Abstract
Agrobacterium tumefaciens is a facultative phytopathogen that causes crown gall disease. For successful plant transformation A. tumefaciens requires the membrane lipid phosphatidylcholine (PC), which is produced via the methylation and the PC synthase (Pcs) pathways. The latter route is dependent on choline. Although choline uptake has been demonstrated in A. tumefaciens, the responsible transporter(s) remained elusive. In this study, we identified the first choline transport system in A. tumefaciens. The ABC-type choline transporter is encoded by the chromosomally located choXWV operon (ChoX, binding protein; ChoW, permease; and ChoV, ATPase). The Cho system is not critical for growth and PC synthesis. However, [14C]choline uptake is severely reduced in A. tumefaciens choX mutants. Recombinant ChoX is able to bind choline with high affinity (equilibrium dissociation constant [KD] of ≈2 μM). Since other quaternary amines are bound by ChoX with much lower affinities (acetylcholine, KD of ≈80 μM; betaine, KD of ≈470 μM), the ChoXWV system functions as a high-affinity transporter with a preference for choline. Two tryptophan residues (W40 and W87) located in the predicted ligand-binding pocket are essential for choline binding. The structural model of ChoX built on Sinorhizobium meliloti ChoX resembles the typical structure of substrate binding proteins with a so-called “Venus flytrap mechanism” of substrate binding.
INTRODUCTION
Choline, a quaternary amine (trimethyl-β-hydroxy-ethylammonium), is widespread in nature. As a precursor of various metabolites, it fulfills numerous biological functions in eukaryotes and prokaryotes (4, 31). In many Rhizobiaceae, choline can serve as a sole carbon/nitrogen source (5, 46) and is a direct precursor of glycine betaine, one of the most potent osmoprotectants (23, 33). Since choline uptake from exogenous sources is energetically more favorable than its de novo synthesis, bacteria have evolved different uptake mechanisms. Most often, protein-dependent ATP-binding cassette (ABC) transporters (8, 13, 50) are used, but uptake via secondary transporters has also been described (7, 24).
Two choline uptake systems are known in Escherichia coli: the ABC transporter ProU and the proton motive force-driven, high-affinity uptake system BetT. At low external concentrations, choline is mainly taken up by BetT, whereas at higher concentrations choline is also transported by ProU (24, 30, 50). In Bacillus subtilis, two closely related high-affinity ABC transport systems (OpuB and OpuC) for choline uptake were identified. Expression of the opuB and opuC operons is regulated in response to increasing osmolality (19). OpuC transports various osmoprotectants, whereas OpuB is highly specific for choline (19). The structural reasons for this discrimination have recently been revealed (37).
Several choline uptake activities have been examined in the plant symbiont Sinorhizobium meliloti. One constitutive system exhibits low affinity. The other two systems have high affinity and are either inducible by choline or constitutively expressed (38). Recently, two choline ABC-type transporters have been identified and characterized in S. meliloti. The Prb system (PrbABCD) belongs to the oligopeptide subfamily and mediates the uptake of proline betaine as well as glycine betaine and choline (2) while the Cho system (ChoXWV) is specific for choline (13). The Cho system has been extensively characterized, and crystal structures of the substrate binding protein ChoX are now available (35, 36).
Apart from its function as nutrient or osmoprotectant, choline is also a direct precursor of phosphatidylcholine (PC), a typical eukaryotic membrane component present in only about 10% of all bacterial species, in particular, in bacteria interacting with eukaryotes (1, 48). Recent studies revealed that PC plays a fundamental role in pathogenic and symbiotic microbe-plant interactions (1, 11, 12, 34, 53). Like many members of the Rhizobiaceae family, Agrobacterium tumefaciens contains PC as a major membrane component (45, 53). An A. tumefaciens mutant lacking PC shows a dramatic virulence defect due to the lack of the type IV secretion system, essential for virulence. In addition, PC is critical for motility, biofilm formation, and stress resistance in A. tumefaciens (20, 53).
Two pathways are known to produce PC in A. tumefaciens: the S-adenosyl-l-methionine-consuming methylation pathway and the energetically more favorable choline-dependent PC synthase (Pcs) pathway. In the latter, PC is formed directly by Pcs-catalyzed condensation of choline with CDP-diacylglycerol (CDP-DAG). Since there is no clear evidence that A. tumefaciens can synthesize choline de novo, this pathway might rely on exogenously provided choline. In early studies radioactively labeled choline uptake from the medium and incorporation into phospholipids have been demonstrated (45). However, a corresponding choline transporter(s) was not revealed. Recently, we analyzed whether the predicted ABC transporter (Atu1792, Atu1791, Atu1790, and Atu1789) encoded immediately downstream of the pcs gene is responsible for choline uptake in A. tumefaciens (21). Our data suggested that either this predicted transporter is not responsible for choline uptake or that additional choline uptake systems exist in A. tumefaciens. Six putative ABC-type transporters for quaternary amines are predicted in A. tumefaciens (40, 54). One of these putative transporters is highly similar to the well-characterized ChoXWV system in S. meliloti. Atu2281 is similar to ChoX (85% similarity), the periplasmic binding protein of the Cho transport system. Genes for a putative permease (Atu2280) and an ATPase (Atu2279) are encoded downstream of atu2281. Based on the functional analysis reported in this study, these genes were designated choXWV.
MATERIALS AND METHODS
Chemicals.
Choline dihydrogen citrate, acetylcholine chloride, and betaine were purchased from Sigma-Aldrich. Radioactively labeled [methyl-14C]choline chloride (0.1 mCi/ml and 55 mCi/mmol) was obtained from Hartmann Analytic. HAWP 02500 filters (0.45-μm pore size) for [14C]choline-binding assays were purchased from Millipore GmbH. The HPTLC silica gel 60 plates were from Merck, and Molybdenum Blue Spray reagent was purchased from Sigma-Aldrich. All other reagents were of the highest standard commercially available.
Bacterial strains, plasmids, and growth conditions.
All strains, plasmids, and oligonucleotides used in this work are listed in Tables 1 and 2. E. coli cells were grown at 37°C in Luria-Bertani (LB) or M9 medium (42) supplemented with ampicillin (Ap; 100 μg/ml), kanamycin (Km; 50 μg/ml), tetracycline (Tc; 10 μg/ml), and streptomycin or spectinomycin (Sm/Sp; 50 μg/ml) if appropriate. E. coli DH5α was used as host for all cloning procedures. E. coli BL21(DE3) served as host for overproduction of the ChoX wild type and mutated variants from the corresponding pET24b-based expression plasmids. A. tumefaciens strain C58 (wild type) and derivatives (choX mutants) were routinely grown at 30°C in yeast extract broth (YEB) complex, AB minimal medium (pH 5.5; 1% [wt/vol] glucose) (44) or M9 minimal medium supplemented with 1.5 μg/ml Tc, 10 μg/ml gentamicin (Gm), and 100 μg/ml Sm and/or 300 μg/ml Sp, if necessary. For β-galactosidase assays choline was added to the AB medium at a final concentration of 0.1 mM.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) or descriptiona | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | Cloning host | 16 |
| BL21(DE3) | Expression host | 49 |
| A. tumefaciens strains | ||
| C58 | Wild type | C. Baron, Montreal, Canada |
| C58 choX::Gm> | choX::Gm> insertion mutant of C58 | This study |
| C58 choX::Gm< | choX::Gm< insertion mutant of C58 | This study |
| C58 ΔpmtA | pmtA deletion mutant of C58 | 53 |
| C58 ΔpmtA choX::Gm> | pmtA choX::Gm> double mutant of C58 | This study |
| C58 ΔpmtA choX::Gm< | pmtA choX::Gm< double mutant of C58 | This study |
| C58 Δpcs | pcs deletion mutant of C58 | 53 |
| C58 Δpcs choX::Gm< | pcs choX::Gm< double mutant of C58 | This study |
| Plasmids | ||
| pBluescriptKS(+) | E. coli cloning vector, lacZ, Ampr | Stratagene, Santa Clara, CA |
| pAC01 | Transcriptional lacZ fusion vector containing promoterless lacZ gene; Tcr Apr | 26 |
| pET24b(+) | Expression vector, T7 promoter; 3′ His codon, Kmr | Novagen, Darmstadt, Germany |
| pVSBADNco | A. tumefaciens expression vector; Spr Smr | 55 |
| pK19mobscacB | Suicide plasmid; Kmr | 43 |
| pYP5 | pBSL15 derivative carrying Gm> | 18 |
| pBO893 | pVSBADNco carrying choX | This study |
| pBO2002 | pET24b carrying choX | This study |
| pBO2017 | pAC01 carrying a choX-lacZ fusion | This study |
| pBO890 | pAC01 carrying a choW-lacZ fusion | This study |
| pBO891 | pAC01 carrying an atu2278-lacZ fusion | This study |
| pBO2004 | pBluescript-choX::Gm> | This study |
| pBO2005 | pBluescript-choX::Gm< | This study |
| pBO2006 | pK19mobsacB-choX::Gm> | This study |
| pBO2007 | pK19mobsacB-choX::Gm< | This study |
Ap, ampicillin; Gm, gentamicin; Km, kanamycin; Sm, streptomycin; Sp, spectinomycin; Tc, tetracycline.
Table 2.
Oligonucleotides used in this study
| Oligonucleotide function and name | Sequence (5′→3′)a |
|---|---|
| Transcriptional lacZ fusions | |
| choX-Kpn_fw | CCCCGGTACCGTCTGCGGACGCTTCGC |
| choX-Xho_rv | CCCCCTCGAGTGTTTGCTCCCTGTTTTTTC |
| choW-up-Kpn-fw | CCCCGGTACCCGAATGTCGGCGCGTTTCTTAAG |
| choW-up-Xho-rv | CCCCCTCGAGGGCCTTCGATGCGAGAAAAATTC |
| nolR_up_Kpn_fw | CCCCGGTACCGGTAATAGATTGAAGCGCCA |
| nolR_up_Xho_rv | CCCCCTCGAGACGCTCAAGTCTCCTGATGAC |
| Construction of pVSBAD-choX expression vector | |
| ChoX_Eco_fw | CCCGAATTCTTGATGGCCGGAAGATATTGATA |
| ChoX_Sal-rv | CCCGTCGACCTAAAGGCCGAGAGCCTTT |
| Construction of pET24b-choX expression vector | |
| choX_Nde_fw | GGAATTCCATATGTTTGCAAATAGAAGTCGC |
| choX_Sal-Xho_rv | CCGGTCGACTCACTCGAGAAGGCCGAGAGCC |
| QuikChange mutagenesis of choX | |
| ChoX-W41A-fw | TGATGTCGGCGCGACCGATATCACCGCC |
| ChoX-W41A-rv | GGCGGTGATATCGGTCGCGCCGACATCA |
| ChoX-W88A-fw | CCTCGGCAACGCGATGCCGACAATGGAAGG |
| ChoX-W88A-rv | CCTTCCATTGTCGGCATCGCGTTGCCGAGG |
| ChoX-W283A-fw | CCGCAACCGCGGCGCTGAAGGCCAATCC |
| ChoX-W283A-rv | GGATTGGCCTTCAGCGCCGCGGTTGCGG |
| ChoX-W294A-fw | GATCGAGCCCGCGTTCGCCAACGTTAAGACAA |
| ChoX-W294A-rv | TTGTCTTAACGTTGGCGAACGCGGGCTCGATC |
| Construction of A. tumefaciens choX mutants | |
| choX_Kpn_fw | GCAGGGTACCAGGCGGCGATTTTCACG |
| choX_Eco_rv | CCGGAATTCGTATTTCCCACTGCATCCG |
Restriction sites in the oligonucleotides are underlined.
Plasmid and mutant construction.
Recombinant DNA work was carried out according to standard protocols (42). For overproduction of ChoX in E. coli, the A. tumefaciens C58 choX gene was PCR amplified with chromosomal DNA as a template and appropriate primers (Table 2). The PCR product was digested with NdeI and XhoI and cloned into pET24b(+) treated with the same enzymes, resulting in hybrid plasmid pBO2002 coding for a C-terminally His-tagged ChoX protein.
For overproduction of ChoX in A. tumefaciens, the coding region, including its own ribosome binding site, was amplified via PCR. The PCR product was digested with EcoRI and SalI and cloned into the corresponding site of the arabinose-inducible vector pVSBADNco (55), resulting in plasmid pBO893.
To generate ChoX with the mutations W41A, W88A, W283A, and W294A and the double mutation W41A W88A, site-directed mutagenesis was conducted using a QuikChange mutagenesis kit (Stratagene) following the supplier's protocol. The vector pBO2002 (pET24b containing wild-type choX) was subjected to site-directed mutagenesis. Mutated choX variants were verified by sequencing.
To construct transcriptional fusions to the lacZ gene, PCR-generated fragments of the promoter regions of choX, choW, and atu2278 (14, 54) were digested with KpnI and XhoI and ligated into pAC01 treated with the same enzymes.
To construct choX insertion mutants, a 1,155-bp A. tumefaciens DNA fragment was PCR amplified using the oligonucleotide pair choX_Kpn_fw and choX_Eco_rv (Table 2). The PCR product was cloned into the Kpn- and EcoRV-restricted pBluescript KS(+) vector. Subsequently, a SphI fragment carrying the Gm resistance cassette from plasmid pYP5 was inserted into the single SphI site within choX, leading to the hybrid plasmids pBO2004 and pBO2005. The 1,703-bp HindIII-choX::Gm-EcoRI fragment from pBO2004 and pBO2005 was then excised with EcoRI and HindIII and cloned into the suicide vector pK19mobsacB (43), resulting in the plasmids pBO2006 and pBO2007. The plasmids were transferred into wild-type A. tumefaciens C58 or pmtA or pcs mutants via electroporation. Single-crossover integration mutants were selected on LB plates containing kanamycin. Single colonies were grown overnight in liquid LB medium without antibiotics and plated on LB medium containing 10% (wt/vol) sucrose to select for plasmid excision by double crossover. Sucrose- and gentamicin-resistant and kanamycin-sensitive clones were analyzed by colony PCR and Southern blot analysis (42).
β-Galactosidase assays.
The β-galactosidase activity of A. tumefaciens cells grown in liquid AB minimal medium in the presence or absence of 1 mM choline was measured according to standard protocols (32). Cells from the exponential growth phase were taken for the β-galactosidase assays. Under these conditions, the addition of 1 mM choline did not influence growth of the tested strains. The plasmid pAC01 containing the promoterless lacZ gene was used as a negative control.
Choline transport assays.
For initial [14C]choline uptake analyses, overnight cultures of A. tumefaciens wild-type and choX mutant strains grown in M9 minimal medium with 0.1 mM choline were washed and adjusted to an optical density at 600 nm (OD600) of 1.0 with M9 medium. Reactions were initiated by addition of [methyl-14C]choline chloride (55 mCi/mmol) to a final concentration of 0.1 mM, and samples were incubated at 30°C. Fifty-microliter aliquots were taken at different time points (5 to 60 min), and reactions were terminated by rapid filtration of samples on HAWP 02500 filters. For the complementation assays, the strains were grown in M9 minimal medium to an OD600 of 0.5 to 0.8. Subsequently, the OD600s of the cells were adjusted with M9 medium to 0.5. In the case of the choX mutants carrying the pVSBAD derivatives, cells were induced at an OD600 of 0.3 with 0.2% arabinose. Reactions were initiated by addition of [methyl-14C]choline chloride (55 mCi/mmol) to a final concentration of 5 μM. Samples (1 ml) were incubated for 10 min at 30°C. Reactions were terminated by rapid filtration as mentioned above. Filters were washed four times with 1 ml of M9 medium, and the amount of radioactively labeled choline in the cells was quantified by liquid scintillation spectrometry (Beckman Counter LS-6000 TA).
Lipid analysis by TLC.
The lipid composition of A. tumefaciens strains was determined via thin-layer chromatography (TLC). Cells were cultivated in M9 medium in the absence or presence of 1 mM choline for 24 h. Two milliliters of the cultures with identical optical densities was harvested by centrifugation, washed with 500 μl of water, and resuspended in 100 μl of water. The lipids were extracted according to the method of Bligh and Dyer (3), separated by one-dimensional TLC using high-performance TLC (HPTLC) silica gel 60 plates (Merck, Darmstadt, Germany), and stained with Molybdenum Blue Spray reagent (Sigma-Aldrich). As running solvent, n-propanol-propionate-chloroform-water (3:2:2:1) was used.
Overproduction and purification of ChoX and mutated variants.
E. coli BL21(DE3) cells carrying the pBO2002 plasmid (pET24b_choX) or derivatives were grown in 1 liter of M9 minimal medium with glucose as the carbon source (42) and kanamycin (50 μg/ml) at 37°C. Protein production was induced at an OD580 of 0.5 by addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 0.1 mM before the culture was incubated for 24 h at 20°C and harvested by centrifugation (6,000 × g for 10 min at 4°C). ChoX contains an N-terminal signal sequence for targeting it into the periplasm. To release the recombinant ChoX protein from the periplasmic space, the cells were subjected to rapid osmotic shock by resuspending the cells in wash buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole [pH 8.0]) containing 500 mM saccharose, followed by incubation for 30 min on ice. Insoluble material was removed by centrifugation (20,000 × g for 40 min at 4°C). Soluble fractions were applied to nickel-iminodiacetic acid (Ni-IDA) columns, and purification was performed as described by the supplier (Macherey and Nagel). ChoX was eluted from the column with 200 mM imidazole in wash buffer. Protein purity was assessed by Coomassie staining of sodium dodecyl sulfate (SDS)-polyacrylamide gels (12.5%).
Size exclusion chromatography.
Size exclusion chromatography was performed using a Superdex 75 10/30 column run with 50 mM NaH2PO4 and 50 mM NaCl (pH 8.0) at a flow rate of 0.5 ml/min. The column was calibrated with the globular proteins RNase (13.7 kDa), chymotrypsinogen A (25 kDa), ovalbumin (43 kDa), and albumin (67 kDa), obtaining an R2 value of 0.99 for the calibration curve. A 250-μl aliquot of protein at a concentration of 0.8 mg/ml was injected. Eluates (0.5 ml) were pooled and concentrated in Amicon Ultra concentrators (molecular-weight cutoff, 10,000; Millipore), and the buffer was exchanged against binding buffer (10 mM Tris-HCl, 200 mM NaCl [pH 7.4]). Protein concentrations were determined from A280 values with a calculated extinction coefficient (ε) of 39,545 M−1 cm−1.
CD spectroscopy.
The circular dichroism (CD) spectra of recombinant ChoX variants were recorded 10 times between 190 and 320 nm with a Jasco 715 spectropolarimeter at 20°C in 50 mM potassium phosphate buffer, pH 8. The final spectra obtained were the average results of the 10 scans, normalized against buffer. Analyses were performed in duplicate using 10 μM enzyme.
In vitro [14C]choline-binding studies.
For choline-binding assays, 5 μM recombinant ChoX protein was incubated with 0 to 50 μM [methyl-14C]choline chloride (55 mCi/mmol) in binding buffer for 5 min at room temperature ([RT] total assay volume, 50 μl). For competition experiments, unlabeled choline chloride (0 to 50 μM) was added to the samples and incubated for 5 min at 30°C before the addition of 5 μM [14C]choline.
Binding assay mixtures were passed over HAWP 02500 filters on a filtration funnel, and unbound [methyl-14C]choline chloride was removed by washing samples four times with 300 μl of binding buffer. Bound [methyl-14C]choline chloride was quantified by liquid scintillation spectrometry (Beckman Counter LS-6000 TA). The Michaelis-Menten equation was used for calculation of choline-binding affinities of the wild type and mutated ChoX variants. The data were fitted by nonlinear regression using SigmaPlot, version 9.0.
Fluorescence-based ligand-binding assays.
The affinity of recombinant ChoX protein for choline acetylcholine and betaine was determined by intrinsic tryptophan fluorescence spectroscopy according to Oswald et al. (36). The excitation wavelength was set to 295 nm, and fluorescence was monitored from 300 to 450 nm using a Thermo Amicon Bowman II Luminescence Spectrometer at 22°C. Different amounts of ligands (0 to 100 μM) in binding buffer (200 mM NaCl, 10 mM Tris-HCl, pH 7.4) were titrated to ChoX (10 μM; assay volume, 100 μl) in binding buffer, and after equilibration (5 min) fluorescence was measured. To account for background fluorescence, spectra with and without protein were subtracted from each other. Upon ligand titration the emission maximum shifted toward shorter wavelengths (blue shift) (see Fig. 6). The changes in the emission maximum (Δλ, in nm) were plotted against the ligand concentration, and the Michaelis-Menten equation was used for calculation of ligand-binding affinities. The data were fitted by nonlinear regression using SigmaPlot, version 9.0.
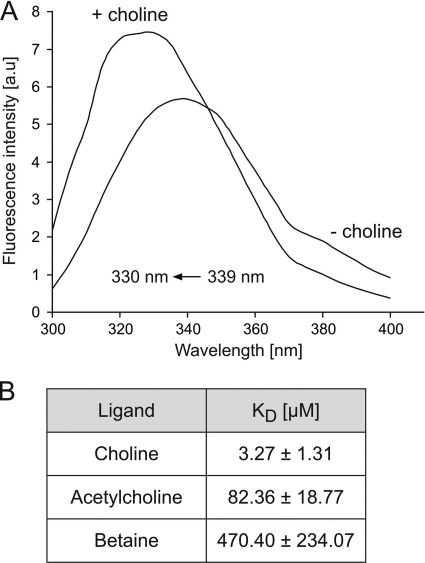
Fig. 6.
Fluorescence-based ligand-binding analysis of ChoX. (A) Emission spectra of ChoX (10 μM) in the absence or presence of 100 μM choline. au, arbitrary units. (B) Binding affinities (KDs) of ChoX for choline, acetylcholine, and betaine were determined via intrinsic tryptophan quenching after addition of the ligands (0 to 100 μM) to ChoX protein (10 μM). Changes in the emission maximum (Δλ in nm) were plotted against the ligand concentration (data not shown), and the Michaelis-Menten equation was used for calculation of ligand-binding affinities. The data were fitted by nonlinear regression using SigmaPlot, version 9.0. Values are the means of three independent experiments ± standard deviations.
Development of a homology model for ChoX.
The three-dimensional structure of ChoX was predicted by the threading method using the I-TASSER online server (41, 56, 57). Structures of proteins with the following Protein Data Bank identifiers (PDB IDs) were chosen by I-TASSER as the templates in the modeling procedure: 2rf1A, 2rinA, and 2regA (S. meliloti ChoX); 1r91A (E. coli ProX); and 3L6gA (lactococcal OpuAC). This server produced five possible models for ChoX. The first model with the best quality of prediction (confidence [C] score, −0.33; template-modeling [TM] score, 0.67 ± 0.13; and root mean square deviation [RMSD], 7.0 ± 4.14 Å) was used here.
RESULTS
Identification of a ChoXWV-like transporter system in A. tumefaciens.
Using the Transporter Protein Analysis Database (40), we identified atu2281, atu2280, and atu2279 encoding a putative choline ABC-transporter similar to the ChoXWV system from S. meliloti. Atu2281 is highly similar in the amino acid sequence (85% similarity and 80% identity) to the periplasmic choline-binding protein ChoX. Atu2280 and Atu2279 are highly similar (87% similarity and 76% identity) to ChoV (permease) and ChoW (ATPase), respectively (13). Accordingly, atu2281, atu2280, and atu2279 were tentatively designated choXWV (Fig. 1).
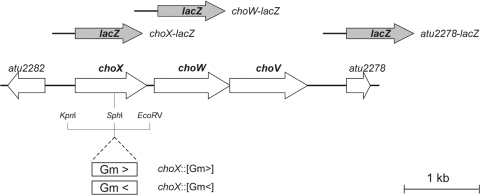
Fig. 1.
Genetic organization of the choXWV locus. The cho genes are flanked by two ORFs: atu2282, encoding a putative thymidine kinase, and atu2278, a NolR-like regulator. choX encodes a periplasmic choline-binding protein, choW encodes a permease, and choV encodes an ATPase. The positions of the gentamicin (Gm) cassette insertion and corresponding mutants (choX::Gm> and choX::Gm<) are indicated. The upper part shows the constructed lacZ fusions.
Genetic organization of the choXWV genes.
The choXWV genes are located on the circular chromosome of A. tumefaciens C58. They are flanked by two open reading frames (ORF), namely, atu2282, transcribed in the opposite direction, and atu2278, oriented in the same direction as the cho genes (Fig. 1). The ORF atu2282 encodes a putative thymidine kinase, and atu2278 codes for a putative NolR regulator. NolR is present in species belonging to the Rhizobium and Sinorhizobium genera and belongs to the AsrR family of regulators. In S. meliloti NolR is a global regulatory protein involved in nodulation, bacterial growth and survival, and conjugative transfer of plasmid (9, 10, 22, 25). The putative A. tumefaciens NolR protein had no significant effect on choXWV expression in the absence or presence of choline (data not shown).
Within the cho locus, choX and choW are separated by a 109-bp noncoding region while the translational start codon of choV overlaps with the translational stop codon of choW, suggesting translational coupling between these two genes.
Expression of the cho operon.
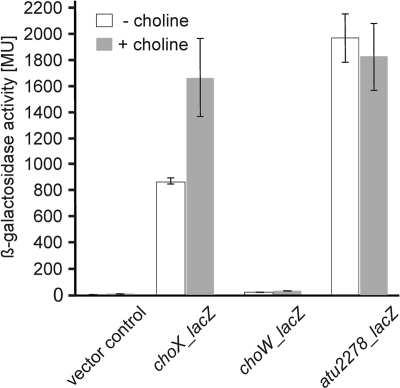
To examine whether choXWV was expressed, transcriptional lacZ fusions were constructed (Fig. 1). A choW-lacZ fusion was used to test whether choWV expression was originated from a separate promoter or was driven from the choX promoter. The resulting reporter plasmids, pBO2017 (choX-lacZ) and pBO890 (choW-lacZ), were electroporated into wild-type A. tumefaciens. To examine the effect of choline on choXWV expression, all reporter strains were grown in AB minimal medium in the presence or absence of 1 mM choline. Clear choX expression was detected in both the absence and presence of choline. Expression was moderately but reproducibly enhanced when cells were grown with choline (Fig. 2). To test whether this effect of choline was specific, an atu2278-lacZ fusion was measured as a control. In contrast to choX, expression of atu2278 was not affected by choline at all (Fig. 2).
Fig. 2.
β-Galactosidase activities of plasmid-encoded transcriptional choX-lacZ, choW-lacZ, and atu2278-lacZ fusions in A. tumefaciens C58. Cells were grown in AB minimal medium in the absence or presence of 1 mM choline at 30°C. The plasmid pAC01 containing the promoterless lacZ gene was used as a negative control. Values are the mean of three independent experiments ± standard deviations.
The choW-lacZ fusion was not expressed under all conditions tested (Fig. 2), excluding a possible promoter in the intergenic region upstream of the choW gene. The conclusion that choXWV genes are organized in an operon is supported by recent RNA sequencing data from our laboratory (data not shown).
Construction and growth phenotype of choX mutants.
To study the role of the choXWV operon in choline uptake, we constructed choX mutants by insertion of a gentamicin (Gm) resistance cassette into the choX coding region, as described in Materials and Methods. The Gm resistance cassette was inserted in either the same (Gm>) or the opposite (Gm<) orientation of the choX gene, resulting in the two A. tumefaciens choX::Gm> and choX::Gm< mutants, respectively. As shown previously (29), Gm cassettes induce polar or nonpolar mutations depending on their orientations. Thus, the choX::Gm< mutant is expected to be defective for choX, choW, and choV, whereas expression of choW and choX should be driven by the Gm promoter in the choX::Gm> mutant.
To test for obvious phenotypic defects, the growth behavior of the choX mutants was investigated under different conditions. Growth of the mutants was indistinguishable from that of the A. tumefaciens wild type at 30°C in minimal medium under low- and high-osmolarity conditions (without added NaCl or with NaCl concentrations ranging from 0.2 to 0.8 M) in the presence of 0.1 mM or 1 mM choline. Likewise, the strains showed no differences when they were cultivated in AB minimal medium without choline (data not shown), demonstrating that choX mutants of A. tumefaciens grow normally in liquid culture in either the absence or presence of choline. Choline enhanced growth of wild-type and mutant strains in the presence of 0.2 to 08 M NaCl, suggesting that choline has an osmoprotective role in A. tumefaciens C58 and that this effect is not dependent on the Cho system.
ChoXWV is not required for choline-dependent PC biosynthesis.
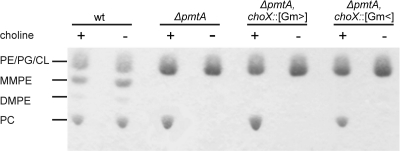
PC formation in an A. tumefaciens pmtA mutant occurs via the Pcs pathway, which is dependent on choline in the growth medium (53). To address whether the ChoXWV transporter is necessary for PC biosynthesis, we analyzed PC production in pmtA choX double mutants grown in minimal medium in the presence or absence of 1 mM choline by TLC. As expected, wild-type A. tumefaciens accumulated phosphatidylethanolamine (PE), monomethyl-PE (MMPE), dimethyl-PE (DMPE), and PC under both tested conditions (Fig. 3). The pmtA mutant contained PC produced via the Pcs pathway in the presence of choline. However, in the absence of choline, none of the methylated lipids were present. The pmtA choX::Gm> mutant strain produced PC amounts comparable to those of the parental strain, indicating that the Pcs pathway is efficiently supplied with its substrate choline. The pmtA choX::Gm< mutant (expected to be deficient for choX, choW, and choV) accumulated slightly reduced PC amounts compared to parental strain levels.
Fig. 3.
PC formation in A. tumefaciens wild-type (wt) and mutant strains. Cells were grown in M9 minimal medium in the absence (−) or presence (+) of 1 mM choline for 24 h. Lipids were extracted, separated by one-dimensional TLC, and visualized by Molybdenum Blue staining. PE, phosphatidylethanolamine; PG, phosphatidylglycerol; CL, cardiolipin; MMPE, monomethyl-PE; DMPE, dimethyl-PE; PC, phosphatidylcholine.
A. tumefaciens choX mutants are severely impaired in choline uptake.
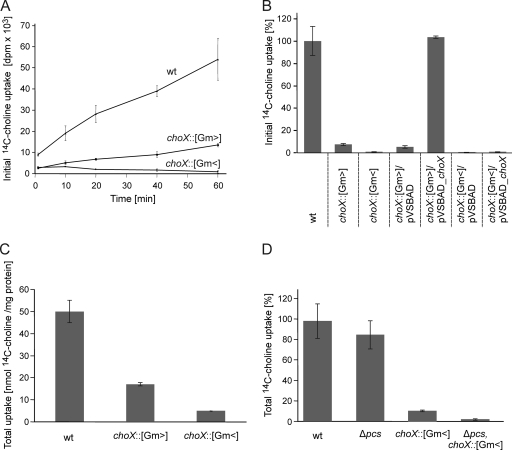
To analyze the role of ChoXWV in A. tumefaciens choline uptake, radioactively labeled choline transport assays were carried out. For this purpose A. tumefaciens cells were incubated with 0.1 mM [14C]choline, and initial choline uptake at different time points was analyzed as described in Materials and Methods. Choline uptake by the A. tumefaciens wild type increased with incubation time (Fig. 4A). The choX::Gm> mutant strain (expressing choWV from the Gm promoter) retained some residual choline uptake activity (∼25%). Since choW and choV are still expressed in this strain from the Gm promoter, other ChoX-like choline-binding proteins might partially compensate for the loss of ChoX. The choX::Gm< mutant (deficient for the entire choXWV system) was completely defective in choline transport (Fig. 4A).
Fig. 4.
[14C]choline uptake analyses of the A. tumefaciens wild type (wt) and choX mutants. (A) Initial choline uptake of the A. tumefaciens wild type and choX mutants with a final choline concentration of 0.1 mM [methyl-14C]choline chloride (55 mCi/mmol) at the time indicated. The points shown are the mean values of three individual experiments. (B) Complementation of the choX mutants by plasmid (pVSBAD)-encoded choX expression. Uptake was assayed over 10 min with a final [14C]choline concentration of 5 μM. Total [14C]choline uptake of the A. tumefaciens wild type and choX mutants (C) and the Δpcs choX::[Gm<] double mutant (D) was assayed using a final choline concentration of 10 μM [14C]choline. The accumulation of radioactivity was quantified after a 24-h incubation. The values shown are the means of two individual experiments.
Choline uptake deficiency of the choX::Gm> mutant was completely restored by expression of vector-encoded choX (pVSBAD_choX) (Fig. 4B). As expected, complementation of the choline uptake deficiency of the choX::Gm< mutant was not possible with plasmid-encoded choX (Fig. 4B) since this strain still lacks the corresponding permease and ATPase (ChoWV).
Although only little or no initial choline uptake was detected in A. tumefaciens choX mutants, they still produced PC via the Pcs pathway (Fig. 3). Two scenarios of how Pcs is provided with choline are possible. First, choline might be taken up by a thus far unidentified low-affinity transporter(s). Second, Pcs itself might bind choline from the periplasm before it converts it to PC. In this case, the Pcs enzyme would be independent from choline uptake systems. To test whether alternative choline uptake systems might exist in A. tumefaciens, we analyzed the accumulation of [14C]choline in the choX mutants by incubating the cells overnight with 10 μM radioactively labeled choline. Compared to the wild type, approximately 35% and 10% choline uptake was measured in the choX::Gm> and choX::Gm< mutant strains, respectively (Fig. 4C). To differentiate whether this long-time accumulation of [14C]choline was due to PC synthesis via Pcs or low-affinity uptake systems, we analyzed [14C]choline accumulation in a pcs choX::Gm< double mutant. Only marginal amounts of [14C]choline were taken up by the pcs choX::Gm< double mutant (Fig. 4D), demonstrating an involvement of Pcs in [14C]choline accumulation.
Purification of A. tumefaciens ChoX.
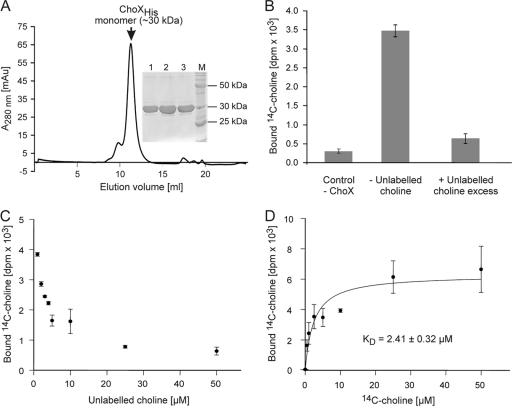
C-terminally His6-tagged ChoX was produced in E. coli BL21(DE3) cells and purified to homogeneity by nickel chelate chromatography and size exclusion chromatography. As judged by SDS-polyacrylamide gel electrophoresis (PAGE), ChoX protein of ∼95% purity was obtained after size exclusion chromatography (Fig. 5A, inset). Consistent with the calculated mass of the His-tagged protein (∼32.2 kDa, without its N-terminal signal peptide), recombinant ChoX migrated as a band with an apparent molecular mass of ∼32 kDa. The major ChoX peak after gel filtration corresponded to a molecular mass of about 30 kDa (Fig. 5A), indicating that ChoX is a monomer.
Fig. 5.
Purification and in vitro choline-binding properties of recombinant ChoX. (A) Size exclusion chromatography of Ni-IDA-purified PmtA. The peak fractions 1, 2, and 3 were analyzed by SDS-PAGE as depicted on the right. AU, absorbance units. Lane M (inset), BenchMark protein standard (Invitrogen). (B) Radioactive choline-binding activity of recombinant ChoX. Choline-binding activity was analyzed with 5 μM ChoX and 5 μM [14C]choline over 5 min. The displacement assay (+unlabeled choline excess) contained, in addition, 10-fold unlabeled choline. (C) Competitive displacement of radioactively labeled choline by unlabeled choline (0 to 50 μM). The assay mixture contained 5 μM recombinant ChoX and 5 μM [14C]choline. (D) Plots of choline-binding data for wild-type ChoX. A reaction volume of 50 μl contained 5 μM recombinant ChoX and 0 to 50 μM [14C]choline. Changes in bound [14C]choline (y axis) were plotted against choline concentration (x axis). All data sets were fitted to the equation for one-site binding (see Materials and Methods) by nonlinear regression using SigmaPlot, version 9.0. The points shown are the mean values of three individual experiments.
ChoX is a high-affinity choline-binding protein.
In order to demonstrate that choX encodes a choline-binding protein, filter-binding assays were performed with radioactively labeled choline and recombinant ChoX protein. Without ChoX, only negligible background radioactivity was detected on the filter (Fig. 5B, Control −ChoX). In the presence of ChoX, significant amounts of [14C]choline were bound, demonstrating that ChoX is a choline-binding protein. A 10-fold molar excess of unlabeled choline (50 μM) efficiently competed with [14C]choline (Fig. 5B, +Unlabeled choline excess). About 50% of the radioligand was exchanged in the presence of equal molar amounts (5 μM) of unlabeled choline (Fig. 5C). A 10-fold molar excess displaced the radioligand to less than 15%.
To determine the kinetics for choline binding of ChoX, we performed [14C]choline-binding experiments with various choline concentrations and calculated a binding constant for choline in the low-micromolar range (2.4 μM) (Fig. 5D). Thus, we concluded ChoX is as a high-affinity choline-binding protein.
As a second approach, intrinsic tryptophan fluorescence-based binding assays were used to quantitate substrate binding. A representative fluorescence spectrum in the absence and presence of choline is shown in Fig. 6A. Binding of choline and acetylcholine to ChoX resulted in a blue shift of the emission spectrum of 9 nm (Fig. 6A) and 5 nm (data not shown), respectively. Binding of betaine to ChoX induced only a marginal blue shift (3 nm) of the emission spectrum (data not shown). These shifts were used to determine the binding affinity constants according to the Michaelis-Menten equation (Fig. 5B). These data demonstrate that ChoX recognizes choline, acetylcholine, and betaine with high, medium, and low affinity, respectively. The equilibrium dissociation constant (KD) of 3.3 μM for choline is in good agreement with the binding constant of 2.4 μM determined via the [14C]choline-binding assay.
Site-directed mutagenesis of residues forming the predicted ligand-binding pocket.
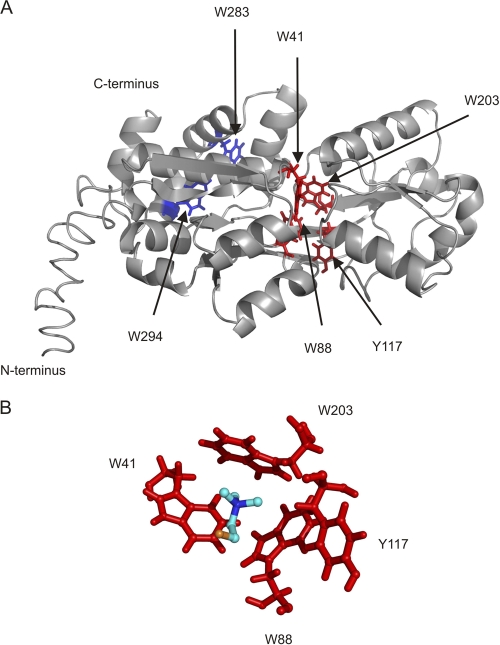
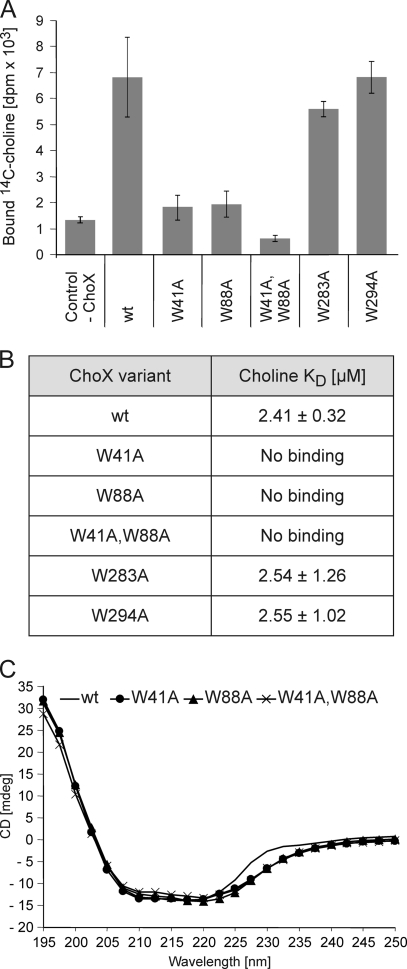
To gain more insights into residues forming the ligand-binding pocket, we generated a model of ChoX using the I-TASSER server. A. tumefaciens ChoX exhibits a fold highly similar to that of S. meliloti ChoX (Fig. 7). The overall structure resembles the well-known substrate binding protein (SBP) fold composed of two domains, which are connected via two β-strands. The predicted choline-binding site is formed by an aromatic box, which is located in a deep cleft between the two domains. This box is composed of four aromatic amino acids, W41, W88, Y117, and W203 (Fig. 7A and B). To examine the role of these amino acid residues for ligand binding, two of these amino acids (W41 and W88) were exchanged to alanine, and the effect of the mutation on choline binding was analyzed. Both individual mutations resulted in a nearly complete loss of choline binding and the double mutant (W41A W88A) was completely unable to bind choline (Fig. 8A and B). As a control, point mutations in two tryptophan residues outside the binding region (W283 and W294) (Fig. 7A) had no influence on choline binding. The structural integrity of the mutated ChoX variants was ascertained using CD spectroscopy. Like the wild-type protein, all mutated variants were structurally intact (Fig. 4C).
Fig. 7.
Homology model of A. tumefaciens ChoX. (A) ChoX model was generated using the online server I-TASSER (41) and visualized by PyMOL (http://www.pymol.org). The fold is characterized by a bilobal organization into two lobes connected via a linker region. The proposed ligand-binding site is located within the two lobes. Proposed choline-binding residues are shown in red, and two aromatic residues (W283 and W294) outside the binding pocket, depicted as negative controls, are shown in blue. (B) Detailed view of the ligand-binding site of A. tumefaciens ChoX. Proposed residues participating in ligand binding and the ligand choline are shown in stick and in ball-and-stick representation, respectively.
Fig. 8.
In vitro choline-binding properties of point-mutated ChoX proteins. (A) [14C]choline binding was assayed with 5 μM ChoX and 5 μM [14C]choline. (B) Choline affinity constants (KDs) of ChoX variants. To calculate affinity constants, 5 μM ChoX protein was titrated with 0 to 50 μM [14C]choline, and bound [14C]choline (in disintegrations per minute) was plotted against the [14C]choline concentration (data not shown). The Michaelis-Menten equation was used for calculation of ligand-binding affinities. The data were fitted by nonlinear regression using SigmaPlot, version 9.0. Values are the means of three independent experiments ± standard deviations. (C) Circular dichroism analysis of the wild type and inactive ChoX variants. The spectrum of each protein with a concentration of 10 μM is plotted as ellipticity in millidegrees (mdeg) versus wavelength.
DISCUSSION
A. tumefaciens contains PC as a major phospholipid in both its inner and outer membranes (21). PC synthesis via the Pcs pathway is dependent on exogenous choline, which is a positively charged quaternary amine and requires a protein-mediated mechanism to effectively pass the membrane lipid barrier (31). In this study, we have characterized a cho-like operon involved in the high-affinity uptake of choline in A. tumefaciens. This system belongs to the ABC-transporter family involving multiple components: a periplasmic choline-binding protein (ChoX), an integral inner membrane protein (ChoW), and an ATPase (ChoV).
The purified A. tumefaciens ChoX is a high-affinity choline-binding protein with a medium affinity for acetylcholine. Binding affinities in the low-micromolar range are typical for periplasmic substrate-binding proteins in Gram-negative bacteria. The determined affinity constants for choline (2.4 μM and 3.3 μM) and for acetylcholine (82.4 μM) are in good agreement with the KD values of 2.7 μM and 2.3 μM for choline and 100 μM for acetylcholine reported previously for the S. meliloti ChoX protein (13, 36). In contrast to the S. meliloti ChoX protein, which binds betaine with a KD of 77 μM, A. tumefaciens ChoX bound betaine with very low affinity (470 μM). Thus, A. tumefaciens ChoX is a substrate-binding protein with a preference for choline.
In silico predictions backed up by site-directed mutagenesis revealed a ligand-binding pocket and mode of substrate binding of A. tumefaciens ChoX highly similar to the S. meliloti ChoX and other glycine/betaine/proline substrate-binding proteins (28, 35, 36, 39). The high-resolution crystal structure of the S. meliloti ChoX protein, with its ligands (choline and acetylcholine) and a ligand-free state, was determined recently (35, 36). Choline is bound via cation-π interaction of the trimethylammonium moiety and four aromatic residues (W43, W90, Y119, and W205) and hydrogen bonds between the hydroxyl group and N156 and D157 (35, 36). These residues are also present in the A. tumefaciens ChoX protein, and as shown here, the conserved aromatic residues W41 and W88 located in the predicted ligand-binding pocket are essential for choline binding of A. tumefaciens ChoX.
Two choX mutants generated via insertion of a Gm cassette in the same or opposite orientation to the cho operon showed a wild-type-like growth behavior in minimal medium in the absence or presence of choline. Interestingly, two different choline uptake phenotypes were observed due to polar effects of the Gm cassette. The choX::Gm> mutant (expression of choWV from Gm promoter possible) transported marginal amounts of choline, whereas the choX::Gm< mutant, expected to be deficient in choXWV, was dramatically reduced in choline uptake. A possible explanation for the residual choline uptake in the choX::Gm> mutant may be that other ChoX-like binding proteins can partially compensate for the loss of ChoX in delivering choline to the core transporter ChoWV. A. tumefaciens encodes two candidates, namely, Atu2060 and Atu4647 sharing 64% similarity to ChoX. Cross talk between ABC transporters via their substrate-binding proteins is not uncommon (8, 51). Recently, a choline-betaine-carnitine (Cbc) transporter in Pseudomonas syringae and Pseudomonas aeruginosa was identified that recruits multiple substrate-binding proteins. In addition to CbcX, the core CbcWV also interacts with the carnitine-specific binding protein CaiX and the betaine-specific binding protein BetX (8, 51). The CbcXWV transport system shares approximately 70% similarity with the A. tumefaciens ChoXWV system. The histidine ABC transporter (HisQMP) from Salmonella enterica serovar Typhimurium is able to recognize two distinct SBPs (17). Furthermore, the oligopeptide transporter (Opp) from Borrelia burgdorferi uses multiple SBPs whose genes are differentially expressed under various environmental conditions (52). The observation that small amounts of choline are taken up when the choX::Gm< mutant is incubated with choline overnight (Fig. 4C) suggests that A. tumefaciens contains at least one further pathway for a low-affinity choline uptake system. At least one of the five remaining putative quaternary amine transporters (40, 54) encoded in A. tumefaciens can be considered for this activity in the choX mutants. Multiple choline uptake systems are common in bacteria. P. aeruginosa encodes two betaine-choline-carnitine transporters ([BCCT] BetT1 to BetT3) and an ABC-type choline transporter (CbcXWV) with different roles (27). In P. syringae one BCCT-type transporter (BetT) and two ABC-type choline transporters (CbcXWV and OpuC) were identified (6–8).
Choline transport systems in A. tumefaciens may be important for transport of choline as a carbon and nitrogen source or for use in osmoregulation as in many other bacteria. Although it was reported that the A. tumefaciens strains can use choline or betaine as a sole N or C source (5, 47), attempts to grow A. tumefaciens C58 in minimal medium with choline as the sole N and/or C source failed in our laboratory (data not shown). Thus, choline seems not to be an efficient N and/or C source for A. tumefaciens C58. Does choline instead act as a precursor for the osmoprotectant glycine betaine in A. tumefaciens C58? Osmoprotection by choline and evidence for a choline dehydrogenase and betaine aldehyde dehydrogenase activity (for two-step enzymatic oxidation of choline to betaine) in A. tumefaciens GMI 9023 crude extracts have been reported (5). In A. tumefaciens C58, the strain used in this study, mannosucrose is the major osmolyte, but a slight osmoprotective effect of betaine was also observed (47). In the absence of osmotic stress, betaine is metabolized by A. tumefaciens C58, but in stressed cultures betaine accumulates and confers enhanced osmotic stress tolerance (47). A putative choline dehydrogenase (Atu0830) and a betaine aldehyde dehydrogenase (Atu0829) are present in the A. tumefaciens C58 genome (14, 54). An osmoprotective effect of choline in this strain was also observed in our laboratory (data not shown). Since this effect was similar in both the wild type and the choX::Gm< mutant (data not shown), the osmoprotective effect of choline seems not to be dependent upon the Cho system.
Remarkably, the ChoXWV system is not essential for choline-dependent PC biosynthesis in A. tumefaciens since both choX mutants were still able to produce PC when choline was supplied in the growth medium. The ΔpmtA choX::Gm> mutant produced wild-type-like PC amounts and the ΔpmtA choX::Gm< mutant showed only slightly reduced PC synthesis (Fig. 3) compared to the parental strain. Thus, both strains are supplied with sufficient amounts of choline for PC synthesis. It is unclear whether Pcs obtains choline from the cytoplasm or from the periplasm. Our data suggest that Pcs obtains choline from the periplasm, and, thus, PC biosynthesis is independent from choline uptake systems in A. tumefaciens. Preliminary experiments with the choline uptake-deficient E. coli MKH13 strain (15, 20) demonstrated that plasmid-encoded pcs is sufficient to enable this strain to produce PC when choline is supplied in the growth medium (data not shown). These lines of evidence suggest that the Pcs enzyme obtains choline from the periplasmic face of the inner membrane. Future biochemical studies on this integral membrane protein will solve this open question.
ACKNOWLEDGMENTS
We thank Sarah Kizilirmak for assistance. We are grateful to Stephanie Hacker for drawing our attention to the choline transporter and Jer-Sheng Lin and Erh-Min Lai (Taiwan) for providing an A. tumefaciens nolR mutant. We thank Erhard Bremer (Marburg, Germany) for discussions in the initial phase of this project and Bernd Masepohl for constructive input.
The study was funded in part by a grant from the German Research Foundation (DFG grant NA 240/7) to F.N. and a fellowship from the Promotionskolleg der Ruhr-Universität Bochum to M.A.
Footnotes
Published ahead of print on 29 July 2011.
REFERENCES
- 1. Aktas M., et al. 2010. Phosphatidylcholine biosynthesis and its significance in bacteria interacting with eukaryotic cells. Eur. J. Cell Biol. 89:888–894 [DOI] [PubMed] [Google Scholar]
- 2. Alloing G., Travers I., Sagot B., Le Rudulier D., Dupont L. 2006. Proline betaine uptake in Sinorhizobium meliloti: characterization of Prb, an Opp-like ABC transporter regulated by both proline betaine and salinity stress. J. Bacteriol. 188:6308–6317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917 [DOI] [PubMed] [Google Scholar]
- 4. Blusztajn J. K. 1998. Choline, a vital amine. Science 281:794–795 [DOI] [PubMed] [Google Scholar]
- 5. Boncompagni E., Osteras M., Poggi M. C., le Rudulier D. 1999. Occurrence of choline and glycine betaine uptake and metabolism in the family Rhizobiaceae and their roles in osmoprotection. Appl. Environ. Microbiol. 65:2072–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen C., Beattie G. A. 2007. Characterization of the osmoprotectant transporter OpuC from Pseudomonas syringae and demonstration that cystathionine-beta-synthase domains are required for its osmoregulatory function. J. Bacteriol. 189:6901–6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen C., Beattie G. A. 2008. Pseudomonas syringae BetT is a low-affinity choline transporter that is responsible for superior osmoprotection by choline over glycine betaine. J. Bacteriol. 190:2717–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen C., Malek A. A., Wargo M. J., Hogan D. A., Beattie G. A. 2010. The ATP-binding cassette transporter Cbc (choline/betaine/carnitine) recruits multiple substrate-binding proteins with strong specificity for distinct quaternary ammonium compounds. Mol. Microbiol. 75:29–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen H., Gao K., Kondorosi E., Kondorosi A., Rolfe B. G. 2005. Functional genomic analysis of global regulator NolR in Sinorhizobium meliloti. Mol. Plant Microbe Interact. 18:1340–1352 [DOI] [PubMed] [Google Scholar]
- 10. Chen H., et al. 2000. Identification of nolR-regulated proteins in Sinorhizobium meliloti using proteome analysis. Electrophoresis 21:3823–3832 [DOI] [PubMed] [Google Scholar]
- 11. Conde-Alvarez R., et al. 2006. Synthesis of phosphatidylcholine, a typical eukaryotic phospholipid, is necessary for full virulence of the intracellular bacterial parasite Brucella abortus. Cell. Microbiol. 8:1322–1335 [DOI] [PubMed] [Google Scholar]
- 12. Conover G. M., et al. 2008. Phosphatidylcholine synthesis is required for optimal function of Legionella pneumophila virulence determinants. Cell Microbiol. 10:514–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dupont L., et al. 2004. The Sinorhizobium meliloti ABC transporter Cho is highly specific for choline and expressed in bacteroids from Medicago sativa nodules. J. Bacteriol. 186:5988–5996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goodner B., et al. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294:2323–2328 [DOI] [PubMed] [Google Scholar]
- 15. Haardt M., Kempf B., Faatz E., Bremer E. 1995. The osmoprotectant proline betaine is a major substrate for the binding-protein-dependent transport system ProU of Escherichia coli K-12. Mol. Gen. Genet. 246:783–786 [DOI] [PubMed] [Google Scholar]
- 16. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 17. Higgins C. F., Ames G. F. 1981. Two periplasmic transport proteins which interact with a common membrane receptor show extensive homology: complete nucleotide sequences. Proc. Natl. Acad. Sci. U. S. A. 78:6038–6042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gisin J., et al. 2010. A Rhodobacter capsulatus member of a universal permease family imports molybdate and other oxyanions. J. Bacteriol. 192:5943–5952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kappes R. M., et al. 1999. Two evolutionarily closely related ABC transporters mediate the uptake of choline for synthesis of the osmoprotectant glycine betaine in Bacillus subtilis. Mol. Microbiol. 32:203–216 [DOI] [PubMed] [Google Scholar]
- 20. Kempf B., Bremer E. 1995. OpuA, an osmotically regulated binding protein-dependent transport system for the osmoprotectant glycine betaine in Bacillus subtilis. J. Biol. Chem. 270:16701–16713 [DOI] [PubMed] [Google Scholar]
- 21. Klüsener S., Aktas M., Thormann K. M., Wessel M., Narberhaus F. 2009. Expression and physiological relevance of Agrobacterium tumefaciens phosphatidylcholine biosynthesis genes. J. Bacteriol. 191:365–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kondorosi E., et al. 1989. Positive and negative control of nod gene expression in Rhizobium meliloti is required for optimal nodulation. EMBO J. 8:1331–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kortstee G. J. 1970. The aerobic decomposition of choline by microorganisms. I. The ability of aerobic organisms, particularly coryneform bacteria, to utilize choline as the sole carbon and nitrogen source. Arch. Mikrobiol. 71:235–244 [PubMed] [Google Scholar]
- 24. Lamark T., et al. 1991. DNA sequence and analysis of the bet genes encoding the osmoregulatory choline-glycine betaine pathway of Escherichia coli. Mol. Microbiol. 5:1049–1064 [DOI] [PubMed] [Google Scholar]
- 25. Li F., Hou B., Hong G. 2008. Symbiotic plasmid is required for NolR to fully repress nodulation genes in Rhizobium leguminosarum A34. Acta Biochim. Biophys. Sin. (Shanghai) 40:901–907 [PubMed] [Google Scholar]
- 26. Liu A. C., Shih H. W., Hsu T., Lai E. M. 2008. A citrate-inducible gene, encoding a putative tricarboxylate transporter, is downregulated by the organic solvent DMSO in Agrobacterium tumefaciens. J. Appl. Microbiol. 105:1372–1383 [DOI] [PubMed] [Google Scholar]
- 27. Malek A. A., Chen C., Wargo M. J., Beattie G. A., Hogan D. A. 2011. Roles of three transporters, CbcXWV, BetT1, and BetT3, in Pseudomonas aeruginosa choline uptake for catabolism. J. Bacteriol. 193:3033–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mao B., Pear M. R., McCammon J. A., Quiocho F. A. 1982. Hinge-bending in l-arabinose-binding protein. The “Venus's-flytrap” model. J. Biol. Chem. 257:1131–1133 [PubMed] [Google Scholar]
- 29. Masepohl B., et al. 2001. Urea utilization in the phototrophic bacterium Rhodobacter capsulatus is regulated by the transcriptional activator NtrC. J. Bacteriol. 183:637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. May G., Faatz E., Villarejo M., Bremer E. 1986. Binding protein dependent transport of glycine betaine and its osmotic regulation in Escherichia coli K-12. Mol. Gen. Genet. 205:225–233 [DOI] [PubMed] [Google Scholar]
- 31. Michel V., Yuan Z., Ramsubir S., Bakovic M. 2006. Choline transport for phospholipid synthesis. Exp. Biol. Med. (Maywood) 231:490–504 [DOI] [PubMed] [Google Scholar]
- 32. Miller J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 33. Miller K. J., Wood J. M. 1996. Osmoadaptation by rhizosphere bacteria. Annu. Rev. Microbiol. 50:101–136 [DOI] [PubMed] [Google Scholar]
- 34. Minder A. C., et al. 2001. Phosphatidylcholine levels in Bradyrhizobium japonicum membranes are critical for an efficient symbiosis with the soybean host plant. Mol. Microbiol. 39:1186–1198 [PubMed] [Google Scholar]
- 35. Oswald C., Smits S. H., Hoing M., Bremer E., Schmitt L. 2009. Structural analysis of the choline-binding protein ChoX in a semi-closed and ligand-free conformation. Biol. Chem. 390:1163–1170 [DOI] [PubMed] [Google Scholar]
- 36. Oswald C., et al. 2008. Crystal structures of the choline/acetylcholine substrate-binding protein ChoX from Sinorhizobium meliloti in the liganded and unliganded-closed states. J. Biol. Chem. 283:32848–32859 [DOI] [PubMed] [Google Scholar]
- 37. Pittelkow M., Tschapek B., Smits S. H., Schmitt L., Bremer E. 1 June 2011, posting date The crystal structure of the substrate-binding protein OpuBC from Bacillus subtilis in complex with choline. J. Mol. Biol. doi:10.1016/j.jmb.2011.05.037 [DOI] [PubMed] [Google Scholar]
- 38. Pocard J. A., Bernard T., Smith L. T., Le Rudulier D. 1989. Characterization of three choline transport activities in Rhizobium meliloti: modulation by choline and osmotic stress. J. Bacteriol. 171:531–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Quiocho F. A., Ledvina P. S. 1996. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol. Microbiol. 20:17–25 [DOI] [PubMed] [Google Scholar]
- 40. Ren Q., Chen K., Paulsen I. T. 2007. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res. 35:D274–D279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roy A., Kucukural A., Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5:725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 43. Schäfer A., Tauch A., J.äger W., Kalinowski J., Thierbach G., Pühler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73 [DOI] [PubMed] [Google Scholar]
- 44. Schmidt-Eisenlohr H., et al. 1999. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J. Bacteriol. 181:7485–7492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sherr S. I., Law J. H. 1965. Phosphatidylcholine synthesis in Agrobacterium tumefaciens. II. Uptake and utilization of choline. J. Biol. Chem. 240:3760–3765 [PubMed] [Google Scholar]
- 46. Smith L. T., Pocard J. A., Bernard T., Le Rudulier D. 1988. Osmotic control of glycine betaine biosynthesis and degradation in Rhizobium meliloti. J. Bacteriol. 170:3142–3149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Smith L. T., Smith G. M., Madkour M. A. 1990. Osmoregulation in Agrobacterium tumefaciens: accumulation of a novel disaccharide is controlled by osmotic strength and glycine betaine. J. Bacteriol. 172:6849–6855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sohlenkamp C., López-Lara I. M., Geiger O. 2003. Biosynthesis of phosphatidylcholine in bacteria. Prog. Lipid Res. 42:115–162 [DOI] [PubMed] [Google Scholar]
- 49. Studier F. W., Moffatt B. A. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113–130 [DOI] [PubMed] [Google Scholar]
- 50. Styrvold O. B., et al. 1986. Selection, mapping, and characterization of osmoregulatory mutants of Escherichia coli blocked in the choline-glycine betaine pathway. J. Bacteriol. 165:856–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thomas G. H. 2010. Homes for the orphans: utilization of multiple substrate-binding proteins by ABC transporters. Mol. Microbiol. 75:6–9 [DOI] [PubMed] [Google Scholar]
- 52. Wang X. G., et al. 2004. Analysis of differences in the functional properties of the substrate binding proteins of the Borrelia burgdorferi oligopeptide permease (Opp) operon. J. Bacteriol. 186:51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wessel M., et al. 2006. Virulence of Agrobacterium tumefaciens requires phosphatidylcholine in the bacterial membrane. Mol. Microbiol. 62:906–915 [DOI] [PubMed] [Google Scholar]
- 54. Wood D. W., et al. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317–2323 [DOI] [PubMed] [Google Scholar]
- 55. Yeo H. J., Yuan Q., Beck M. R., Baron C., Waksman G. 2003. Structural and functional characterization of the VirB5 protein from the type IV secretion system encoded by the conjugative plasmid pKM101. Proc. Natl. Acad. Sci. U. S. A. 100:15947–15952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang Y. 2009. I-TASSER: fully automated protein structure prediction in CASP8. Proteins 77(Suppl. 9):100–113 [DOI] [PMC free article] [PubMed] [Google Scholar]