Abstract
During transmembrane signaling by Escherichia coli Tsr, changes in ligand occupancy in the periplasmic serine-binding domain promote asymmetric motions in a four-helix transmembrane bundle. Piston displacements of the signaling TM2 helix in turn modulate the HAMP bundle on the cytoplasmic side of the membrane to control receptor output signals to the flagellar motors. A five-residue control cable joins TM2 to the HAMP AS1 helix and mediates conformational interactions between them. To explore control cable structural features important for signal transmission, we constructed and characterized all possible single amino acid replacements at the Tsr control cable residues. Only a few lesions abolished Tsr function, indicating that the chemical nature and size of the control cable side chains are not individually critical for signal control. Charged replacements at I214 mimicked the signaling consequences of attractant or repellent stimuli, most likely through aberrant structural interactions of the mutant side chains with the membrane interfacial environment. Prolines at residues 214 to 217 also caused signaling defects, suggesting that the control cable has helical character. However, proline did not disrupt function at G213, the first control cable residue, which might serve as a structural transition between the TM2 and AS1 helix registers. Hydrophobic amino acids at S217, the last control cable residue, produced attractant-mimic effects, most likely by contributing to packing interactions within the HAMP bundle. These results suggest a helix extension mechanism of Tsr transmembrane signaling in which TM2 piston motions influence HAMP stability by modulating the helicity of the control cable segment.
INTRODUCTION
Chemoreceptors known as methyl-accepting chemotaxis proteins (MCPs) mediate the adaptive locomotor behaviors of many bacterial and archaeal cells (1, 70, 75). The MCPs of Escherichia coli are the best studied and offer tractable models for elucidating molecular mechanisms of transmembrane signaling (26, 27). The serine (Tsr), aspartate (Tar), ribose/galactose (Trg), and dipeptide/pyrimidine (Tap) transmembrane receptors all contain periplasmic ligand-binding domains that communicate stimulus information to a cytoplasmic kinase control domain (Fig. 1). Changes in ligand occupancy promote small (∼2-Å) displacements of the membrane-spanning TM2 helix in one subunit of the receptor homodimer (18, 25, 43). This asymmetric piston motion impinges on a HAMP domain at the cytoplasmic side of the membrane, which translates that conformational input into symmetric structural changes of an extended four-helix bundle to modulate activity of the receptor-associated CheA autokinase (26, 46). Attractant (ATT) stimuli promote inward TM2 displacements that inhibit CheA activity, and repellent (REP) stimuli promote outward piston movements that stimulate CheA activity (25). The kinase-off state favors counterclockwise (CCW) rotation of the cell's flagellar motors, producing forward swimming, and the kinase-on state promotes clockwise (CW) motor rotation, which causes tumbling episodes that randomly reorient the cell's heading (37). By modulating the direction of motor rotation in response to changing chemoeffector levels, the cell tracks attractant and repellent gradients in its environment.
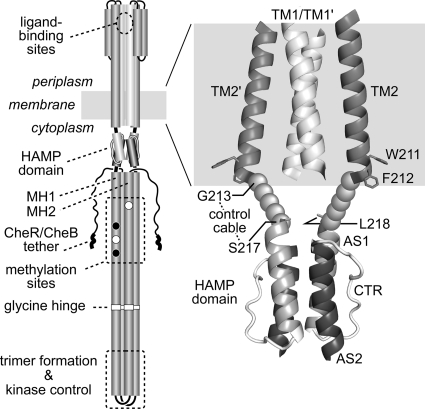
Fig. 1.
Tsr structure and location of the control cable. (Left) The native Tsr molecule is a homodimer of 551-residue subunits. Cylindrical segments represent α-helical regions, drawn approximately to scale. Methylation sites shown as black circles indicate glutamine residues that must be deamidated to glutamates by CheB before they can accept methyl groups, and open circles represent glutamate residues that are direct substrates for the CheR methyltransferase. Thickened regions at the C terminus of each subunit represent a pentapeptide sequence (NWETF) to which CheB and CheR bind. (Right) Expanded view of the TM2-HAMP region of Tsr. The structure of the 4-helix TM bundle is based on modeled coordinates for the TM bundle of the related receptors Trg and Tar (48). Side chains for aromatic residues W211 and F212 are shown as stick representations and mark the cytoplasmic end of TM2. The HAMP bundle is modeled on the structural coordinates for Af1503 HAMP (28). L218 is the first functionally critical residue of the Tsr HAMP bundle (74). The structure of the control cable, defined as the segment from G213 to S217, is unknown.
A sensory adaptation system enables E. coli chemoreceptors to detect temporal changes in chemoeffector concentrations as the cell moves about (49). The cytoplasmic portion of each receptor subunit contains 4 to 6 glutamyl residues, interposed between the HAMP domain and the kinase control tip of the receptor molecule, that undergo reversible methylation (Fig. 1) (30, 31, 63). CheR, a methyltransferase, attaches methyl groups to MCPs, while CheB, a methylesterase, removes MCP methyl groups (56, 58). The signaling state of the receptor determines the relative activities of CheR and CheB. Receptor molecules in the kinase-off state are better substrates for CheR; kinase-on receptors are better substrates for CheB (32, 62). Thus, the modification levels of receptor molecules reflect their recent signaling and stimulus history. An attractant increase, for example, produces an imbalance between receptor occupancy and modification state, triggering a kinase-off output signal. This response, in turn, leads to a net increase in receptor methylation that offsets its ligand occupancy, canceling the kinase-off response (55). The methylation changes that produce sensory adaptation also reverse the piston displacements triggered by the initial chemoeffector stimulus (36). Thus, the receptor methylation state provides an alternative readout of receptor conformation and signaling activity. Conversely, mutationally imposed modifications can shift receptor signaling states (11, 57).
The HAMP domain handles the receptor's input/output transactions (46). Each HAMP subunit is about 50 amino acids in length, comprising two amphipathic helices (AS1, AS2) joined by a nonhelical connector (CTR) (5, 67). The four HAMP helices in a homodimeric receptor molecule organize as a parallel four-helix bundle (28, 61, 66). The HAMP bundle conveys conformational information to the methylation helix (MH) bundle through the adjoined AS2 and MH1 helices to modulate receptor output. The alternative signaling states of HAMP could correspond to discrete conformations, as proposed in the gearbox (28) and scissors (61) models. In contrast, recent evidence suggests that HAMP signaling states may correspond to structural ensembles with different dynamic behaviors (46, 73, 74). The dynamic bundle model proposes that the packing stabilities of the HAMP and MH bundles are oppositionally coupled and regulated by the interplay of piston inputs to HAMP and methylation changes to the MH bundle (74).
In Tsr, two aromatic residues, W211 and F212, reside at the membrane core-head group interface and define the cytoplasmic ends of the TM2 helices (Fig. 1) (9, 22, 34). A five-residue segment joins F212 to L218, the first critical residue of the HAMP AS1 helices (Fig. 1) (74). This so-called control cable mediates conformational interactions between the TM2 and AS1 helices during transmembrane signaling (44, 46, 68, 74). In the dynamic bundle model of input/output signaling, the role of the control cable is to modulate the structural stability of HAMP in response to piston displacements of TM2 (74). It could conceivably act by relaxing or tightening structural tension on the HAMP helix packing interactions. In the crankshaft-gearbox and pushrod-scissors models of HAMP action, the control cable backbone would presumably need to have a rigid structure to transmit rotary or push-pull motions to HAMP (46).
Little is known about the structure of the control cable and its mechanism of conformational communication (46). Wright et al. investigated the signaling properties of mutant Tar receptors with alterations at the control cable-AS1 junction and concluded that the control cable had a helical character but that “a strong, permanent helical connection … is not necessary for normal transmembrane signaling” (68). Molecular dynamics simulations performed by Park et al. also indicate that the control cable could have a helical secondary structure (44). To gain further insight into the role of the control cable in transmembrane signaling, we constructed all possible single amino acid replacements at the five control cable residues of Tsr. We then characterized the signaling properties of the mutant receptors with respect to their steady-state kinase activity and whether their outputs changed in response to a serine stimulus or to actions of the sensory adaptation enzymes. Our findings shed new light on structural features of the control cable that are important for transmembrane signaling.
MATERIALS AND METHODS
Bacterial strains.
Strains were derivatives of E. coli K-12 strain RP437 (47), and their relevant genotypes are the following: UU1250 [Δaer-1 Δtsr-7028 Δ(tar-tap)5201 Δtrg-100] (3); UU1535 [Δaer-1 Δtsr-7028 Δ(tar-cheB)2234 Δtrg-100] (8); UU1623 [Δtsr-7028 Δtap-3654 Δtrg-100] (4); UU2377 [tsr-R69E Δaer-1 Δ(tar-tap)5201 Δtrg-4543 ΔrecA-SstII/EcoRI] (4); UU2378 [tsr-T156K Δaer-1 Δ(tar-tap)5201 Δtrg-4543 ΔrecA-SstII/EcoRI] (4); UU2610 [Δaer-1 Δ(tar-cheB)4346 Δtsr-5547 Δtrg-4543] (73); UU2611 [Δaer-1 Δ(tar-cheR)4283 Δtsr-5547 Δtrg-4543] (73); UU2612 [Δaer-1 Δ(tar-tap)4530 Δtsr-5547 Δtrg-4543] (73); and UU2632 [Δaer-1 Δ(tar-tap)4530 ΔcheB-4345 Δtsr-5547 Δtrg-4543] (73).
Plasmids.
Plasmids used included pKG116, a derivative of pACYC184 (16) that confers chloramphenicol resistance and has a sodium salicylate-inducible expression/cloning site (13); pPA114, a relative of pKG116 that carries wild-type tsr under salicylate control (3); pRR48, a derivative of pBR322 (10) that confers ampicillin resistance and has an expression/cloning site with a tac promoter and an ideal (perfectly palindromic) lac operator under the control of a plasmid-carried lacI repressor, inducible by isopropyl-β-d-thiogalactopyranoside (IPTG) (60); and pRR53, a derivative of pRR48 that carries wild-type tsr under IPTG control (60).
The plasmids used in receptor clustering assays were pVS49, a derivative of pACYC184 (16) that makes a functional yellow fluorescent protein (YFP)-CheZ fusion protein under inducible arabinose control (54); pVS102, a relative of pVS49 that makes a functional YFP-CheR fusion protein under inducible arabinose control (33); pPA789, a relative of pRR48 that expresses a functional cyan fluorescent protein (CFP)-CheZ fusion protein under inducible IPTG control (2); and pPA803, a pRR48 derivative that expresses a functional YFP-CheR fusion protein under inducible IPTG control (73).
Construction of control cable mutants.
Mutations in plasmids pPA114 and pRR53 were generated by QuikChange PCR mutagenesis, using either degenerate codon or site-specific primers, as previously described (3, 4). QuikChange products were introduced into UU1250 by either electroporation or CaCl2 transformation and tested for the ability to support Tsr function on soft agar plates (see below). Candidate plasmids were verified by sequencing the entire tsr coding region. Some derivatives of pPA114 and pRR53 carrying a missense mutation at tsr codon 215, 216, or 217 were obtained in a previous study (74).
Chemotaxis assays.
Host strains carrying tsr plasmids were assessed for chemotactic ability on tryptone soft agar plates (45) containing the appropriate antibiotics (ampicillin [50 μg/ml] or chloramphenicol [12.5 μg/ml]) and inducers (100 μM IPTG or 0.6 μM sodium salicylate). Plates were incubated at 30°C or 32.5°C for 7 to 10 h or at 24°C for 15 to 20 h.
Expression levels of mutant Tsr proteins.
Tsr expression from pRR53 and pPA114 derivatives was analyzed in strains UU1535 and UU2610 (to avoid multiple modification states) as described previously (3, 42).
Receptor clustering assays.
Mutant pPA114 derivatives were introduced by transformation into UU2612 cells already harboring either pPA789 or pPA803, and mutant pRR53 derivatives were introduced into UU2612 cells harboring pVS49 or pVS102. Cells containing each pair of compatible plasmids were grown at 30°C in tryptone broth containing 50 μg/ml ampicillin and 12.5 μg/ml chloramphenicol. Tsr expression from pRR53 derivatives was induced with 100 μM IPTG, YFP-CheZ (pVS49) was induced with 0.005% l-(+)-arabinose, and YFP-CheR (pVS102) was induced with 0.01% l-(+)-arabinose. Tsr expression from pPA114 derivatives was induced with 0.6 μM sodium salicylate, and CFP-CheZ (pPA789) and YFP-CheR (pPA803) were induced with 100 μM IPTG. Cells at mid-exponential phase were examined by fluorescence microscopy and analyzed as previously described (3, 42).
Assay of receptor modification state.
Cells harboring pRR53 derivatives were grown in tryptone broth containing 50 μg/ml ampicillin and 100 μM IPTG, while cells harboring pPA114 derivatives were grown in tryptone broth containing 12.5 μg/ml chloramphenicol and 0.6 μM sodium salicylate. Cells were grown at 30°C to mid-exponential phase, and 1-ml samples were pelleted by centrifugation, washed twice with KEP (10 mM K-PO4, 0.1 mM K-EDTA; pH 7.0), and lysed by boiling in sample buffer (35). Tsr bands were resolved by electrophoresis in 8% polyacrylamide gels containing sodium dodecyl sulfate and visualized by immunoblotting with a polyclonal rabbit anti-Tsr antiserum, as described previously (42).
To assess methylation responses to a serine stimulus, UU2612 cells containing test plasmids were grown and prepared as described above. The washed cells were divided into two 500-μl aliquots and incubated at 30°C for 20 min, after which l-serine (Sigma) was added to one aliquot to a final concentration of 10 mM. Both samples were incubated at 30°C for an additional 20 min and then analyzed by SDS-PAGE, as described above.
Flagellar rotation assays.
Flagellar rotation patterns of plasmid-containing cells were analyzed by antibody tethering as described previously (40, 42, 51). We classified cells into 5 categories according to their pattern of flagellar rotation, as follows: exclusively CCW, CCW reversing, balanced CCW-CW, CW reversing, and exclusively CW. The fraction of CW rotation time for a population of tethered cells was computed by a weighted sum of these rotation classes, as described previously (3, 73).
Protein modeling and structural display.
Atomic coordinates for the Tsr HAMP domain were generated from the Af1503 HAMP coordinates (Protein Data Bank [PDB] accession number 2ASW) (4). Coordinates for the TM bundle of Salmonella enterica serovar Typhimurium Tar were based on the modeled TM structure of Trg (48) and provided by Gerald Hazelbauer (University of Missouri). Structure images were prepared with MacPyMOL software (http://www.pymol.org).
RESULTS
Mutational scan of the Tsr control cable.
To identify functionally critical features of the control cable residues, we constructed single-amino-acid-replacement mutants in Tsr by all-codon mutagenesis of tsr codons 213 to 217 in plasmids pRR53 and pPA114. At optimal inducer concentrations (see Materials and Methods), these plasmids confer robust Tsr function to receptorless host strain UU2612, which carries chromosomal deletions of all five E. coli MCP family genes (tsr, tar, tap, trg, and aer). We tested UU2612 cells carrying mutant tsr plasmids for Tsr function on tryptone soft agar plates at 30°C. The control cable mutants fell into the following three general classes (Fig. 2): full function (colony size and ring morphology similar to those of the wild type) (Fig. 2A, white circles), impaired function (decreased colony size and/or mutant ring morphology) (Fig. 2A, gray circles), and null (comparable to the vector control phenotype) (Fig. 2A, black circles). Representative examples of these mutant phenotypes are shown in Fig. 2B.
Fig. 2.
Amino acid replacements and chemotaxis phenotypes of Tsr control cable mutants. (A) Summary of the mutational survey. Plus symbols denote the wild-type residues. The Tsr phenotypes produced by various amino acid replacements at each of the five control cable residues are indicated by circles: Tsr+ (white), Tsr+/− and Tsr−/+ (gray), and Tsr− (black). See the footnotes to Table 1 for an explanation of these phenotypes. (B) Examples of mutant receptor phenotypes. Tsr plasmids were introduced into strain UU2612, and transformant colonies were transferred to tryptone soft agar plates with toothpicks. The top plate contained 50 μg/ml ampicillin and 100 μM IPTG. The bottom plate contained 12.5 μg/ml chloramphenicol and 0.6 μM sodium salicylate. Plates were incubated for 7 h at 30°C. The wild-type parental plasmids used are pRR53 and pPA114. The corresponding empty vectors are pRR48 and pKG116. Tsr−: pRR48, I214D, I214P, I214R; pKG116, K215P. Tsr−/+: I214E, I214K, A216P. Tsr+/−: G213Y, I214H, I214L; S217I, S217V, S217L, S217M. Tsr+: pRR53, G213P; pPA114, S217A.
We characterized the functional properties of control cable mutants, particularly those with partial or complete loss-of-function chemotaxis phenotypes, with a number of additional tests, described below and summarized in Table 1.
Table 1.
Properties of Tsr control cable mutants
| Amino acid changea | Tsr phenotypeb | Tsr*c | % CW rotation time in hostd: |
Modified bye: |
SER CH3 responsef | ||||
|---|---|---|---|---|---|---|---|---|---|
| UU2610 (R− B−) | UU2611 (R− B+) | UU2612 (R+ B+) | UU2632 (R+ B−) | CheB (UU2611) | CheR (UU2632) | ||||
| Wild type | + | 1.00 | 75 | 29 | 25 | 81 | + | + | + |
| ATT mimic [1] | |||||||||
| G213P | + | 1.25 | 89 | 0 | 22 | 57 | + | ++ | + |
| G213Y | +/− | 0.90 | 49 | 0 | 24 | 81 | + | ++ | + |
| I214H | +/− | 1.45 | 80 | 0 | 26 | 90 | + | ++ | + |
| I214L | +/− | 1.30 | 74 | 0 | 28 | 86 | + | ++ | + |
| I214N | +/− | 1.20 | 81 | 0 | 25 | 91 | +/− | ++ | + |
| A216P | −/+ (CS) | 0.55 | 0 | 0 | 4 | 18 | +/− | ++ | + |
| S217A | + | 1.25 | 72 | 0 | 22 | 60 | + | ++ | + |
| S217I | +/− (CS) | 1.70 | 72 | 0 | 8 | 35 | +/− | ++ | + |
| S217L | +/− | 1.80 | 87 | 0 | 9 | 80 | +/− | ++ | + |
| S217M | +/− (CS) | 2.00 | 0 | 0 | 5 | 75 | +/− | ++ | + |
| S217V | +/− (CS) | 1.65 | 75 | 0 | 9 | 72 | +/− | + | + |
| ATT mimic [2] | |||||||||
| I214D | − | 1.30 | 0 | 1 | 1 | 9 | − | − | − |
| I214P | − | 0.95 | 0 | 1 | 2 | 7 | − | − | − |
| I214R | − | 0.95 | 0 | 0 | 5 | 7 | +/− | +++ | − |
| REP mimic | |||||||||
| I214E | −/+ (HS) | 1.30 | 88 | 74 | 18 | 86 | +/− | +/− | +/− |
| K215I | −/+ | 1.00 | 78 | 80 | 92 | 49 | + | − | − |
| Unclassified | |||||||||
| I214K | −/+ (HS) | 1.15 | 84 | 1 | 8 | 87 | +/− | +/− | + |
| K215P | − | 0.60 | 38 | 6 | 14 | 38 | − | − | +/− |
| K215Q | +/− (HS) | 1.15 | 84 | 0 | 18 | 56 | + | + | + |
| S217P | − (HS) | 1.00 | 77 | 0 | 25 | 40 | +/− | +/− | +/− |
Mutational changes are denoted by the amino acid replacement they cause in the following format: wild-type residue, residue number, mutant residue. Lightface text indicates mutant genes carried by plasmid pRR53, and italics indicate mutant genes carried by plasmid pPA114.
Tsr function was assessed on tryptone soft agar plates at 30°C, as follows: colony size and morphology comparable to those of the wild type (+), reduced colony size and/or aberrant ring morphology (+/−), colony size greater than that of the vector control but no evident ring at the border (−/+), and colony size and morphology comparable to those of the vector control (−). Heat-sensitive (HS) and cold-sensitive (CS) defects were identified by additional soft agar tests at 24°C and 36°C.
Expression level of the mutant protein relative to that of wild-type Tsr. See Materials and Methods for measurement details.
CW rotation times were calculated from the flagellar rotation profiles (see Materials and Methods) and rounded to the nearest whole number. Underlined values are averages of two or more independent measurements.
Modification of the mutant protein by CheB and CheR was assessed by band patterns in SDS-PAGE analyses (see Materials and Methods), as follows: no evident modification (−), some modification but less extensive than that for the wild-type protein (+/−), extent of modification comparable to that for the wild type (+); more extensive modification than that for wild-type Tsr (++); and much more extensive modification than that for wild-type Tsr (+++).
Increase in methylation state induced by a serine stimulus (SER) in host UU2612. Symbols are defined above in footnote e.
Expression level and stability of the mutant proteins.
Mutant plasmids were transferred to a receptorless strain that also lacks the CheR methyltransferase and CheB deamidase/esterase enzymes of the sensory adaptation system. Owing to the absence of MCP covalent modifications in this background, the mutant Tsr molecules migrated as a single band in SDS-PAGE, thereby facilitating quantitative analysis (see Materials and Methods). All mutant proteins had steady-state expression levels that differed from the wild type by no more than 2-fold (Table 1). We conclude that the mutant proteins adopt native or near-native structures and that their functional defects are not due to poor expression or accelerated degradation.
Dominance and signaling symmetry of control cable defects.
Mutant plasmids were tested for dominance effects in tsr strains UU2377 and UU2378, which have recessive lesions at the serine-binding determinants in the Tsr periplasmic domain (R69E and T156K, respectively). Coexpression of binding-defective and mutant control cable (Tsr*) subunits produced fully functional heterodimers for all combinations of tested mutants (data not shown). R69E/Tsr* molecules are expected to transmit serine-binding information through the R69E subunit, whereas T156K/Tsr* heterodimers should signal through the Tsr* subunit (4, 72). The absence of functional asymmetry in these tests indicates that one normal control cable suffices for signaling, regardless of the transmission path. Moreover, the recessive character of the control cable defects indicates that the mutant subunits have normal membrane topology and dimerization ability.
Functional interactions of Tsr control cable mutants with the aspartate receptor.
Mutant plasmids were tested for functional rescue and jamming effects in UU1623, which carries a wild-type aspartate receptor gene (tar). Tar and Tsr molecules can function in mixed signaling teams based on trimers of dimers (2, 3, 59). Mixed-team formation enables wild-type Tar to restore function to some Tsr mutants (rescue); conversely, some Tsr defects block wild-type Tar function (jamming). Of the nine null or near-null Tsr control cable defects, six (I214D, I214E, I214K, I214P, K215I, S217P) exhibited neither rescue nor jamming behavior, two (K215P, A216P) were rescued by Tar, and one (I214R) jammed Tar (data not shown). We conclude that at least the three rescuable or jamming receptors are able to form mixed trimers of dimers with Tar molecules.
Temperature effects on mutant control cable function.
According to the dynamic bundle model of HAMP signal control, attractant (kinase-deactivating) stimuli enhance HAMP stability, whereas repellent (kinase-activating) stimuli reduce HAMP stability (74). If Tsr control cable lesions perturb HAMP stability, their functional defects might respond to temperature shifts. Accordingly, we assessed the chemotactic behavior of control cable mutants, initially defined at 30°C, at lower (24°C) and higher (36°C) temperatures. Four mutants (S217V, S217I, S217M, A216P) exhibited somewhat more function at 36°C than at 24°C; we consider their defects to be cold sensitive (Table 1). Similarly, four mutants (I214E, I214K, K215Q, S217P) exhibited somewhat more function at 24°C than at 36°C; we consider their defects to be heat sensitive (Table 1).
If we assume that lower temperature enhances HAMP stability, then control cable lesions with cold-sensitive behavior might mimic the attractant-stimulated condition, because low temperature (i.e., increased HAMP stability) exacerbates their signaling defect. Conversely, if higher temperatures reduce HAMP stability, then lesions that cause heat-sensitive behavior might mimic a repellent-stimulated condition, because high temperature (i.e., decreased HAMP stability) exacerbates their signaling defect. To explore the relationship between temperature effects and signal output, we next examined the CheR and CheB substrate properties of mutant Tsr molecules and their steady-state output signals at different modification states. We interpreted the mutant behaviors in the context of a biphasic relationship between HAMP stability and CheA kinase activity, a consequence of dynamic bundle control of HAMP output (73) (Fig. 3A).
Fig. 3.
Output signals of wild-type Tsr molecules in different modification states. (A) CheA activity as a function of HAMP or MH bundle structural stability. The output curve is based on recent studies that posit a biphasic, dynamic bundle mechanism for Tsr-HAMP input-output control (73, 74). Black arrowheads denote stabilizing effects, and white arrowheads denote destabilizing effects. The flagellar rotation patterns of Tsr strains containing different combinations of CheR and CheB proteins were assessed by cell tethering (73). Tsr molecules in the absence of both CheB and CheR have a QEQE modification state, approximating a half-methylated condition (dark gray circle). In the CheB-only strain, Tsr molecules are deamidated to EEQE, QEEE, and EEEE states, corresponding to a mostly unmethylated condition (white circle). In the CheR-only strain, Tsr molecules can be methylated to QEmQE, QEQEm, and QEmQEm states, corresponding to a nearly fully methylated condition (black circle). In strains containing both CheR and CheB, Tsr molecules undergo deamidation, methylation, and demethylation reactions that drive the receptor ensemble to the adaptation set point, corresponding to a low effective methylation state (light gray circle). (B) SDS-PAGE assessment of Tsr modification states. Tsr proteins from the four hosts used for panel A were analyzed by gel electrophoresis and visualized by immunoblotting, as described in Materials and Methods. The triplet bands are a mixture of Tsr reference molecules in the EEEE, QEQE, and QQQQ modification states.
Assessing the signaling behaviors of Tsr molecules.
The flagellar rotation patterns of receptor-containing cells reflect their steady-state CheA activity: high kinase activity produces CW rotation, and low kinase activity produces CCW rotation. Attractant stimuli, such as serine increases, shift wild-type Tsr molecules toward a kinase-off signaling state [CCW(A)], while repellent stimuli shift output toward a kinase-on state (CW) (Fig. 3A). In adaptation-proficient cells, the interplay of CheR and CheB modifications returns receptor molecules to their prestimulus set point, characterized by intermediate kinase activity (Fig. 3A). The dynamic bundle model proposes that these output behaviors reflect opposed stability interactions between the HAMP and methylation helix (MH) bundles (74). In the receptor's physiological operating regime, kinase activity varies inversely with the stability of the HAMP bundle and directly with the stability of the MH bundle. The biphasic character of the output relationship is evident when the stability of the system is driven far from the adaptation set point into a reversed output control regime with a kinase-off state [CCW(B)] that results from a very unstable HAMP bundle and a very stable MH bundle (73).
To manipulate the receptor modification state, we expressed mutant Tsr molecules in strains that had different combinations of the CheR and CheB enzymes. To assess the modification states of those Tsr molecules, we examined their SDS-PAGE band patterns under conditions in which the effective methylation level influences electrophoretic mobility (12, 17, 24) (Fig. 3B). In strain UU2610, which lacks both modification enzymes (i.e., CheR− CheB− [R− B−]), Tsr molecules have glutamine residues at two of the four principal methylation sites (29). Wild-type Tsr molecules in this QEQE state behave as though half methylated, producing an intermediate SDS-PAGE migration rate (Fig. 3B) and high kinase activity (Fig. 3A). In UU2612, which has both adaptation enzymes (R+ B+), the wild-type receptor population is heterogeneous, but the average modification state maintains overall output at the set point position (∼25% CW) (Fig. 3A) (73). In low-serine environments, the set point modification state is low, corresponding to about one Q or methylated E (Em) residue per subunit (Fig. 3B). In a high-serine environment, the average methylation state at the set point is considerably higher (Fig. 3B). The UU2611 host (R− B+) can only deamidate Q sites, shifting Tsr modification toward the fully unmethylated EEEE state, which has the lowest SDS-PAGE migration rate (Fig. 3B) and produces set point-level kinase activity (Fig. 3A). In contrast, the UU2632 host (R+ B−) can only methylate E sites, shifting Tsr modification toward the QEmQEm state, which has a much higher SDS-PAGE migration rate (Fig. 3B). Highly modified wild-type Tsr molecules produce inverted aerotactic responses (21), implying that they lie in the reversed regime of the biphasic output curve.
The biphasic dynamic bundle model predicts that highly methylated receptors could exhibit less kinase activity than QEQE receptors if the system is driven far enough toward the CCW(B) state. This paradoxical behavior is not readily apparent with wild-type Tsr, whose CW output was comparably high in both UU2610 and UU2632 (Fig. 3A), but was evident with some control cable mutants (see below). Biphasic output curves provide the simplest explanation for reduced CW output at high modification states and are used below to summarize the relationship between the modification state and signal output in various mutant receptors. In the output diagrams, the degree of shading for the four host symbols reflects the effective methylation states those hosts support for wild-type receptors, as follows: unmethylated (UU2611, white symbols), some methylation (UU2612, light gray symbols), QEQE modification (UU2610, dark gray symbols), and mostly methylated (UU2632, black symbols).
Control cable lesions that mimic an attractant stimulus.
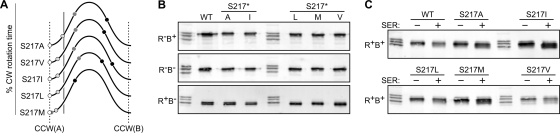
Our working model predicts that receptors with ATT-mimic lesions should have outputs shifted toward the CCW(A) kinase-off state. As a consequence, the sensory adaptation system should increase the steady-state methylation level of the mutant receptors. Eleven control cable mutants met these criteria (ATT mimic [1]) (Table 1), and those with hydrophobic amino acid replacements at S217 serve to illustrate these traits (Fig. 4). The S217V, S217I, and S217L receptors produced no CW output in the R− B+ host, some CW output in the R+ B+ host but below the wild-type set point, and high CW output in the R− B− host, comparable to that of the wild type in the QEQE state. In the R+ B− host, these mutant receptors were extensively methylated (Fig. 4B), but their outputs were no more, and often less, CW than those in the R− B− host, which we interpret as evidence of entry into the reversed output regime (Fig. 4A). Tsr(S217A) had a similar output pattern except in the R+ B+ host, where its CW output reached the wild-type set point (Fig. 3A). In contrast, Tsr(S217M) produced very little CW output in all but the R+ B− host (Fig. 4A). The S217V, S217I, and S217M defects also exhibited cold-sensitive character, a feature consistent with ATT-mimic defects (Table 1). We suggest that the hydrophobic replacement S217* receptors represent a range of ATT-mimic behaviors, with the severity of their signaling defect related to the size and hydrophobicity of their mutant amino acid. S217A output is close to that of the wild type but more easily driven to the CCW(A) state by CheB action. At the opposite extreme, S217M output is driven away from the CCW(A) state only by CheR action. The modification patterns of the S217* proteins support this interpretation. All of these mutant receptors were more highly modified than the wild type in both the R+ B− host (Fig. 4B) and the R+ B+ host (Fig. 4C). The increased methylation levels of the S217* receptors are consistent with an intrinsic signal output shifted toward the CCW(A) state, necessitating extensive methylation to drive them toward the adaptation set point. Other control cable mutants (G213P, G213Y, I214H, I214L, I214N, A216P) had similar properties (Table 1).
Fig. 4.
Examples of Tsr control cable mutants with ATT-mimic [1] signaling defects. (A) Output curves for Tsr(S217) mutants. These curves use the same shading convention for modification states and hosts as the curve for wild-type Tsr shown in Fig. 3A. y axis, extent of CW rotation time; x axis, dynamic range of the HAMP and MH bundles; vertical gray line, wild-type adaptation set point. The curves for different mutant receptors are stacked simply to conserve space. (B) Modification profiles of the S217 mutant receptors in various host strains. Unlabeled lanes are EEEE/QEQE/QQQQ markers (Fig. 3B). WT, wild type. (C) Modification profiles of the S217 mutant receptors in an adaptation-competent (CheR+ CheB+) host before (−) and after (+) experiencing a serine stimulus (SER). Unlabeled lanes are EEEE/QEQE/QQQQ markers (Fig. 3B).
All mutants assigned to the ATT-mimic [1] class exhibited partial to nearly full Tsr function in soft agar assays (Table 1). In all cases, however, the mutant colonies had sharper (thinner, denser) bands of chemotactic cells at their leading edge than did wild-type colonies (Fig. 2B). The sharp-edge phenotype has been seen previously in Tar mutants (22, 68) and is most likely a symptom of the elevated methylation status of ATT-mimic receptors. Extensively methylated Tsr molecules should have a reduced dynamic range of serine sensing (38, 39), so the sharp cell band probably reflects congregation of the migrating cells in a narrow concentration range at the high end of the serine gradient.
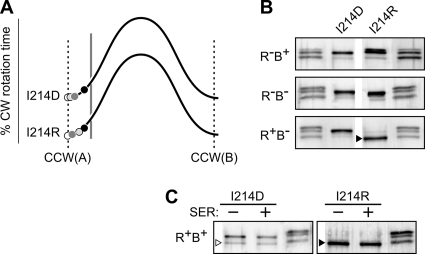
Three control cable mutants with null chemotaxis phenotypes (I214D, I214P, I214R) appear to have more extreme ATT-mimic [2] defects (Table 1). These mutant receptors produced very low levels of CW output in all hosts, but their CW output was highest in the R+ B− host, consistent with the CCW(A) end of the biphasic output curve (Table 1 and Fig. 5A; see also Fig. 7A). Tsr(I214R) underwent some deamidation in the R− B+ host and extensive methylation in the R+ B− host (Fig. 5B). In contrast, the I214D and I214P receptors were not appreciably modified in any host (Fig. 5B; see also Fig. 7B). Consistent with their null chemotaxis phenotypes, none of these mutant receptors showed any methylation increase in response to a serine stimulus (Fig. 5C and Table 1; see also Fig. 7C). We suggest that Tsr(I214R) has a strong ATT-mimic defect that elicits extensive methylation, but with little effect on signal output. Tsr(I214D) and Tsr(I214P) may have an even stronger ATT-mimic defect that locks the receptor in the CCW(A) state, making it refractory to modification by both CheB and CheR.
Fig. 5.
Examples of Tsr control cable mutants with ATT-mimic [2] signaling defects. (A) Output curves for Tsr(I214D) and Tsr(I214R). See Fig. 4A for further explanation. (B) Modification profiles of the I214D and I214R mutant receptors in various hosts. The band marked with a black triangle corresponds to the faster of the two heavily methylated bands produced by wild-type Tsr in the same host strain. Unlabeled lanes are EEEE/QEQE/QQQQ markers (Fig. 3B). (C) Modification profiles of the I214D and I214R mutant receptors in an adaptation-competent (CheR+ CheB+) host before (−) and after (+) experiencing a serine stimulus. The white triangle indicates a chromosomally encoded nonreceptor protein that cross-reacts with the anti-Tsr antibody. The black triangle marks the same band as that described above in panel B. Unlabeled lanes are EEEE/QEQE/QQQQ markers (Fig. 3B).
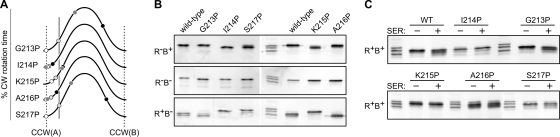
Fig. 7.
Signaling properties of Tsr control cable mutants with proline replacements. (A) Output curves for mutant receptors with a single proline replacement at each control cable residue. See Fig. 4A for further explanation. (B) Modification profiles of the proline replacement mutant receptors in various hosts. (C) Modification profiles of the proline replacement mutant receptors in an adaptation-competent (CheR+ CheB+) host before (−) and after (+) experiencing a serine stimulus.
Cluster formation by ATT-mimic receptors.
Five control cable mutants with ATT-mimic lesions (S217M, A216P, I214R, I214P, I214D) showed no evidence of CheA activation in the R− B− host (Table 1). In the QEQE modification state, these receptors might be defective in ternary complex assembly; alternatively, they might form ternary complexes that are locked in a kinase-off signaling state. To distinguish these possibilities, we examined the clustering properties of these ATT-mimic receptors in the R− B− host, using two different fluorescence microscopy reporters: YFP-CheR binds to the NWETF pentapeptide sequence at the C terminus of each receptor subunit (50, 54, 69), and YFP-CheZ binds to CheAS, which is an alternate cheA translation product and a component of ternary signaling complexes (15, 52, 54). Receptors that fail to assemble trimers of dimers, a prerequisite for both ternary complex and cluster formation, do not exhibit clusters with either of the reporters (K. K. Gosink, Y. Zhao, and J. S. Parkinson, unpublished data). Receptors that form trimers of dimers but cannot assemble ternary complexes show clusters detectable with the CheR reporter but not with the CheZ reporter (42). Finally, receptors that are able to form ternary complexes will show clustering with both reporters (42; Gosink et al., unpublished). The S217M, A216P and I214R receptors formed polar clusters detectable with both reporters (data not shown). In the QEQE state these receptors evidently assemble ternary complexes that cannot activate CheA. In contrast, the I214P and I214D receptors failed to form clusters detectable with either of the reporters (data not shown). In the QEQE state, these receptors evidently have a conformation or dynamic behavior that precludes trimer-of-dimer formation, consistent with their classification as the most extreme of the ATT-mimic control cable lesions (Table 1).
Control cable lesions that mimic a repellent stimulus.
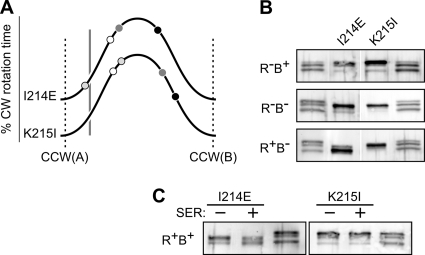
Two control cable mutants (I214E and K215I) had signaling properties that may represent repellent (REP)-mimic defects (Table 1). The K215I receptor produced considerable CW output in all hosts, but its kinase activity was lowest in the R+ B− host, implying an overall output shift toward the CCW(B) end of the stability range (Fig. 6A). Notably, Tsr(K215I) showed much higher CW output than the wild type in the R− B+ host, despite undergoing extensive CheB modification (Fig. 6B). Moreover, the K215I receptor was a poor CheR substrate (Fig. 6B) and showed no methylation response to a serine stimulus (Fig. 6C). These properties are consistent with a REP-mimic lesion that shifts HAMP stability toward the high CW portion of the biphasic output curve.
Fig. 6.
Examples of Tsr control cable mutants with REP-mimic signaling defects. (A) Output curves for Tsr(I214E) and Tsr(K215I). See Fig. 4A for further explanation. (B) Modification profiles of the I214E and K215I mutant receptors in various hosts. Unlabeled lanes are EEEE/QEQE/QQQQ markers (Fig. 3B). (C) Modification profiles of the I214E and K215I mutant receptors in an adaptation-competent (CheR+ CheB+) host before (−) and after (+) experiencing a serine stimulus. Unlabeled lanes are EEEE/QEQE/QQQQ markers (Fig. 3B).
Tsr(I214E) exhibited similar, but less extreme, behavior: it underwent some CheR modification (Fig. 6B), showed a modest methylation response to serine (Fig. 6C), and had a heat-sensitive character, consistent with a conditional REP-mimic defect (Table 1). The I214E receptor produced high CW output in three of the four host strains but low CW output in the R+ B+ host (Fig. 6A). Perhaps the interplay of CheB and CheR action in the R+ B+ host enables I214E molecules to attain modification states that cannot be achieved with either enzyme alone. In addition, the heterogeneity of modification states in adaptation-competent strains can influence the signaling behavior of the receptor ensemble (53). Conceivably, these factors shift Tsr(I214E) output close to the adaptation set point in the R+ B+ host. We discount the possibility that the I214E receptor has been driven into the reversed regime because it was not as extensively modified in the R+ B+ host as it was in the R+ B− host (Fig. 6B and C).
Control cable lesions with ambiguous signaling defects.
Four control cable mutants (I214K, K215P, K215Q, and S217P) exhibited less easily classified behaviors (Table 1). Three of these mutant receptors (I214K, K215Q, S217P) showed heat-sensitive characters and high CW output in R− B− and R+ B− hosts, properties consistent with REP-mimic defects (Table 1). Moreover, two of them (I214K and S217P) were poor substrates for both CheR and CheB modifications, similar to the I214E REP-mimic mutant described above (Table 1; Fig. 7). However, all three receptors produced low CW output in CheB+ hosts, suggesting that CheB action drives them close to the CCW(A) end of the output range, behavior that is difficult to reconcile with a simple REP-mimic defect.
The K215P receptor produced CW output near the set point level in both the R− B− and R+ B+ hosts, unlike any other control cable mutants (Fig. 7A). Moreover, its output in the R+ B− host varied from one experiment to another (data not shown), implying that K215P molecules might be inefficient substrates for CheR-mediated methylation. In fact, SDS-PAGE analyses showed that this mutant receptor underwent essentially no modification by CheR and little modification by CheB (Fig. 7B). Thus, K215P signal activity lies near the CCW(A) end of the output curve, reminiscent of an ATT-mimic [1] defect. However, its inefficient modification by CheR and CheB implies a more extreme ATT-mimic [2] defect (Table 1).
Signaling consequences of proline replacements in the Tsr control cable.
Most amino acid replacements in the control cable residues had no detrimental effects on Tsr signaling (Fig. 2A). In contrast, proline replacements at four of the five control cable residues either seriously impaired (A216P) or abrogated (I214P, K215P, S217P) Tsr function (Fig. 2). The unique side-chain character of proline could conceivably disrupt a control cable secondary structure, perhaps an α-helix, that is critical for signaling. However, proline replacements caused somewhat different signaling behaviors at each control cable residue (Fig. 7).
The G213P receptor was almost fully functional but exhibited modest ATT-mimic [1] behavior in output (Fig. 7A) and modification (Fig. 7B and C) tests. The I214P receptor had severe ATT-mimic [2] output behavior (Fig. 7A) and was not subject to either CheR or CheB modification (Fig. 7B and C). The K215P receptor was also an ineffective substrate for CheR and CheB modifications (Fig. 7B and C) but generated moderate levels of CW output in all host strains (Fig. 7A) and regained Tsr function in the presence of aspartate (Tar) receptors (data not shown). The A216P receptor was also functionally rescued by Tar (data not shown) but had conventional ATT-mimic [1] output (Fig. 7A) and modification (Fig. 7B and C) behaviors. Finally, the S217P receptor had a nearly normal output pattern, similar to that of G213P (Fig. 7A), but lacked Tsr signaling function, was a poor substrate for CheB, and was an even poorer substrate for CheR modifications (Fig. 7B and C).
DISCUSSION
Symmetric versus asymmetric conformational inputs to HAMP.
Stimulus-induced conformational changes in Tsr molecules are asymmetric; the TM2 helix in one subunit moves relative to its counterpart in the other subunit (25). Some amino acid replacements in the Tsr control cable residues altered the steady-state signal output of the receptor and appeared to mimic the signaling consequences of attractant or repellent stimuli. Unlike the asymmetric TM2 piston displacements that accompany sensory stimuli, control cable lesions necessarily cause symmetric structural changes in the Tsr homodimer. However, symmetric structural changes (amino acid replacements, disulfide bonds) in the periplasmic sensing domain or TM bundle can also mimic chemoeffector signaling effects (shifted outputs, altered adaptation responses) (6, 7, 22, 23, 25, 71). Moreover, the hybrid transducer Nart, which carries the periplasmic sensing through the HAMP domains of NarX joined to the methylation and kinase control domains of Tar (64, 65), mediates chemotactic responses to nitrate, whose binding causes quasisymmetric piston displacements (19). We conclude that symmetric conformational inputs most likely modulate HAMP signaling by the same mechanism as that used for asymmetric TM2 piston displacements. In both cases, control cable input influences HAMP conformation or structural stability to regulate signal output.
Dynamic control of receptor modification reactions.
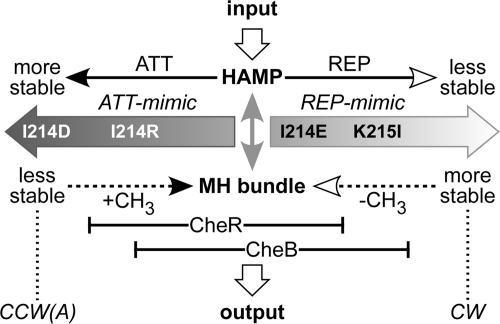
The signaling properties of Tsr control cable mutants are readily interpretable in terms of the dynamic bundle model (74). The most severe ATT-mimic lesions, I214D and I214P, blocked assembly of ternary signaling complexes, most likely due to the inability to form trimers of dimers. This behavior could reflect a lack of dynamic motion at the signaling hairpin tip (Fig. 1), whose stability bears an opposed, yin-yang relationship to that of the MH bundle (62). The I214D and I214P receptors were ineffective substrates for both CheR and CheB modifications, suggesting that neither enzyme can act on a highly dynamic MH bundle. The I214R receptor exhibited less drastic ATT-mimic behavior: it assembled ternary signaling complexes but could not activate CheA. Tsr(I214R) molecules were inefficiently modified by CheB but underwent extensive CheR-mediated methylation, implying that CheR can act on more dynamic MH bundles than CheB. The substrate preferences of CheR and CheB also seem to differ at the kinase-on end of the physiological dynamic range (Fig. 8). The REP-mimic K215I receptor, for example, was subject only to CheB modification, whereas the I214E REP-mimic receptor underwent modification by both of the enzymes. K215I caused more severe CW-shifted output than did I214E, suggesting that CheB can act on more stable MH bundles than CheR (Fig. 8). We conclude that the dynamic properties of the MH bundle probably determine the substrate preferences of the adaptation enzymes (73), which in turn account for net methylation increases in response to attractant stimuli and for net methylation decreases in response to repellent stimuli (Fig. 8).
Fig. 8.
Interplay of stimulus and modification effects in the physiological operating range. The opposed stability interactions of the HAMP and MH bundles, which are central to the dynamic bundle model of HAMP signaling, are denoted by the gray, double-headed vertical arrow. Horizontal solid lines indicate input controls on HAMP bundle stability; dashed lines indicate modification influences on MH bundle packing. Black arrowheads denote stabilizing effects, and white arrowheads denote destabilizing effects. Most control cable lesions mimic the HAMP-stabilizing effects of an attractant stimulus, but some mimic the HAMP-destabilizing effects of a repellent stimulus. The modification properties of extreme ATT-mimic (I214D, I214R) and REP-mimic (I214E, K215I) lesions indicate that CheR operates most effectively on MH bundles at the unstable end of the dynamic range but that CheB operates most effectively on MH bundles that are shifted toward the more stable end of the dynamic range. The substrate ranges for the two enzymes overlap at intermediate MH bundle stabilities around the adaptation set point. Tsr(I214D) is not modified by either enzyme, Tsr(I214R) is modified only by CheR, Tsr(I214E) is modified by both CheR and CheB, and Tsr(K215I) is modified only by CheB.
Structural features of the TM2-AS1 junction.
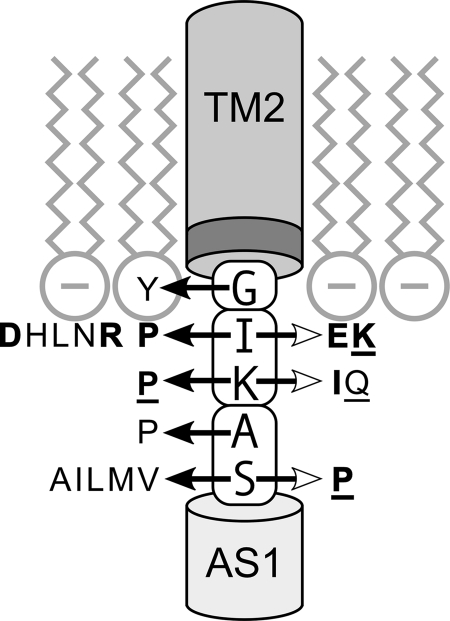
The Tsr control cable resides at the membrane-cytoplasm interface (Fig. 9). TM2 residues W211 and F212 comprise an aromatic belt that probably partitions between the membrane head groups and fatty acid tails to anchor the cytoplasmic end of the TM2 helix at the interfacial region (34). G213 most likely resides in the same membrane environment as the aromatic belt residues, because only tyrosine, perhaps acting as an additional aromatic anchor, impaired function at this position. I214 probably lies near the membrane head groups because this position was sensitive to both basic and acidic replacements, which could conceivably perturb control cable structure through attractive or repulsive interactions with the negatively charged phospholipids or with the positively charged residues in the cytoplasmic N terminus of Tsr. (The two subunits in a Tsr molecule probably lie too far apart at the TM2-AS1 junction for substantive intersubunit structural interactions between the control cable residues [14, 25].) K215, in contrast, tolerated both positive and negative charge replacements, suggesting that it lies outside the influence of the membrane head groups and the N terminus. Finally, A216 and S217 tolerated a wide variety of amino acid replacements, suggesting that these residues are located well away from the membrane interfacial region.
Fig. 9.
Signaling properties of control cable lesions. The cartoon depicts the inner leaflet of the cytoplasmic membrane and the portions of the TM2 and AS1 helices that adjoin the control cable. The darker segment at the cytoplasmic end of TM2 denotes the aromatic belt residues (W211, F212) that partition at the lipid tail-head group interface. Amino acid replacements in the control cable that impair (lightface type) or abrogate (boldface type) signaling function are indicated by horizontal arrows: white arrowheads denote lesions with REP-mimic properties, and black arrowheads denote lesions with ATT-mimic properties. Underlined replacements produce some, but not all, of the signaling characteristics of ATT- or REP-mimic defects. Proline replacements, which impair or abrogate function at all but the residue 213 position, probably do so through effects on control cable helicity. The control cable residues fall into three functionally distinct groups, indicated by the enclosing boxes: G213 (G), structural swivel; I214 (I) and K215 (K), membrane proximal; and A216 (A) and S217 (S), membrane distal, possibly part of the AS1 helix (28).
This view of the TM2-AS1 junction in Tsr agrees with that of Boldog and Hazelbauer, who reported that residues comprising the aromatic belt and control cable in Trg were fully solvent exposed (9). In contrast, Miller and Falke suggested that the corresponding segment of Tar was embedded in the hydrophobic core of the membrane (41). Conceivably, control cable topology varies a bit from one receptor type to another, although Tar and Tsr are mechanistically indistinguishable in most other respects.
Proline replacements at 4/5 control cable residues disrupted function, suggesting that the control cable has some helical character or requires backbone flexibility, structural attributes that would be compromised by proline (20). However, if the five-residue control cable were strictly helical, the axial faces of the TM2 and AS1 helices would be nearly 180° out of register (25, 61). We suggest that the control cable must have some flexibility in order to moderate structural conflict between the TM2 and AS1 helices. G213 was the only control cable position that tolerated a proline replacement, suggesting that it is not an important structural element of a control cable helix. Moreover, because prolines typically destabilize helices in the N-terminal direction (20), it follows that G213 is not a critical part of the TM2 helix. Rather, residue 213 may serve as a structural swivel between the TM2 helix and a control cable helix comprised of residues 214 to 217, all of which are sensitive to proline replacements.
Helix extension model of TM2 piston signaling.
If the TM2 control cable connection at G213 has structural flexibility, it seems unlikely that TM2 piston motions could propagate through the control cable to modulate HAMP. Moreover, chemoreceptor HAMP domains typically have a proline residue near the start of the AS1 helix (P221 in Tsr), which probably serves to discourage a rigid helical connection between the control cable and AS1. These structural features suggest that TM2 movements might influence HAMP stability by regulating control cable helicity. Perhaps piston-induced changes in the local environment of residue 213 modulate the flexibility of the TM2 control cable junction. HAMP-destabilizing repellent signals might stiffen the connection, extending the TM2 helix into the control cable to exacerbate the structural clash with HAMP. HAMP-stabilizing attractant signals might relax the connection to relieve the structural strain on the HAMP bundle.
Park et al. recently reported molecular dynamics simulations of the Tar TM bundle joined to the Af1503 HAMP domain that show how stimulus signals might regulate control cable structure (44). They suggested that an attractant-induced piston displacement causes bending and lateral sliding of the signaling TM2 helix in the plane of the membrane, which in turn modulate control cable helicity.
A helix extension mechanism of TM2 signaling is fully consistent with the properties of Tsr control cable mutants and with the earlier study by Wright et al. (68). Proline replacements at control cable residues 214 to 216 caused ATT-mimic behaviors, consistent with a reduction of control cable helicity and a consequent relaxation of the structural conflict between TM2 and HAMP. However, the S217P mutant receptor had some REP-mimic character, perhaps reflecting direct destabilization of the AS1 helix. Other than prolines, the Tsr control cable tolerated a variety of amino acid replacements with no apparent detriment in transmembrane signaling function. Glycine, alanine, cysteine, serine, threonine, phenylalanine, and tryptophan all supported full Tsr function at each control cable position, indicating that the chemical nature and size of the control cable side chains are not individually critical for signal control. At residue 213, only tyrosine caused a signaling defect, perhaps by partitioning at the aromatic anchor position. At the membrane-proximal residues 214 and 215, some polar and nonpolar amino acid replacements impaired function (Fig. 9). The I214 signaling defects most likely reflect aberrant structural interactions of the mutant side chains with the interfacial environment; the K215 defects might arise through interactions of the mutant side chains with the adjacent 214 residue. Those interactions might change control cable helicity or render it insensitive to piston control. The membrane-distal residues, 216 and 217, were impervious to the interfacial environment (Fig. 9). Except for proline, the side-chain character of residue 216 was irrelevant to signaling proficiency. The S217 position was similarly tolerant of most amino acid replacements, except for those with hydrophobic side chains, which caused ATT-mimic behavior. The severity of these signaling defects scaled with the length of the aliphatic side chain: alanine caused modest defects, and methionine caused severe defects. We suggest that a hydrophobic residue at the S217 position stabilizes the HAMP bundle by contributing to nonpolar packing forces between the AS1 helices.
The helix extension model makes a number of testable predictions about control cable structural properties that are important for signaling. For example, the 5-residue length of the wild-type Tsr control cable might not be a critical factor if helix potential is the key variable. However, if the control cable transmits piston or rotary motions to HAMP, then precise length should be critical for signal control. Moreover, if helix potential is paramount, then a variety of artificial control cables with comparable helicity might be expected to retain signaling ability. These and other tests of the helix extension model will be reported in a follow-up study.
ACKNOWLEDGMENTS
We thank Jerry Hazelbauer (University of Missouri) for providing atomic coordinates, Danyelle Strehlow (University of Utah) for technical assistance, and Claudia Studdert (National University, Mar del Plata, Argentina), Mike Manson (Texas A&M University), and Roger Draheim (Goethe University, Frankfurt, Germany) for providing comments on the manuscript.
This work was supported by research grant GM19559 from the National Institute of General Medical Sciences. The Protein-DNA Core Facility at the University of Utah (oligonucleotide synthesis and DNA sequencing) receives support from National Cancer Institute grant CA42014 to the Huntsman Cancer Institute.
Footnotes
Published ahead of print on 29 July 2011.
REFERENCES
- 1. Alexander R. P., Zhulin I. B. 2007. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. U. S. A. 104:2885–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ames P., Parkinson J. S. 2006. Conformational suppression of inter-receptor signaling defects. Proc. Natl. Acad. Sci. U. S. A. 103:9292–9297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ames P., Studdert C. A., Reiser R. H., Parkinson J. S. 2002. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:7060–7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ames P., Zhou Q., Parkinson J. S. 2008. Mutational analysis of the connector segment in the HAMP domain of Tsr, the Escherichia coli serine chemoreceptor. J. Bacteriol. 190:6676–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aravind L., Ponting C. P. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 176:111–116 [DOI] [PubMed] [Google Scholar]
- 6. Baumgartner J. W., Hazelbauer G. L. 1996. Mutational analysis of a transmembrane segment in a bacterial chemoreceptor. J. Bacteriol. 178:4651–4660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beel B. D., Hazelbauer G. L. 2001. Signalling substitutions in the periplasmic domain of chemoreceptor Trg induce or reduce helical sliding in the transmembrane domain. Mol. Microbiol. 40:824–834 [DOI] [PubMed] [Google Scholar]
- 8. Bibikov S. I., Miller A. C., Gosink K. K., Parkinson J. S. 2004. Methylation-independent aerotaxis mediated by the Escherichia coli Aer protein. J. Bacteriol. 186:3730–3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boldog T., Hazelbauer G. L. 2004. Accessibility of introduced cysteines in chemoreceptor transmembrane helices reveals boundaries interior to bracketing charged residues. Protein Sci. 13:1466–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bolivar F., et al. 1977. Construction and characterization of new cloning vehicles. Gene 2:95–113 [PubMed] [Google Scholar]
- 11. Bornhorst J. A., Falke J. J. 2001. Evidence that both ligand binding and covalent adaptation drive a two-state equilibrium in the aspartate receptor signaling complex. J. Gen. Physiol. 118:693–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boyd A., Simon M. I. 1980. Multiple electrophoretic forms of methyl-accepting chemotaxis proteins generated by stimulus-elicited methylation in Escherichia coli. J. Bacteriol. 143:809–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Buron-Barral M., Gosink K. K., Parkinson J. S. 2006. Loss- and gain-of-function mutations in the F1-HAMP region of the Escherichia coli aerotaxis transducer Aer. J. Bacteriol. 188:3477–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Butler S. L., Falke J. J. 1998. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochemistry 37:10746–10756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cantwell B. J., et al. 2003. CheZ phosphatase localizes to chemoreceptor patches via CheA-short. J. Bacteriol. 185:2354–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chang A. C. Y., Cohen S. N. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic miniplasmid. J. Bacteriol. 134:1141–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chelsky D., Dahlquist F. W. 1980. Structural studies of methyl-accepting chemotaxis proteins of Escherichia coli: evidence for multiple methylation sites. Proc. Natl. Acad. Sci. U. S. A. 77:2434–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chervitz S. A., Falke J. J. 1996. Molecular mechanism of transmembrane signaling by the aspartate receptor: a model. Proc. Natl. Acad. Sci. U. S. A. 93:2545–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cheung J., Hendrickson W. A. 2009. Structural analysis of ligand stimulation of the histidine kinase NarX. Structure 17:190–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chou P. Y., Fasman G. D. 1978. Empirical predictions of protein conformation. Annu. Rev. Biochem. 47:251–276 [DOI] [PubMed] [Google Scholar]
- 21. Dang C. V., Niwano M., Ryu J., Taylor B. L. 1986. Inversion of aerotactic response in Escherichia coli deficient in cheB protein methylesterase. J. Bacteriol. 166:275–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Draheim R. R., Bormans A. F., Lai R. Z., Manson M. D. 2005. Tryptophan residues flanking the second transmembrane helix (TM2) set the signaling state of the Tar chemoreceptor. Biochemistry 44:1268–1277 [DOI] [PubMed] [Google Scholar]
- 23. Draheim R. R., Bormans A. F., Lai R. Z., Manson M. D. 2006. Tuning a bacterial chemoreceptor with protein-membrane interactions. Biochemistry 45:14655–14664 [DOI] [PubMed] [Google Scholar]
- 24. Engstrom P., Hazelbauer G. L. 1980. Multiple methylation of methyl-accepting chemotaxis proteins during adaptation of E. coli to chemical stimuli. Cell 20:165–171 [DOI] [PubMed] [Google Scholar]
- 25. Falke J. J., Hazelbauer G. L. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26:257–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hazelbauer G. L., Falke J. J., Parkinson J. S. 2008. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem. Sci. 33:9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hazelbauer G. L., Lai W. C. 2010. Bacterial chemoreceptors: providing enhanced features to two-component signaling. Curr. Opin. Microbiol. 13:124–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hulko M., Berndt F., Gruber M., Linder J. U., Truffault V., Schultz A., Martin J., Schultz J. E., Lupas A. N., Coles M. 2006. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell 126:929–940 [DOI] [PubMed] [Google Scholar]
- 29. Kehry M. R., Bond M. W., Hunkapiller M. W., Dahlquist F. W. 1983. Enzymatic deamidation of methyl-accepting chemotaxis proteins in Escherichia coli catalyzed by the cheB gene product. Proc. Natl. Acad. Sci. U. S. A. 80:3599–3603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kehry M. R., Dahlquist F. W. 1982. Adaptation in bacterial chemotaxis: CheB-dependent modification permits additional methylations of sensory transducer proteins. Cell 29:761–772 [DOI] [PubMed] [Google Scholar]
- 31. Kehry M. R., Dahlquist F. W. 1982. The methyl-accepting chemotaxis proteins of Escherichia coli. Identification of the multiple methylation sites on methyl-accepting chemotaxis protein I. J. Biol. Chem. 257:10378–10386 [PubMed] [Google Scholar]
- 32. Kehry M. R., Doak T. G., Dahlquist F. W. 1984. Stimulus-induced changes in methylesterase activity during chemotaxis in Escherichia coli. J. Biol. Chem. 259:11828–11835 [PubMed] [Google Scholar]
- 33. Kentner D., Thiem S., Hildenbeutel M., Sourjik V. 2006. Determinants of chemoreceptor cluster formation in Escherichia coli. Mol. Microbiol. 61:407–417 [DOI] [PubMed] [Google Scholar]
- 34. Killian J. A., von Heijne G. 2000. How proteins adapt to a membrane-water interface. Trends Biochem. Sci. 25:429–434 [DOI] [PubMed] [Google Scholar]
- 35. Laemmli U. K. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 36. Lai W. C., Beel B. D., Hazelbauer G. L. 2006. Adaptational modification and ligand occupancy have opposite effects on positioning of the transmembrane signalling helix of a chemoreceptor. Mol. Microbiol. 61:1081–1090 [DOI] [PubMed] [Google Scholar]
- 37. Larsen S. H., Reader R. W., Kort E. N., Tso W. W., Adler J. 1974. Change in direction of flagellar rotation is the basis of the chemotactic response in Escherichia coli. Nature 249:74–77 [DOI] [PubMed] [Google Scholar]
- 38. Levit M. N., Stock J. B. 2002. Receptor methylation controls the magnitude of stimulus-response coupling in bacterial chemotaxis. J. Biol. Chem. 277:36760–36765 [DOI] [PubMed] [Google Scholar]
- 39. Li G., Weis R. M. 2000. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell 100:357–365 [DOI] [PubMed] [Google Scholar]
- 40. Liu J. D., Parkinson J. S. 1989. Role of CheW protein in coupling membrane receptors to the intracellular signaling system of bacterial chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 86:8703–8707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miller A. S., Falke J. J. 2004. Side chains at the membrane-water interface modulate the signaling state of a transmembrane receptor. Biochemistry 43:1763–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mowery P., Ostler J. B., Parkinson J. S. 2008. Different signaling roles of two conserved residues in the cytoplasmic hairpin tip of Tsr, the Escherichia coli serine chemoreceptor. J. Bacteriol. 190:8065–8074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ottemann K. M., Xiao W., Shin Y. K., Koshland D. E., Jr 1999. A piston model for transmembrane signaling of the aspartate receptor. Science 285:1751–1754 [DOI] [PubMed] [Google Scholar]
- 44. Park H., Im W., Seok C. 2011. Transmembrane signaling of chemotaxis receptor Tar: insights from molecular dynamics simulation studies. Biophys. J. 100:2955–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Parkinson J. S. 1976. cheA, cheB, and cheC genes of Escherichia coli and their role in chemotaxis. J. Bacteriol. 126:758–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Parkinson J. S. 2010. Signaling mechanisms of HAMP domains in chemoreceptors and sensor kinases. Annu. Rev. Microbiol. 64:101–122 [DOI] [PubMed] [Google Scholar]
- 47. Parkinson J. S., Houts S. E. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peach M. L., Hazelbauer G. L., Lybrand T. P. 2002. Modeling the transmembrane domain of bacterial chemoreceptors. Protein Sci. 11:912–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Segall J. E., Block S. M., Berg H. C. 1986. Temporal comparisons in bacterial chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 83:8987–8991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shiomi D., Zhulin I. B., Homma M., Kawagishi I. 2002. Dual recognition of the bacterial chemoreceptor by chemotaxis-specific domains of the CheR methyltransferase. J. Biol. Chem. 277:42325–42333 [DOI] [PubMed] [Google Scholar]
- 51. Slocum M. K., Parkinson J. S. 1985. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: null phenotypes of the tar and tap genes. J. Bacteriol. 163:586–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Smith R. A., Parkinson J. S. 1980. Overlapping genes at the cheA locus of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 77:5370–5374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sourjik V., Berg H. C. 2004. Functional interactions between receptors in bacterial chemotaxis. Nature 428:437–441 [DOI] [PubMed] [Google Scholar]
- 54. Sourjik V., Berg H. C. 2000. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol. Microbiol. 37:740–751 [DOI] [PubMed] [Google Scholar]
- 55. Springer M. S., Goy M. F., Adler J. 1979. Protein methylation in behavioural control mechanisms and in signal transduction. Nature 280:279–284 [DOI] [PubMed] [Google Scholar]
- 56. Springer W. R., Koshland D. E., Jr 1977. Identification of a protein methyltransferase as the cheR gene product in the bacterial sensing system. Proc. Natl. Acad. Sci. U. S. A. 74:533–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Starrett D. J., Falke J. J. 2005. Adaptation mechanism of the aspartate receptor: electrostatics of the adaptation subdomain play a key role in modulating kinase activity. Biochemistry 44:1550–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stock J. B., Koshland D. E., Jr 1978. A protein methylesterase involved in bacterial sensing. Proc. Natl. Acad. Sci. U. S. A. 75:3659–3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Studdert C. A., Parkinson J. S. 2004. Crosslinking snapshots of bacterial chemoreceptor squads. Proc. Natl. Acad. Sci. U. S. A. 101:2117–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Studdert C. A., Parkinson J. S. 2005. Insights into the organization and dynamics of bacterial chemoreceptor clusters through in vivo crosslinking studies. Proc. Natl. Acad. Sci. U. S. A. 102:15623–15628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Swain K. E., Falke J. J. 2007. Structure of the conserved HAMP domain in an intact, membrane-bound chemoreceptor: a disulfide mapping study. Biochemistry 46:13684–13695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Swain K. E., Gonzalez M. A., Falke J. J. 2009. Engineered socket study of signaling through a four-helix bundle: evidence for a yin-yang mechanism in the kinase control module of the aspartate receptor. Biochemistry 48:9266–9277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Terwilliger T. C., Koshland D. E., Jr 1984. Sites of methyl esterification and deamination on the aspartate receptor involved in chemotaxis. J. Biol. Chem. 259:7719–7725 [PubMed] [Google Scholar]
- 64. Ward S. M., Bormans A. F., Manson M. D. 2006. Mutationally altered signal output in the Nart (NarX-Tar) hybrid chemoreceptor. J. Bacteriol. 188:3944–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ward S. M., Delgado A., Gunsalus R. P., Manson M. D. 2002. A NarX-Tar chimera mediates repellent chemotaxis to nitrate and nitrite. Mol. Microbiol. 44:709–719 [DOI] [PubMed] [Google Scholar]
- 66. Watts K. J., Johnson M. S., Taylor B. L. 2008. Structure-function relationships in the HAMP and proximal signaling domains of the aerotaxis receptor Aer. J. Bacteriol. 190:2118–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Williams S. B., Stewart V. 1999. Functional similarities among two-component sensors and methyl-accepting chemotaxis proteins suggest a role for linker region amphipathic helices in transmembrane signal transduction. Mol. Microbiol. 33:1093–1102 [DOI] [PubMed] [Google Scholar]
- 68. Wright G. A., Crowder R. L., Draheim R. R., Manson M. D. 2011. Mutational analysis of the transmembrane helix 2-HAMP domain connection in the Escherichia coli aspartate chemoreceptor Tar. J. Bacteriol. 193:82–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wu J., Li J., Li G., Long D. G., Weis R. M. 1996. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry 35:4984–4993 [DOI] [PubMed] [Google Scholar]
- 70. Wuichet K., Zhulin I. B. 2010. Origins and diversification of a complex signal transduction system in prokaryotes. Sci. Signal. 3:ra50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yaghmai R., Hazelbauer G. L. 1992. Ligand occupancy mimicked by single residue substitutions in a receptor: transmembrane signaling induced by mutation. Proc. Natl. Acad. Sci. U. S. A. 89:7890–7894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yang Y., Park H., Inouye M. 1993. Ligand binding induces an asymmetrical transmembrane signal through a receptor dimer. J. Mol. Biol. 232:493–498 [DOI] [PubMed] [Google Scholar]
- 73. Zhou Q., Ames P., Parkinson J. S. 2011. Biphasic control logic of HAMP domain signalling in the Escherichia coli serine chemoreceptor. Mol. Microbiol. 80:596–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhou Q., Ames P., Parkinson J. S. 2009. Mutational analyses of HAMP helices suggest a dynamic bundle model of input-output signalling in chemoreceptors. Mol. Microbiol. 73:801–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhulin I. B. 2001. The superfamily of chemotaxis transducers: from physiology to genomics and back. Adv. Microb. Physiol. 45:157–198 [DOI] [PubMed] [Google Scholar]