Abstract
Xylose is rarely described as a component of bacterial glycans. UDP-xylose is the nucleotide-activated form necessary for incorporation of xylose into glycans and is synthesized by the decarboxylation of UDP-glucuronic acid (UDP-GlcA). Enzymes with UDP-GlcA decarboxylase activity include those that lead to the formation of UDP-xylose as the end product (Uxs type) and those synthesizing UDP-xylose as an intermediate (ArnA and RsU4kpxs types). In this report, we identify and confirm the activities of two Uxs-type UDP-GlcA decarboxylases of Bacteroides fragilis, designated BfUxs1 and BfUxs2. Bfuxs1 is located in a conserved region of the B. fragilis genome, whereas Bfuxs2 is in the heterogeneous capsular polysaccharide F (PSF) biosynthesis locus. Deletion of either gene separately does not result in the loss of a detectable phenotype, but deletion of both genes abrogates PSF synthesis, strongly suggesting that they are functional paralogs and that the B. fragilis NCTC 9343 PSF repeat unit contains xylose. UDP-GlcA decarboxylases are often annotated incorrectly as NAD-dependent epimerases/dehydratases; therefore, their prevalence in bacteria is underappreciated. Using available structural and mutational data, we devised a sequence pattern to detect bacterial genes encoding UDP-GlcA decarboxylase activity. We identified 826 predicted UDP-GlcA decarboxylase enzymes in diverse bacterial species, with the ArnA and RsU4kpxs types confined largely to proteobacterial species. These data suggest that xylose, or a monosaccharide requiring a UDP-xylose intermediate, is more prevalent in bacterial glycans than previously appreciated. Genes encoding BfUxs1-like enzymes are highly conserved in Bacteroides species, indicating that these abundant intestinal microbes may synthesize a conserved xylose-containing glycan.
INTRODUCTION
The pentose sugar xylose is a common component of the cell walls of plants (44), fungal glycans (33), and glycosaminoglycans of higher organisms (26). Xylose is rarely described as a component of bacterial glycans. Bacterial molecules shown to contain xylose include the O antigen of Pseudomonas aeruginosa serotype O7 (10, 25), the core oligosaccharide of the lipopolysaccharide (LPS) of Sinorhizobium meliloti (20), the LPS core oligosaccharide of Ralstonia solanacearum (19), the LPS of Leptospira species (23, 35), and an S-layer glycoprotein of Parabacteroides distasonis (15).
The incorporation of xylose into glycans requires its addition from the nucleotide-activated precursor UDP-xylose. UDP-xylose was first shown to be synthesized from UDP-glucuronic acid (UDP-GlcA) by a UDP-GlcA decarboxylase in the fungus Cryptococcus laurentii (5), and this enzymatic activity has since been identified in plants and animals. The UDP-xylose synthase (Uxs) family enzymes found in animals produce UDP-xylose by decarboxylation of UDP-GlcA using NAD (NAD+). Similarly, the enzymes of the UDP-apiose/UDP-xylose synthase (Uaxs) family found in plants convert UDP-GlcA to UDP-apiose and UDP-xylose (21).
Only three bacterial enzymes catalyzing oxidative decarboxylation of UDP-GlcA to UDP-xylose have been described. SmUxs1 from Sinorhizobium meliloti (20) leads to the synthesis of UDP-xylose as an end product. ArnA of Escherichia coli is a bifunctional enzyme that has a C-terminal portion with UDP-GlcA decarboxylase activity, catalyzing the C-4 oxidation and C-6 decarboxylation of UDP-GlcA. This enzyme's N-terminal portion also has formyl transferase activity, and both activities participate in a pathway leading ultimately to the synthesis of UDP-4-amino-4-deoxy-l-arabinose (UDP-l-Ara4N). RsU4kpxs, from Ralstonia solanacearum (19), is similar to the C-terminal portion of ArnA, has UDP-GlcA decarboxylase activity, and produces UDP-xylose as an end product. However, much of the UDP-xylose of this organism is further converted to UDP-l-Ara4N by a formyl transferase, similar to the N-terminal portion of ArnA, encoded by an adjacent gene.
UDP-GlcA decarboxylases are members of the short-chain dehydrogenase/reductase (SDR) superfamily that also include epimerases, dehydratases, and reductases that use nucleotide-activated monosaccharides as substrates. Enzymes in this large family of proteins share a Rossmann-fold NAD(P)H/NAD(P)(+) binding domain. Genes encoding SDR enzymes are often annotated solely as NAD-dependent epimerases/dehydratases, and therefore, many UDP-GlcA decarboxylase genes are likely incorrectly annotated.
Intestinal Bacteroidales species are the most abundant Gram-negative bacteria of the human colonic microbiota, reaching densities of more than 1010 bacteria per gram of feces (45). Intestinal Bacteroidales species encode an extensive number of proteins dedicated to the synthesis of glycosylated molecules, including many capsular polysaccharides (27), glycoproteins (17), extracellular polysaccharides (11), and glycosylated S-layer proteins (15). Many of these molecules are necessary for the organism to colonize the mammalian intestine (12, 13, 17), and one has been shown to provide beneficial properties to the host in the intestine (30) and abscess-promoting properties in extraintestinal sites (40).
These bacteria have a tremendous capacity to degrade the numerous plant and host polysaccharides that are abundant in the colonic ecosystem. Bacteroides genomes in general encode a large number of xylosidases, which allow the bacteria to degrade xylose-containing polymers as an energy source. We have found that these bacteria often include monosaccharides that are abundant in the intestinal ecosystem in their own glycosylated molecules (13). Therefore, we sought to explore whether xylose was likely present in the glycans of intestinal Bacteroidales species.
Elucidating the structures of glycosylated molecules of Bacteroides is a difficult process, as the glycosylated molecule first must be purified from other glycans that often are of similar size and have similar charge properties. Despite the importance of these molecules to the biology of the bacteria and to their interactions with the host in both symbiotic and pathogenic relationships, the structures of very few glycans of the intestinal Bacteroidales are known, nor is there much information regarding their compositions. Therefore, we searched for genes encoding UDP-GlcA decarboxylases as an indication that xylose might be a constituent of the glycans of these bacteria.
In this report, we characterize two UDP-glucuronic acid (UDP-GlcA) decarboxylases of Bacteroides fragilis. Using a residue pattern to distinguish UDP-GlcA decarboxylases from other enzymes of the short-chain dehydrogenase/reductase (SDR) superfamily, we found similar proteins encoded by species throughout the Bacteroides genus and by numerous other bacterial species. Therefore, xylose is likely present in the glycans of most Bacteroides species and is more common than previously appreciated in bacterial glycans in general.
MATERIALS AND METHODS
All primers used in this study are listed in Table S1 in the supplemental material.
Cloning BF2189 and BF1555.
BF2189 was PCR amplified as a 998-bp product using high-fidelity Taq polymerase (Invitrogen). The product was digested with BamHI (Invitrogen) and cloned into the BamHI site of vector pET16b to create an N-terminal His-tagged protein. BF1555 was amplified and cloned in a similar manner. BF1555 is annotated in the B. fragilis NCTC 9343 genome as having an alternate start codon, followed immediately by an ATG codon, which we considered to be the start codon for this study as each of the four major bacterial gene prediction programs (Prodigal 2.50, Glimmer 3, GeneMarkHMM 2.6r, and GeneMark 2.5m) predict that this gene starts with the ATG codon. The sequence fidelity and correct orientation of both clones were confirmed by sequencing, and each was transformed into E. coli BL21(DE3) (37) for protein purification. As controls, the pET16b vector alone and pDC22.6 (a pET28a-based clone containing the Cryptococcus neoformans UXS1 gene [6]) were also transformed into E. coli BL21(DE3).
Enzyme purifications.
E. coli BL21(DE3) cells containing recombinant vectors were grown at 37°C in 3 ml of Luria broth supplemented with either ampicillin (100 μg/ml) or kanamycin (30 μg/ml, for pDC22.6) for 1.5 h and then used to inoculate 100 ml of the same medium. These cultures were grown until an optical density at 600 nm of ∼0.4 was reached, and each was split into 50-ml aliquots. One aliquot of each culture was induced by adding 0.4 mM isopropyl β-d-1-thiogalactopyranoside (IPTG; Invitrogen), and incubation was continued for 2.5 h. The cells were recovered by centrifugation at 4°C, and each pellet was resuspended in 2.5 ml of 50 mM cold Tris-HCl (pH 7.8) and stored at −20°C.
Thoroughly resuspended Talon Dynabeads (50 μl, 2 mg; Invitrogen), which are precharged with Co2+, were equilibrated for use according to the manufacturer's directions, except that the binding/wash buffer contained 50 mM sodium phosphate buffer (pH 7.0), 500 mM NaCl, and 0.01% (vol/vol) Tween 20. Cells from 25 ml of IPTG-induced culture (see above) were resuspended in 1.5 ml of ice-cold binding/wash buffer and lysed by sonication. DNase (Worthington Biochemical Corporation, Lakewood, NJ) was added to each sample at a final concentration of 1 μg/ml, and the suspensions were held on ice. The lysates were briefly (15 s) centrifuged in a bench-top centrifuge to pellet the bulk of the cell debris, and 100 μl of each lysate was added to 50 μl of the washed and equilibrated paramagnetic beads. The tubes were allowed to stand on ice for 15 min with occasional resuspension and then incubated end over end for 30 min at 4°C. The beads were collected magnetically and washed four times in 700 μl of the binding and wash buffer. The enzyme-coated magnetic beads retain the enzymes in their native form and facilitate their removal postreaction and were thus used directly in the enzymatic activity assays.
Enzymatic reactions.
Each bead preparation was resuspended in 100 μl of enzyme reaction mixture consisting of 65 mM morpholinepropanesulfonic acid (MOPS) (pH 7.4), 1 mM NAD (NAD+; Sigma), and 1 mM UDP-glucuronic acid (UDP-GlcA; Sigma) or 1 mM UDP-galacturonic acid (UDP-GalA; CarboSource Services, Atlanta, GA). The reaction mixtures were incubated at 30°C while rotating end over end for 1 h. The enzymes were removed magnetically after the reaction.
High-performance anion-exchange chromatography.
Samples (25 μl) of the reactions and standards were analyzed by high-performance anion-exchange chromatography using a Dionex (Sunnyvale, CA) pulsed amperometric detector and a CarboPac PA1 carbohydrate column. The detector sensitivity was set at 300 nA with a 0.05-V applied pulse potential. Compounds were eluted with 825 mM sodium acetate in 20 mM sodium hydroxide, with a flow rate of 1 ml per minute. Standards included UDP-xylose, UDP-arabinose, UDP-glucuronic acid, and UDP-galacturonic acid (CarboSource Services).
Creation of single and double deletion mutants.
Deletions of Bfuxs1 (BF2189) and Bfuxs2 (BF1555) were constructed such that 834 bp of the 936-bp Bfuxs1 and 826 bp of the 942-bp Bfuxs2 were removed by allelic replacement. DNA segments upstream and downstream of each region to be deleted were PCR amplified and cloned by three-way ligation into the Bacteroides conjugal suicide vector pJST55 (39). The resulting plasmids were conjugally transferred into wild-type B. fragilis NCTC 9343, and cointegrates were selected on the basis of erythromycin resistance encoded by the vector. The cointegrate strains were passaged, plated on nonselective medium, and replica plated on medium containing erythromycin. Erythromycin-sensitive colonies were screened by PCR to detect colonies that had acquired the mutant genotype. The ΔBfuxs1 ΔBfuxs2 double mutant was constructed by first deleting Bfuxs1 and then Bfuxs2.
SDS-PAGE and Western blot analysis.
Bacterial lysates from 18-h cultures grown in basal medium (32) were separated using NuPAGE 4 to 12% gradient SDS-polyacrylamide gels with morpholineethanesulfonic acid (MES) buffer (Invitrogen). The contents of the gels were transferred to polyvinylidene difluoride membranes (Invitrogen) and probed with rabbit antiserum specific to individual capsular polysaccharides of B. fragilis NCTC 9343 (27), to the glycan of the B. fragilis glycoproteins, or to the protein component of glycoprotein BF2494 (17). The ΔBfuxs1 ΔBfuxs2 adsorbed antiserum was prepared as previously described (27), except that the ΔBfuxs1 ΔBfuxs2 mutant was used to adsorb antibodies to the wild-type strain. Following this adsorption, only antibodies to immunogenic molecules produced by the wild-type strain but not by the mutant remained. By comparing the reactivities of this antiserum to wild-type and mutant extracts, molecules that are synthesized by the wild type but lost in the mutant can be identified. All blots were then probed with alkaline phosphatase-conjugated goat anti-rabbit secondary antibodies and developed colorimetrically using 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium (Kirkegaard & Perry).
Bioinformatic analyses.
All sequences were retrieved from GenBank in January 2011. Similar sequences were retrieved with BLAST (3, 4). Taxonomy assignment is based on NCBI's taxonomy database. Alignments were routinely performed with ClustalW2 (28). Custom Perl scripts were used throughout these analyses. Secondary structure prediction was performed using the SABLE program (1).
To identify proteins encoded by bacterial genomes with predicted UDP-GlcA decarboxylase activity, a BLAST search was performed using the amino acid sequence of BfUxs1 to query NCBI's nonredundant database. Similar proteins (E value, ≤10−3) encoded by bacterial genomes with completed or draft genomic sequences were retained. These proteins were then analyzed for those matching a Perl regular expression, G..G..G.{17,27}(D|E).{76,84}TSE.{26,28}Y.{3}K.{106,127}?R, devised to detect UDP-GlcA decarboxylases, as described above. Each sequence on this list of candidate UDP-GlcA decarboxylases was then classified as closest to one of the three types of bacterial enzymes described to have UDP-GlcA decarboxylase activity: Uxs1, ArnA, and RsU4kpxs. Each UDP-GlcA decarboxylase was further analyzed using a custom BLAST database to determine whether it was more similar (based on the lowest E value) to the C-terminal UDP-GlcA decarboxylase domain of the ArnA protein (NCBI accession no. 2BLL_A, GI:66361562) or to UDP-glucuronic acid decarboxylase 1 (Uxs1) from Homo sapiens (NCBI accession no. NP_079352.2, GI:42516563). As ArnA proteins have a formyl transferase domain at the N terminus, the UDP-GlcA decarboxylases were also analyzed with the Pfam database (version 25.0 of March 2011). If the protein returned a significant match to PF00551.13 (Formyl_trans_N) and/or PF02911.12 (Formyl_trans_C)—two Pfam motifs found in the N-terminal formyl transferase domain of ArnA—it was assigned as an ArnA type of UDP-GlcA decarboxylase. As RsU4kpxs-type UDP-GlcA decarboxylase proteins are more similar to the C terminus of ArnA than to that of Uxs1 but have the formyltransferase activity of ArnA encoded by an adjacent gene, the RsU4kpxs-type proteins were identified as being more similar to ArnA than to Uxs1, lacking a formyltransferase domain, and as having an adjacent gene encoding a protein with a significant match to PF00551.13 (Formyl_trans_N) and/or PF02911.12 (Formyl_trans_C).
RESULTS
Identification of putative UDP-GlcA decarboxylases of B. fragilis.
To determine whether B. fragilis may synthesize glycans containing xylose, we searched the B. fragilis NCTC 9343 genome for genes encoding orthologs of the human UDP-glucuronic acid decarboxylase protein UXS1 (NCBI accession no. AY147934.1). Two B. fragilis orthologs encoded by BF2189 and BF1555 (both 76% similar to the human ortholog and 87% similar to each other) were identified (Fig. 1). Each of these products is annotated as an NAD-dependent epimerase/dehydratase, which is a general category applied to diverse products involved in the synthesis and modification of nucleotide-linked sugars. When the amino acid sequences of BF2189 and BF1555 are used to query the RCSB Protein Data Bank (PDB) (34), the closest structural match is human UDP-glucuronate decarboxylase (PDB entry 2B69; alignment E value, 5.8 × 10−105). BF2189, which we have designated Bfuxs1, is conserved in the B. fragilis genome and is the first gene of a putative two-gene operon. BF1555, designated Bfuxs2, is contained within the polysaccharide F (PSF) capsular polysaccharide biosynthesis locus of strain NCTC 9343, which is a heterogeneous region of the B. fragilis genome.
Fig. 1.
Similarities of BfUxs1 (NCBI accession no. CAH07883) and BfUxs2 (accession no. CAH07260) with the human UDP-GlcA decarboxylase UXS1 (accession no. AAN39844). BOXSHADE alignment of the three proteins with the relevant SDR superfamily residues are highlighted with blue asterisks. Below the primary sequence are the secondary structures predicted by SABLE (1). Helical areas are indicated by a red cylindrical line, while beta sheet areas are indicated by a gold arrow.
The predicted secondary structures of BfUxs1, BfUxs2, and the human USX1 ortholog are remarkably similar (Fig. 1). As SDR superfamily members, UDP-GlcA decarboxylase enzymes conform to the structural characteristics of this superfamily. SABLE (1) predicts that each protein has an alternating series of alpha helices and beta strands (Fig. 1) comprising the structural motif known as a Rossmann fold. The sequence motif GX(X)GXXG, involved in NAD+ binding, is also present (G8G11G14 in both BfUxs1 and BfUxs2), with the third glycine residue appropriately positioned in the first helix (29). Eighteen residues after the glycine-rich sequence pattern, after the second beta strand, there is a conserved aspartic acid residue (D32), which indicates that the enzyme utilizes NAD(H) as a nicotinamide cofactor (24). The conserved catalytic tetrad of N-[S/T]-Y-K (14, 18) is present as well (represented by N98T115Y144K146). These residues catalyze the oxidation of the substrate to a 4′-keto intermediate via hydride transfer between the substrate and the nicotinamide cofactor, the hallmark reaction of all SDR enzymes (14).
Within the large SDR superfamily, there are features of BfUxs1 and BfUxs2 that strongly suggest that they are UDP-GlcA decarboxylases. Analysis of the crystal structure of the human USX1 protein (PDB identification [ID] no. 2B69) is unpublished; however, two research groups have independently published analyses of the structure (PDB ID no. 1U9J and 2BLL) of the C-terminal portion of E. coli ArnA (18, 42). The C-terminal portion of this bifunctional enzyme catalyzes the NAD+-dependent oxidative decarboxylation of UDP-GlcA as part of a pathway leading to production of UDP-4-amino-4-deoxy-l-arabinose (7, 18, 42). Analysis of the crystal structure of the C-terminal decarboxylase region of ArnA revealed that R619 and S433 are important for decarboxylase activity (7, 18, 42). In addition, site-directed mutations of S433A, E434A, and E434Q of ArnA affected UDP-GlcA decarboxylase activity. These three residues are represented by S116, E117, and R276 in BfUxs1 and BfUxs2 and are also conserved in the human USX1 decarboxylase, represented by S203, E204, and R361 (Fig. 2). Alignment of BfUxs1 and BfUxs2 with other functionally characterized UDP-GlcA decarboxylases from diverse species shows that they all retain these Ser, Glu, and Arg residues, whereas other bacterial SDR enzymes with nondecarboxylase activity do not (Fig. 2).
Fig. 2.
Conserved UDP-GlcA decarboxylase residues. All of these SDR enzymes were aligned together for the BOXSHADE analysis. Segments of the proteins corresponding to the conserved SDR and UDP-GlcA decarboxylase residues are highlighted with green (*) and pink (+), respectively. Black and gray shading indicates consensus residues conserved in more than half the sequences and residues considered similar to the consensus residues, respectively. Dots represent gaps in the sequences displayed. The UDP-GlcA decarboxylase sequences shown are Sinorhizobium meliloti 1021 SmUxs1.1 (NCBI accession no. NP_436980) (20), Arabidopsis thaliana AtUxs1 (accession no. NP_850694) (22), Cryptococcus neoformans Uxs1p (accession no. AF385328_1) (6), Homo sapiens UXS1 (accession no. AAN39844) (31), Ralstonia solanacearum GMI1000 RsU4kpxs (accession no. NP_519440.1) (19), and the C-terminal portion of Escherichia coli ArnA (accession no. NP_416758) (42). Nondecarboxylase SDR family enzymes are the UDP-GlcNAc 4,6-dehydratases UngD1 (accession no. YP_211344) and UngD2 (accession no. YP_212462) from B. fragilis NCTC 9343 (12, 13), WbpM from Pseudomonas aeruginosa PAO1 (accession no. AAC45867) (9), RmlB dTDP-glucose 4,6-dehydratase from Streptococcus suis (accession no. CAD49092) (2), GalE UDP-galactose 4-epimerase from Escherichia coli (accession no. BAA35421) (38), and GDP-mannose-4,6-dehydratase (Gmd) (accession no. YP_211522) and GDP-4-keto-6-deoxy-d-mannose-3,5-epimerase-4-reductase (Fcl) (accession no. YP_211521) from B. fragilis NCTC 9343 (12, 13). The activities of all listed enzymes were determined experimentally.
Enzymatic characterization of BfUxs1 and BfUxs2.
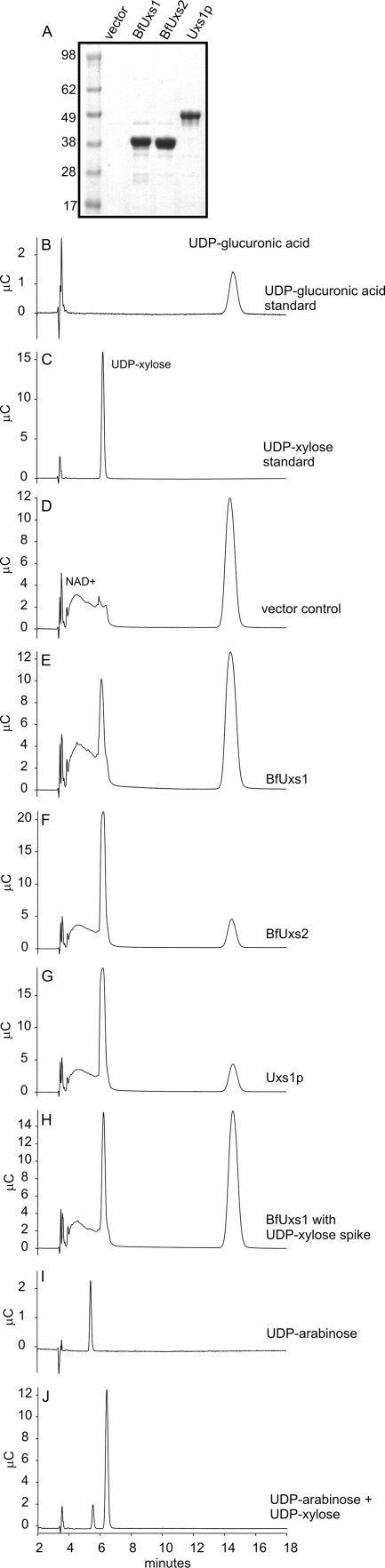
To determine whether BfUxs1 and BfUxs2 have UDP-GlcA decarboxylase activity, we purified the proteins and performed enzymatic analyses. The characterized His-tagged UDP-GlcA decarboxylase of Cryptococcus neoformans (6) was included as a positive control, and the purifications of each of these enzymes are shown in Fig. 3A. The conversion of UDP-GlcA to UDP-xylose was monitored by high-performance anion-exchange chromatography. The products of both the BfUxs1 and BfUxs2 reactions eluted identically to the UDP-xylose control at approximately 6.24 min (Fig. 3), easily distinguishable from the UDP-GlcA substrate, which elutes at approximately 14.5 min. These peaks were also indistinguishable from that produced by the Cryptococcus neoformans USX1p UDP-GlcA decarboxylase (6). To provide further evidence that BfUxs1 and BfUxs2 were catalyzing the conversion of UDP-GlcA to UDP-xylose, the reaction products were spiked with UDP-xylose prior to analysis. For this analysis, there remains a single sharp UDP-xylose peak with no shouldering (Fig. 3H), providing further evidence that UDP-xylose is the reaction product. Arabinose is a five-carbon sugar that is an epimer of xylose and has been detected in plant and bacterial glycans (8, 20). The elution of UDP-arabinose at 5.43 min is easily distinguished from that of UDP-xylose, demonstrating that this detection method can differentiate these two closely related nucleotide-activated pentoses (Fig. 3I). Reactions performed under the same conditions using protein purified in the same manner from E. coli BL21(DE3) containing vector alone failed to produce a UDP-xylose peak (Fig. 3D). No product was formed when UDP-galacturonic acid was substituted for UDP-GlcA as the substrate (data not shown), demonstrating that these enzymes are specific for UDP-GlcA.
Fig. 3.
Analysis of reaction substrates and products by high-performance anion-exchange chromatography. (A) Coomassie-stained gel of the purified enzymes used in the reactions or the vector control. Molecular masses of standards are shown to the left in kDa. BfUxs1 and BfUxs2 are approximately 38.7 kDa, whereas Uxs1p is approximately 48.7 kDa. (B) UDP-glucuronic acid standard. (C) UDP-xylose standard. (D to J) Enzymatic reaction mixtures containing the following His-tagged enzymes or controls: pET16b (vector only) (D), BfUxs1 (E), BfUxs2 (F), C. neoformans Uxs1p (G), BfUxs1 spiked with UDP-xylose (H), UDP-arabinose standard (I), and UDP-arabinose and UDP-glucuronic acid (J).
Distribution of UDP-GlcA decarboxylase proteins.
With the confirmation that the BfUxs proteins have UDP-GlcA decarboxylase activity, we sought to determine the prevalence of UDP-GlcA decarboxylases within the domain Bacteria. First, a BLAST search was performed using the amino acid sequence of BfUxs1 to query NCBI's nonredundant database for similar proteins encoded by bacterial genomes with completed or draft genomic sequences. This search returned 9,915 bacterial proteins with an E value of ≤10−3, virtually all of which contain the residues indicative of SDR superfamily enzymes. We then devised a Perl regular expression to detect the pattern of conserved residues of SDR superfamily members combined with those residues that have been shown by structural or mutagenesis analysis to be essential for UDP-GlcA decarboxylase activity. The Perl regular expression used to query the 9,915 proteins returned from the BLAST search is G..G..G.{17,27}(D|E).{76,84}TSE.{26,28}Y.{3}K.{106,127}?R, where ? signifies that only the first R in the range should be considered. This analysis identified 826 potential bacterial UDP-GlcA decarboxylases (see Table S2 in the supplemental material). Proteins of proteobacterial origin predominated, comprising 70.3% of the list (581 proteins), likely due to their relative overrepresentation in the database. Of the 1,116 proteobacterial strains with sequence information available, 467 encode one or more proteins matching the decarboxylase motif.
These 826 predicted bacterial UDP-GlcA decarboxylases were then further categorized into the three types of bacterial proteins with this described activity, Uxs, ArnA, and RsU4kpxs. Of the 826 UDP-GlcA decarboxylase proteins, 518 are the Uxs type, 227 are the ArnA type, and 81 are the RsU4kpxs type. With only two exceptions, the ArnA types are contained exclusively in species within the Proteobacteria phylum. The ArnA types are encoded mostly by gammaproteobacteria, and 94 of the 108 queried E. coli genomes encode a protein with the characteristics of the larger bifunctional ArnA type, suggesting that these bacteria synthesize UDP-4-amino-4-deoxy-l-arabinose. No E. coli genomes analyzed encoded Uxs or RsU4kpxs types. The RsU4kpxs types are encoded largely by betaproteobacteria and are highly represented in Burkholderia species.
Although the Proteobacteria contained nearly all of the ArnA- and RsU4kpxs-type UDP-GlcA decarboxylases, there are also 276 proteins of the Uxs type encoded by proteobacterial genomes. In addition to containing sequences of the RsU4kpxs type, 66 of the 69 Burkholderia species analyzed also encode a Uxs-type enzyme, suggesting that xylose is likely consistently present in a glycan(s) of this genus.
Other bacteria with a high prevalence of Uxs types of UDP-GlcA decarboxylase enzymes include Chloroflexi (9/12 genomes), the Chlamydiae/Verrucomicrobia group (11/14 genomes), Cyanobacteria (50/60 genomes) Fibrobacteres/Acidobacteria (7/8 genomes), Planctomycetes (7/7 genomes), and Leptospira spp. (6/6 genomes). Table S2 in the supplemental material lists the distribution of proteomes with predicted UDP-GlcA decarboxylases within the different phyla for species with draft or completed genomes. Of note is the rarity of these proteins in the phyla Firmicutes (7/799 genomes) and Fusobacteria (0/25 genomes).
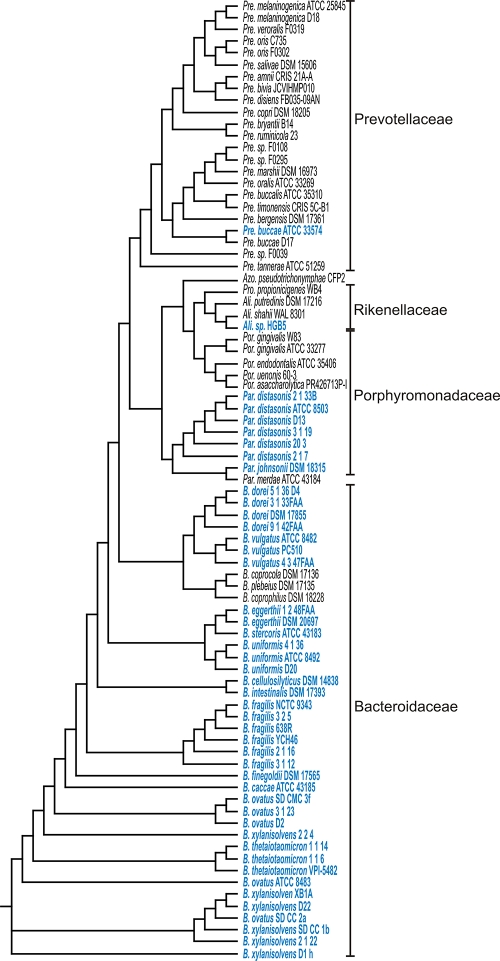
Of the 44 sequenced strains classified by NCBI as belonging to the genus Bacteroides (excluding B. pectinophilus, which 16S rRNA analysis suggests is a member of the Firmicutes phylum), 41 synthesize at least one Uxs-type UDP-GlcA decarboxylase (Fig. 4). The closely related species B. plebeius, B. coprocola, and B. coprophilus are the three exceptions. In each of these 41 strains, the orthologous protein is more similar to BfUxs1 than to BfUxs2 and is present in the same genomic context as Bfuxs1. With the exception of the Parabacteroides species, which—like the Bacteroides—are abundant colonic organisms, other species of the order Bacteroidales, many of which are oral organisms, rarely encode this enzymatic function. Analysis of the prevalence of UDP-GlcA decarboxylases within the other two orders of Bacteroidetes demonstrates that these proteins are encoded consistently by Sphingobacteriales (9/9 genomes) but variably within the order Flavobacteroidales (14/26 genomes).
Fig. 4.
16S rRNA gene cladogram of Bacteroidales strains. Strains with a protein that matches the UDP-GlcA decarboxylase motif are highlighted in blue. Genus abbreviations are as follows: Pre., Prevotella; Azo., Azobacteroides; Pal., Paludibacter; Ali., Alistipes; Por., Porphyromonas; Par., Parabacteroides; and B., Bacteroides.
Phenotypic analysis of Bfuxs mutants.
As Uxs-type UDP-GlcA decarboxylases are predicted to be encoded by nearly all Bacteroides species analyzed, xylose may be present in a conserved glycan of the genus. Bacteroides fragilis synthesizes eight capsular polysaccharides and numerous glycoproteins. To determine whether synthesis of any of these molecules relies on the production of UDP-xylose, we analyzed glycan production by ΔBfuxs1, ΔBfuxs2, and ΔBfuxs1 ΔBfuxs2 mutants.
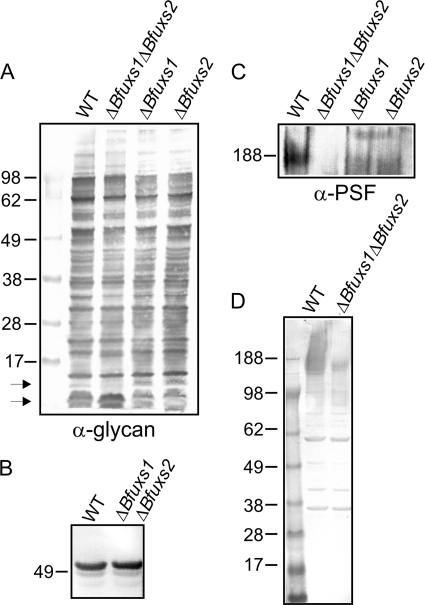
We previously showed that numerous proteins of different molecular sizes are glycosylated with the same glycans (16, 17), and these glycoproteins are present in all extracellular compartments (17) of B. fragilis. As shown by the Western immunoblot in Fig. 5A, an antiserum specific to the glycan portion of the B. fragilis glycoproteins binds numerous glycoproteins produced by all three mutants. There are a few low-molecular-weight molecules that appear to migrate differently in the double mutant; however, the deletion of Bfuxs1 and Bfuxs2 does not result in a general protein glycosylation defect or an alteration in the glycan that is added to proteins. This is also illustrated in Fig. 5B, where the size of a single glycoprotein, BF2494, is shown to be the same from both the wild-type and mutant strains. We previously demonstrated that alteration of the glycan results in a size reduction of every glycoprotein (17), a shift not evident in these mutants.
Fig. 5.
Western immunoblot analysis of ΔBfuxs1, ΔBfuxs2, and ΔBfuxs1 ΔBfuxs2 phenotypes. Whole-cell lysates of the indicated strains were probed with an antiserum specific to the glycan portion of the B. fragilis glycoproteins (A), an antiserum specific to the protein portion of glycoprotein BF2494 (B), an adsorbed antiserum specific to PSF (C), and an adsorbed antiserum specific to immunogenic molecules produced by the wild type (WT) but not by the ΔBfuxs1 ΔBfuxs2 mutant (D). Arrows in panel A indicate molecules with apparent differences in the double mutant.
Analysis of the eight capsular polysaccharides revealed that all but one (PSF) were synthesized, and the defect in synthesis of this molecule was observed only in the Bfuxs double mutant (Fig. 5C and data not shown). These data demonstrate that both Bfuxs1 and Bfuxs2 are expressed under the in vitro conditions used and that they are functional paralogs, as expected based on the analysis of their enzymatic activity (Fig. 3). To determine whether there is an additional xylose-containing glycan synthesized by B. fragilis that we not did detect with these antisera, we made an adsorbed serum. A polyclonal antiserum generated to B. fragilis NCTC 9343 whole cells was adsorbed with the ΔBfuxs1 ΔBfuxs2 mutant to remove antibodies to those molecules common to the wild type and mutant. Therefore, this adsorbed serum will contain antibodies to those molecules synthesized by the wild type but not by the ΔBfuxs1 ΔBfuxs2 mutant. The only apparent difference in the reactivity of this serum between the two strains is to that of a high-molecular-weight polysaccharide consistent with the migration of PSF (Fig. 5D).
DISCUSSION
Very few studies have shown xylose to be a component of bacterial glycans. As there are currently no lectins commercially available that recognized xylose, its detection relies on glycan purification and compositional analysis. Most bacterial glycans that have been compositionally or structurally analyzed are those of pathogens which—with a few exceptions—do not contain xylose. Identification of species that may synthesize xylose-containing glycans is further hindered by the incorrect annotation of the non-ArnA class of UDP-GlcA decarboxylases. These enzymes are frequently annotated as NAD-dependent epimerases/dehydratases.
Based on enzymatic analyses demonstrating that B. fragilis NCTC 9343 synthesizes two UDP-GlcA decarboxylases, we devised methods to detect each of the three types of UDP-GlcA decarboxylases encoded in the genomes of other bacteria. To do this, we first began by finding all proteins similar to BfUxs1 and then used a Perl regular expression to identify those SDR enzymes with a residue pattern characteristic of UDP-GlcA decarboxylases. Each of the three types of UDP-GlcA decarboxylases was then differentiated by comparing the BLAST E values of each to those of known ArnA and Uxs prototypes and by the presence or absence of domains indicative of formyl transferase activity. The list of bacteria encoding proteins similar to BfUxs1 and containing the UDP-GlcA decarboxylases pattern described in this study includes diverse species such as Helicobacter hepaticus, Burkholderia spp., various soil and aquatic bacteria, and cyanobacteria, to name a few. The likely success of identifying true UDP-GlcA decarboxylases using this pattern search is evidenced by our findings in Leptospira species. All six of the Leptospira strains with genomic sequences have Uxs types of UDP-GlcA decarboxylases, and the LPS of Leptospira species has previously been shown to contain xylose (23, 35).
Uxs-type UDP-GlcA decarboxylases are relatively well conserved in the genus Bacteroides, with nearly all species encoding at least one UDP-GlcA decarboxylase. Each of these Bacteroides proteins is very similar to BfUxs1 and less so to BfUxs2. The genetic regions containing the Bfuxs1 orthologs have similar architecture, with Bfuxs1 being the first gene in a two-gene operon in all Bacteroides species analyzed. The downstream gene encodes a predicted hybrid two-component histidine sensor kinase, which is 85 to 94% similar to those of different Bacteroides species. Only two other B. fragilis strains, 2_1_16 and 3_2_5, have two proteins that match the UDP-GlcA decarboxylase pattern. Analysis of the two proteins from both of these strains shows that one of each set is 99% identical to BfUxs1, and the other is 99% identical to BfUxs2. As in B. fragilis NCTC 9343, the Bfuxs2 orthologs of these strains are in capsular polysaccharide loci that are closely related at the DNA level to the B. fragilis NCTC 9343 PSF locus. Thus, it is quite likely that both Bacteroides sp. 2_1_16 and Bacteroides sp. 3_2_5 produce a capsular polysaccharide related or identical to PSF. Bfuxs2 orthologs were likely acquired by horizontal DNA transfer, whereas the Bfuxs1 orthologs are likely endogenous to the genus.
The question remains as to what molecule(s) utilizes the UDP-xylose synthesized by Bacteroides BfUxs1 orthologs or whether the UDP-xylose is further processed by an enzyme encoded by an unlinked gene. The only molecule clearly shown to be impacted by the loss of Bfuxs1 and Bfuxs2 is PSF. As the PSF region is heterogeneous, and most Bacteroides species contain only a Bfuxs1 ortholog, the UDP-xylose produced by these bacteria likely serves another purpose. Analysis of previous transcriptional profiling of in vitro-grown bacteria (12) reveals that Bfuxs1 is actively transcribed in vitro, further confirmed by its ability to provide UDP-xylose for PSF synthesis in the Bfuxs2 mutant (see above). Transcriptional profiling assays of the related species Bacteroides thetaiotaomicron VPI 5482 grown in culture or isolated from the cecum of mice (36) (GEO data set record GDS1849) demonstrate that the Bfuxs1 ortholog BT_1059 (NCBI accession no. AAO76166.1 [43]) is expressed at high levels under both conditions, making it unlikely that the predicted conserved xylose-containing molecule was not detected due to lack of expression. Another possibility is that this molecule is not immunogenic. The phenotypic analyses performed in this study depended on this molecule inducing an antibody response in rabbits for detection. Clearly, further studies are needed to assess the importance of UDP-xylose synthesis within Bacteroides species. Based on the expression profile of these genes in vivo and their widespread occurrence throughout the genus, xylose-containing glycans are likely an important component of the bacterial surface and contribute to their success in the intestinal environment.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tamara Doering for providing pDC22.6.
This work was funded by NIH/NIAID grant AI067711.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 29 July 2011.
REFERENCES
- 1. Adamczak R., Porollo A., Meller J. 2005. Combining prediction of secondary structure and solvent accessibility in proteins. Proteins 59:467–475 [DOI] [PubMed] [Google Scholar]
- 2. Allard S. T., et al. 2002. Toward a structural understanding of the dehydratase mechanism. Structure 10:81–92 [DOI] [PubMed] [Google Scholar]
- 3. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 4. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ankel H., Feingold D. S. 1966. Biosynthesis of UDP d-xylose. II. UDP d-glucuronate carboxy-lyase of Cryptococcus laurentii. Biochemistry 5:182–189 [DOI] [PubMed] [Google Scholar]
- 6. Bar-Peled M., Griffith C. L., Doering T. L. 2001. Functional cloning and characterization of a UDP-glucuronic acid decarboxylase: the pathogenic fungus Cryptococcus neoformans elucidates UDP-xylose synthesis. Proc. Natl. Acad. Sci. U. S. A. 98:12003–12008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Breazeale S. D., Ribeiro A. A., Raetz C. R. 2002. Oxidative decarboxylation of UDP-glucuronic acid in extracts of polymyxin-resistant Escherichia coli. Origin of lipid A species modified with 4-amino-4-deoxy-l-arabinose. J. Biol. Chem. 277:2886–2896 [DOI] [PubMed] [Google Scholar]
- 8. Burget E. G., Verma R., Molhoj M., Reiter W. D. 2003. The biosynthesis of l-arabinose in plants: molecular cloning and characterization of a Golgi-localized UDP-d-xylose 4-epimerase encoded by the MUR4 gene of Arabidopsis. Plant Cell 15:523–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burrows L. L., Charter D. F., Lam J. S. 1996. Molecular characterization of the Pseudomonas aeruginosa serotype O5 (PAO1) B-band lipopolysaccharide gene cluster. Mol. Microbiol. 22:481–495 [DOI] [PubMed] [Google Scholar]
- 10. Bystrova O. V., et al. 2006. Structures of the core oligosaccharide and O-units in the R- and SR-type lipopolysaccharides of reference strains of Pseudomonas aeruginosa O-serogroups. FEMS Immunol. Med. Microbiol. 46:85–99 [DOI] [PubMed] [Google Scholar]
- 11. Chatzidaki-Livanis M., Coyne M. J., Roche-Hakansson H., Comstock L. E. 2008. Expression of a uniquely regulated extracellular polysaccharide confers a large-capsule phenotype to Bacteroides fragilis. J. Bacteriol. 190:1020–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Coyne M. J., Chatzidaki-Livanis M., Paoletti L. C., Comstock L. E. 2008. Role of glycan synthesis in colonization of the mammalian gut by the bacterial symbiont Bacteroides fragilis. Proc. Natl. Acad. Sci. U. S. A. 105:13099–13104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coyne M. J., Reinap B., Lee M. M., Comstock L. E. 2005. Human symbionts use a host-like pathway for surface fucosylation. Science 307:1778–1781 [DOI] [PubMed] [Google Scholar]
- 14. Filling C., et al. 2002. Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J. Biol. Chem. 277:25677–25684 [DOI] [PubMed] [Google Scholar]
- 15. Fletcher C. M., Coyne M. J., Bentley D. L., Villa O. F., Comstock L. E. 2007. Phase-variable expression of a family of glycoproteins imparts a dynamic surface to a symbiont in its human intestinal ecosystem. Proc. Natl. Acad. Sci. U. S. A. 104:2413–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fletcher C. M., Coyne M. J., Comstock L. E. 2011. Theoretical and experimental characterization of the scope of protein O-glycosylation in Bacteroides fragilis. J. Biol. Chem. 286:3219–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fletcher C. M., Coyne M. J., Villa O. F., Chatzidaki-Livanis M., Comstock L. E. 2009. A general O-glycosylation system important to the physiology of a major human intestinal symbiont. Cell 137:321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gatzeva-Topalova P. Z., May A. P., Sousa M. C. 2004. Crystal structure of Escherichia coli ArnA (PmrI) decarboxylase domain. A key enzyme for lipid A modification with 4-amino-4-deoxy-l-arabinose and polymyxin resistance. Biochemistry 43:13370–13379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gu X., et al. 2010. Identification of a bifunctional UDP-4-keto-pentose/UDP-xylose synthase in the plant pathogenic bacterium Ralstonia solanacearum strain GMI1000, a distinct member of the 4,6-dehydratase and decarboxylase family. J. Biol. Chem. 285:9030–9040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gu X., Lee S. G., Bar-Peled M. 2011. Biosynthesis of UDP-xylose and UDP-arabinose in Sinorhizobium meliloti 1021: first characterization of a bacterial UDP-xylose synthase, and UDP-xylose 4-epimerase. Microbiology 157:260–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guyett P., Glushka J., Gu X., Bar-Peled M. 2009. Real-time NMR monitoring of intermediates and labile products of the bifunctional enzyme UDP-apiose/UDP-xylose synthase. Carbohydr. Res. 344:1072–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harper A. D., Bar-Peled M. 2002. Biosynthesis of UDP-xylose. Cloning and characterization of a novel Arabidopsis gene family, UXS, encoding soluble and putative membrane-bound UDP-glucuronic acid decarboxylase isoforms. Plant Physiol. 130:2188–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Isogai E., Isogai H., Kurebayashi Y., Ito N. 1986. Biological activities of leptospiral lipopolysaccharide. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 261:53–64 [DOI] [PubMed] [Google Scholar]
- 24. Kavanagh K. L., Jornvall H., Persson B., Oppermann U. 2008. The SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell. Mol. Life Sci. 65:3895–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Knirel Y. A. 1990. Polysaccharide antigens of Pseudomonas aeruginosa. Crit. Rev. Microbiol. 17:273–304 [DOI] [PubMed] [Google Scholar]
- 26. Kornfeld R., Kornfeld S. 1976. Comparative aspects of glycoprotein structure. Annu. Rev. Biochem. 45:217–237 [DOI] [PubMed] [Google Scholar]
- 27. Krinos C. M., et al. 2001. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature 414:555–558 [DOI] [PubMed] [Google Scholar]
- 28. Larkin M. A., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 29. Lesk A. M. 1995. NAD-binding domains of dehydrogenases. Curr. Opin. Struct. Biol. 5:775–783 [DOI] [PubMed] [Google Scholar]
- 30. Mazmanian S. K., Round J. L., Kasper D. L. 2008. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453:620–625 [DOI] [PubMed] [Google Scholar]
- 31. Moriarity J. L., et al. 2002. UDP-glucuronate decarboxylase, a key enzyme in proteoglycan synthesis: cloning, characterization, and localization. J. Biol. Chem. 277:16968–16975 [DOI] [PubMed] [Google Scholar]
- 32. Pantosti A., Tzianabos A. O., Onderdonk A. B., Kasper D. L. 1991. Immunochemical characterization of two surface polysaccharides of Bacteroides fragilis. Infect. Immun. 59:2075–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reiss E., Huppert M., Cherniak R. 1985. Characterization of protein and mannan polysaccharide antigens of yeasts, moulds, and actinomycetes. Curr. Top. Med. Mycol. 1:172–207 [DOI] [PubMed] [Google Scholar]
- 34. Rose P. W., et al. 2011. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 39:D392–D401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shimizu T., et al. 1987. Chemical properties of lipopolysaccharide-like substance (LLS) extracted from Leptospira interrogans serovar canicola strain Moulton. Microbiol. Immunol. 31:717–725 [DOI] [PubMed] [Google Scholar]
- 36. Sonnenburg J. L., et al. 2005. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307:1955–1959 [DOI] [PubMed] [Google Scholar]
- 37. Studier F. W., Moffatt B. A. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113–130 [DOI] [PubMed] [Google Scholar]
- 38. Thoden J. B., Henderson J. M., Fridovich-Keil J. L., Holden H. M. 2002. Structural analysis of the Y299C mutant of Escherichia coli UDP-galactose 4-epimerase. Teaching an old dog new tricks. J. Biol. Chem. 277:27528–27534 [DOI] [PubMed] [Google Scholar]
- 39. Thompson J. S., Malamy M. H. 1990. Sequencing the gene for an imipenem-cefoxitin-hydrolyzing enzyme (CfiA) from Bacteroides fragilis TAL2480 reveals strong similarity between CfiA and Bacillus cereus beta-lactamase II. J. Bacteriol. 172:2584–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tzianabos A. O., Onderdonk A. B., Rosner B., Cisneros R. L., Kasper D. L. 1993. Structural features of polysaccharides that induce intra-abdominal abscesses. Science 262:416–419 [DOI] [PubMed] [Google Scholar]
- 41. Reference deleted.
- 42. Williams G. J., Breazeale S. D., Raetz C. R., Naismith J. H. 2005. Structure and function of both domains of ArnA, a dual function decarboxylase and a formyltransferase, involved in 4-amino-4-deoxy-l-arabinose biosynthesis. J. Biol. Chem. 280:23000––23008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu J., et al. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074–2076 [DOI] [PubMed] [Google Scholar]
- 44. York W. S., O'Neill M. A. 2008. Biochemical control of xylan biosynthesis—which end is up? Curr. Opin. Plant Biol. 11:258–265 [DOI] [PubMed] [Google Scholar]
- 45. Zitomersky N., Coyne M. J., Comstock L. E. 2011. Longitudinal analysis of the prevalence, maintenance, and IgA response to species of the order Bacteroidales in the human gut. Infect. Immun. 79:2012–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.