochem. Mol. Biol. 40:396–403 [DOI] [PubMed] [Google Scholar]
- 15. Csonka L. N., Hanson A. D. 1991. Prokaryotic osmoregulation: genetics and physiology. Annu. Rev. Microbiol. 45:569–606 [DOI] [PubMed] [Google Scholar]
- 16. Csonka L. N., Leisinger T. 2007, posting date Chapter 34.6.1.4, biosynthesis of proline. In Böck A., et al. (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC: doi:10.1128/ecosal.4.7.4 [Google Scholar]
- 17. Cutting S. M., Vander Horn P. B. 1990. Genetic analysis, p. 27–74 In Harwood C. R., Cutting S. M. (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Inc., Chichester, United Kingdom [Google Scholar]
- 18. Dandekar A. M., Uratsu S. L. 1988. A single base pair change in proline biosynthesis genes causes osmotic stress tolerance. J. Bacteriol. 170:5943–5945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. den Besten H. M., Mols M., Moezelaar R., Zwietering M. H., Abee T. 2009. Phenotypic and transcriptomic analyses of mildly and severely salt-stressed Bacillus cereus ATCC 14579 cells. Appl. Environ. Microbiol. 75:4111–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Earl A. M., Losick R., Kolter R. 2008. Ecology and genomics of Bacillus subtilis. Trends Microbiol. 16:269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fisher M. T. 2006. Proline to the rescue. Proc. Natl. Acad. Sci. U. S. A. 103:13265–13266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fujisawa M., Ito M., Krulwich T. A. 2007. Three two-component transporters with channel-like properties have monovalent cation/proton antiport activity. Proc. Natl. Acad. Sci. U. S. A. 104:13289–13294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fujita T., et al. 2003. Identification of regions of the tomato gamma-glutamyl kinase that are involved in allosteric regulation by proline. J. Biol. Chem. 278:14203–14210 [DOI] [PubMed] [Google Scholar]
- 24. Gotsche S., Dahl M. K. 1995. Purification and characterization of the phospho-α(1,1)glucosidase (TreA) of Bacillus subtilis 168. J. Bacteriol. 177:2721–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hahne H., et al. 2010. A comprehensive proteomics and transcriptomics analysis of Bacillus subtilis salt stress adaptation. J. Bacteriol. 192:870–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harwood C. R., Archibald A. R. 1990. Growth, maintenance and general techniques, p. 1–26 In Harwood C. R., Cutting S. M. (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Inc., Chichester, United Kingdom [Google Scholar]
- 27. Hecker M., Pane-Farre J., Völker U. 2007. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu. Rev. Microbiol. 61:215–236 [DOI] [PubMed] [Google Scholar]
- 28. Helmann J. D. 1995. Compilation and analysis of Bacillus subtilis σA-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 23:2351–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hoffmann T., Boiangiu C., Moses S., Bremer E. 2008. Responses of Bacillus subtilis to hypotonic challenges: physiological contributions of mechanosensitive channels to cellular survival. Appl. Environ. Microbiol. 74:2454–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hoffmann T., Bremer E. 2011. Protection of Bacillus subtilis against cold stress via compatible-solute acquisition. J. Bacteriol. 193:1552–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Holtmann G., Bakker E. P., Uozumi N., Bremer E. 2003. KtrAB and KtrCD: two K+ uptake systems in Bacillus subtilis and their role in adaptation to hypertonicity. J. Bacteriol. 185:1289–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Holtmann G., Bremer E. 2004. Thermoprotection of Bacillus subtilis by exogenously provided glycine betaine and structurally related compatible solutes: involvement of Opu transporters. J. Bacteriol. 186:1683–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Höper D., Bernhardt J., Hecker M. 2006. Salt stress adaptation of Bacillus subtilis: a physiological proteomics approach. Proteomics 6:1550–1562 [DOI] [PubMed] [Google Scholar]
- 34. Höper D., Völker U., Hecker M. 2005. Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis. J. Bacteriol. 187:2810–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ignatova Z., Gierasch L. M. 2006. Inhibition of protein aggregation in vitro and in vivo by a natural osmoprotectant. Proc. Natl. Acad. Sci. U. S. A. 103:13357–13361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jebbar M., von Blohn C., Bremer E. 1997. Ectoine functions as an osmoprotectant in Bacillus subtilis and is accumulated via the ABC-transport system OpuC. FEMS Microbiol. Lett. 154:325–330 [Google Scholar]
- 37. Kappes R., Bremer E. 1998. Response of Bacillus subtilis to high osmolarity: uptake of carnitine, crotonobetaine, and γ-butyrobetaine via the ABC transport system OpuC. Microbiology 144:83–90 [DOI] [PubMed] [Google Scholar]
- 38. Kappes R. M., Kempf B., Bremer E. 1996. Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: characterization of OpuD. J. Bacteriol. 178:5071–5079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kappes R. M., et al. 1999. Two evolutionarily closely related ABC-transporters mediate the uptake of choline for synthesis of the osmoprotectant glycine betaine in Bacillus subtilis. Mol. Microbiol. 32:203–216 [DOI] [PubMed] [Google Scholar]
- 40. Kempf B., Bremer E. 1995. OpuA, an osmotically regulated binding protein-dependent transport system for the osmoprotectant glycine betaine in Bacillus subtilis. JBC. 270:16701–16713 [DOI] [PubMed] [Google Scholar]
- 41. Kempf B., Bremer E. 1998. Uptake and synthesis of compatible solutes as microbial stress responses to high osmolality environments. Arch. Microbiol. 170:319–330 [DOI] [PubMed] [Google Scholar]
- 42. Koch A. L., Higgins M. L., Doyle R. J. 1982. The role of surface stress in the morphology of microbes. J. Gen. Microbiol. 128:927–945 [DOI] [PubMed] [Google Scholar]
- 43. Kuhlmann A. U., Bremer E. 2002. Osmotically regulated synthesis of the compatible solute ectoine in Bacillus pasteurii and related Bacillus spp. Appl. Environ. Microbiol. 68:772–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kunst F., et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249–256 [DOI] [PubMed] [Google Scholar]
- 45. Marco-Marin C., et al. 2007. A novel two-domain architecture within the amino acid kinase enzyme family revealed by the crystal structure of Escherichia coli glutamate 5-kinase. J. Mol. Biol. 367:1431–1446 [DOI] [PubMed] [Google Scholar]
- 46. Measures J. C. 1975. Role of amino acids in osmoregulation of non-halophilic bacteria. Nature 257:398–400 [DOI] [PubMed] [Google Scholar]
- 47. Nau-Wagner G., Boch J., Le Good J. A., Bremer E. 1999. High-affinity transport of choline-O-sulfate and its use as a compatible solute in Bacillus subtilis. Appl. Environ. Microbiol. 65:560–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Oren A. 2010. Thermodynamic limits to life at high salt concentrations. Environ. Microbiol. doi:10.1111/j.1462-2920.2010.02365.x [DOI] [PubMed] [Google Scholar]
- 49. Perez-Arellano I., Carmona-Alvarez F., Gallego J., Cervera J. 2010. Molecular mechanisms modulating glutamate kinase activity. Identification of the proline feedback inhibitor binding site. J. Mol. Biol. 404:890–901 [DOI] [PubMed] [Google Scholar]
- 50. Perez-Arellano I., Carmona-Alvarez F., Martinez A. I., Rodriguez-Diaz J., Cervera J. 2010. Pyrroline-5-carboxylate synthase and proline biosynthesis: from osmotolerance to rare metabolic disease. Protein Sci. 19:372–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Perez-Arellano I., Rubio V., Cervera J. 2006. Mapping active site residues in glutamate-5-kinase. The substrate glutamate and the feed-back inhibitor proline bind at overlapping sites. FEBS Lett. 580:6247–6253 [DOI] [PubMed] [Google Scholar]
- 52. Price C. W. 2011. General stress responses in Bacillus subtilis and related Gram-positive bacteria, p. 301–318 In Storz G., Hengge R. (ed.), Bacterial stress responses, 2nd ed ASM Press, Washington, DC [Google Scholar]
- 53. Sagot B., et al. 2010. Osmotically induced synthesis of the dipeptide N-acetylglutaminylglutamine amide is mediated by a new pathway conserved among bacteria. Proc. Natl. Acad. Sci. U. S. A. 107:12652–12657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sambrook J., Fritsch E. F., Maniatis T. E. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 55. Shabala L., et al. 2009. Ion transport and osmotic adjustment in Escherichia coli in response to ionic and non-ionic osmotica. Environ. Microbiol. 11:137–148 [DOI] [PubMed] [Google Scholar]
- 56. Sleator R. D., Gahan C. G., Hill C. 2001. Mutations in the listerial proB gene leading to proline overproduction: effects on salt tolerance and murine infection. Appl. Environ. Microbiol. 67:4560–4565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Smith C. J., Deutch A. H., Rushlow K. E. 1984. Purification and characteristics of a gamma-glutamyl kinase involved in Escherichia coli proline biosynthesis. J. Bacteriol. 157:545–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Spiegelhalter F., Bremer E. 1998. Osmoregulation of the opuE proline transport gene from Bacillus subtilis: contributions of the σA- and σB-dependent stress-responsive promoters. Mol. Microbiol. 29:285–296 [DOI] [PubMed] [Google Scholar]
- 59. Steil L., Hoffmann T., Budde I., Völker U., Bremer E. 2003. Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity. J. Bacteriol. 185:6358–6370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Street T. O., Bolen D. W., Rose G. D. 2006. A molecular mechanism for osmolyte-induced protein stability. Proc. Natl. Acad. Sci. U. S. A. 103:13997–14002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. von Blohn C., Kempf B., Kappes R. M., Bremer E. 1997. Osmostress response in Bacillus subtilis: characterization of a proline uptake system (OpuE) regulated by high osmolarity and the alternative transcription factor sigma B. Mol. Microbiol. 25:175–187 [DOI] [PubMed] [Google Scholar]
- 63. Wahome P. G., Setlow P. 2008. Growth, osmotic downshock resistance and differentiation of Bacillus subtilis strains lacking mechanosensitive channels. Arch. Microbiol. 189:49–58 [DOI] [PubMed] [Google Scholar]
- 64. Whatmore A. M., Chudek J. A., Reed R. H. 1990. The effects of osmotic upshock on the intracellular solute pools of Bacillus subtilis. J. Gen. Microbiol. 136:2527–2535 [DOI] [PubMed] [Google Scholar]
- 65. Whatmore A. M., Reed R. H. 1990. Determination of turgor pressure in Bacillus subtilis: a possible role for K+ in turgor regulation. J. Gen. Microbiol. 136:2521–2526 [DOI] [PubMed] [Google Scholar]
- 66. Wood J. M., et al. 2001. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 130:437–460 [DOI] [PubMed] [Google Scholar]
- 67. Yancey P. H. 2004. Compatible and counteracting solutes: protecting cells from the Dead Sea to the deep sea. Sci. Prog. 87:1–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.