Abstract
Clostridium difficile is a nosocomial pathogen involved in antibiotic-associated diarrhea. C. difficile expresses a cysteine protease, Cwp84, which has been shown to degrade some proteins of the extracellular matrix and play a role in the maturation of the precursor of the S-layer proteins. We sought to analyze the localization and the maturation process of this protease. Two identifiable forms of the protease were found to be associated in the bacteria: a form of ∼80 kDa and a cleaved one of 47 kDa, identified as the mature protease. They were found mainly in the bacterial cell surface fractions and weakly in the extracellular fraction. The 80-kDa protein was noncovalently associated with the S-layer proteins, while the 47-kDa form was found to be tightly associated with the underlying cell wall. Our data supported that the anchoring of the Cwp84 47-kDa form is presumably due to a reassociation of the secreted protein. Moreover, we showed that the complete maturation of the recombinant protein Cwp8430-803 is a sequential process beginning at the C-terminal end, followed by one or more cleavages at the N-terminal end. The processing sites of recombinant Cwp84 are likely to be residues Ser-92 and Lys-518. No proteolytic activity was detected with the mature recombinant protease Cwp8492-518 (47 kDa). In contrast, a fragment including the propeptide (Cwp8430-518) displayed proteolytic activity on azocasein and fibronectin. These results showed that Cwp84 is processed essentially at the bacterial cell surface and that its different forms may display different proteolytic activities.
INTRODUCTION
Clostridium difficile, a Gram-positive spore-forming anaerobic bacterium, is the leading bacterial cause of nosocomial intestinal infection worldwide and is responsible for illness ranging from mild diarrhea to life-threatening pseudomembranous colitis (8, 16). The clinical relevance of C. difficile has increased significantly during the past few years, particularly since 2003, when hypervirulent PCR-ribotype 027 strains have been involved in outbreaks and have been associated with severe disease in North America and Europe (39).
As in other pathogenic bacteria, C. difficile expresses several virulence factors. The two large clostridial toxins A (TcdA) and B (TcdB) are the most important and the best-characterized virulence factors of C. difficile (24, 26, 30, 37), leading to clinical manifestations by disorganizing the cell actin cytoskeleton. At present, the interactions between C. difficile and the host cell surface are not fully understood, even if several cell surface proteins, including adhesins and flagella, have been shown to mediate bacterial attachment (5, 17, 18, 35, 38). The S layer is a paracrystalline array on the outer cell surface that completely coats the bacterium; it is composed of two proteins, the high-molecular-weight S-layer protein (HMW-SLP) and the low-molecular-weight S-layer protein (LMW-SLP), derived from a common precursor, SlpA (6). These two proteins are the major surface proteins in C. difficile and play a role in the intestinal colonization and in the inflammatory process (1, 5). However, the colonization step needs to be further characterized in order to better understand the whole pathogenesis process of C. difficile.
Proteolytic and hydrolytic enzymes also play a role in the pathogenesis of several diseases (27, 32). Cwp84 is a surface-associated cysteine protease, which displays a proteolytic activity toward several proteins of the extracellular matrix (ECM) such as fibronectin, laminin, and vitronectin (20). As a member of papain-like proteins, Cwp84 possesses the catalytic triad cysteine, histidine, and asparagine (34). It has been recently shown to play a key role in the maturation of the precursor SlpA into two components of the S-layer, the HMW-SLP and the LMW-SLP (10, 23). The translational product of cwp84 gene is a preproenzyme (Cwp841-803, 803 amino acid residues, 84 kDa) containing a hydrophobic signal peptide of 32 amino acid residues, an N-terminal domain of 338 amino acid residues (amino acids 33 to 370) containing the catalytic triad, and a C-terminal domain with three Pfam 04122 motifs, presumed to serve as an anchoring domain to the underlying cell wall. Previously, we have shown that Cwp84 is matured presumably by an autoproteolytic cleavage (20). We also showed that cwp84 is highly conserved in C. difficile strains of different toxinotypes or serotypes (34). Furthermore, this protease induces an immune response during the course of the infection, as shown by the presence of specific antibodies in patients with C. difficile infection (CDI) (29, 41). These observations suggested that Cwp84 could play a role in C. difficile pathophysiology.
The aim of our study was to investigate the localization of Cwp84 in the bacterium and its maturation process, in relation to its putative role in C. difficile virulence.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Clostridium difficile strain 630 was cultured at 37°C in an anaerobic chamber (Jacomex, France) in tryptone-yeast extract infusion broth (pH 7.4), with (TYG) or without (TY) glucose (Difco Laboratories). Bacillus subtilis strain 168 was grown aerobically in brain heart infusion (BHI) broth or agar (Difco Laboratories). The Escherichia coli recombinant strains BL21/pET-28a(+)Ωcwp8430-803 (20), BL21/pET28a(+)Ωcwp8492-518, BL21/pET28a(+)Ωcwp8430-518, and BL21/pET28a(+)Ωcwp84C116A were grown in LB agar or in broth (Difco Laboratories) supplemented with 50 μg of kanamycin/ml to maintain the pET plasmid at 37°C. The cwp84 mutant strain, the 630Δerm cwp84347a::erm strain (a generous gift from Neil Fairweather, Imperial College of London, England), and the 630Δerm fbpA640a::erm strain were cultured in BHI supplemented with 5 μg of erythromycin/ml.
Animal model.
Eight germfree mice (purchased from the CNRS, Orléans, France) were inoculated orally with 5 × 105 vegetative cells of C. difficile strain 630 and were sacrificed at 40 h postchallenge. The cecal contents were collected, and protease inhibitor cocktail (Sigma) was immediately added. Bacteria were pelleted, and bacterial proteins were extracted from different fractions.
C. difficile protein extraction.
C. difficile proteins were extracted from bacteria grown in different media or from bacteria collected from mouse ceca at 40 h postchallenge.
S-layer proteins.
The S-layer proteins (SLPs) were prepared by the low-pH glycine extraction method as previously described (6).
Cell surface-associated proteins.
Surface-associated proteins were extracted using the method of Wexler et al. (40) with some modifications. Briefly, harvested and washed cells were resuspended in 1 mM Tris (pH 6.8) with 60 μg of mutanolysin/ml, followed by incubation at 37°C for 30 min. After mixing with a vortex (30 s) at room temperature, the cells were harvested by centrifugation at 20,000 × g for 20 min, and the supernatant containing the cell wall-associated proteins was retained. In some experiments, trans-epoxysuccinyl-l-leucylamido (4-guanidino) butane (E64; Sigma), a specific cysteine protease inhibitor, was added to the Tris buffer at a final concentration of 10 μM.
Secreted proteins.
To collect the extracellular proteins from broth culture, the bacteria were removed by centrifugation (4,500 × g, 20 min, 4°C), and the supernatant was filtered by passage through a 0.45-μm-pore-size sterile filter. The proteins in the supernatant were precipitated by adding 10% trichloroacetic acid (Invitrogen) with gentle shaking at 4°C for 15 h. The proteins were then pelleted by centrifugation (5,000 × g, 60 min, 4°C) and suspended in 8 M urea.
The protein concentration was determined by the Bradford method using bovine albumin (1 mg/ml) as a standard. Protein extraction for each growth condition was performed in triplicate.
SDS-PAGE and immunoblotting.
The proteins from different extracts were separated by SDS-PAGE on a 12% polyacrylamide separating gel and transferred onto a polyvinylidene difluoride (PVDF) membrane (Amersham Biosciences) for immunoblotting with specific anti-Cwp84 antibodies (used at a 1:5,000 dilution in blocking buffer). Primary antibodies were detected using alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (1:20,000; Sigma) and BCIP/NBT substrate (5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium; Sigma). In experiments detecting the His tag of the recombinant protein Cwp8430-803, anti-His tag antibodies were used as a 1:10,000 dilution in blocking buffer, and detection was performed as indicated above.
Recombinant Cwp84 proteins production and purification.
Four recombinant proteins were produced. They correspond, respectively, to the proprotein form Cwp8430-803, the cleaved mature form Cwp8492-518, the mature form, including the propeptide Cwp8430-518, and the Cwp8430-803 containing the substitution C116A (Cwp84C116A). The fragments corresponding to the processed cleaved form Cwp8492-518 and the partially cleaved form Cwp8430-518 (incorporating the N-terminal propeptide of the protease) were cloned into pET28a(+) (Novagen) and expressed in E. coli BL21 Star(DE3) (Invitrogen) under the control of IPTG (isopropyl-β-d-thiogalactopyranoside). The primers used to amplify by PCR the fragment from nucleotides 274 to 1554 were Cwp8492(F) (5′-CTAGCTAGCTCAAGTGTAGCATACAACCC-3′) and Cwp84518(R) (5′-CCGGAATTCTTAAACTGCTGTTTCATATC-3′), incorporating the NheI and EcoRI restriction sites, respectively (underlined). The Cwp84518(R) primer contained a stop codon (indicated in boldface). The primers used to amplify by PCR the fragment from nucleotides 88 to 1554 were Cwp8430(F) (5′-TGAGCTAGCGCAGAAAACCATAAAACTCTAGATG-3′), incorporating also the NheI restriction site, and Cwp84518(R). The resulting NheI/EcoRI-digested PCR products were each inserted into the linearized pET28a(+) to create the N-terminal His-tag proteins Cwp8492-518 and Cwp8430-518. The resulting constructions were transformed in E. coli BL21 according to the manufacturer's instructions. The nucleotide sequences of the junctions between the vector and the insert were confirmed.
To generate site-directed mutagenesis of Cwp84, we used the oligonucleotides 5′-CAAGGAAGTCTTAATACAGCATGGTCTTTTTCAGGTATG-3′ and 5′-CATACCTGAAAAAGACCATGCTGTATTAAGACTTCCTTG-3′. Briefly, a PCR of 18 cycles was performed using pET28a(+)Ωcwp8430-803 as a template and Phusion polymerase (Finnzymes). The parental plasmid was digested by DpnI, and the PCR product was transformed into E. coli BL21. The active residue Cys-116 was then substituted with an Ala residue.
Recombinant Cwp8430-803 (previously rCwp84) was purified as already described (20); in one experiment, Cwp8430-803 purification was modified by adding E64 in the purification buffers at a final concentration of 50 μM to inhibit, at least partially, the putative automaturation of the protease. The recombinant Cwp8430-518, Cwp8492-518, and Cwp84C116A were purified using the same protocol, with some modifications. Briefly, clones were grown in 1 liter of LB supplemented with 50 μg of kanamycin/ml at 37°C to an optical density at 600 nm of 0.6 to 0.8. Protein expression was then induced by the addition of IPTG at a final concentration of 1 mM at 37°C, and the cells were harvested at 4 h postinduction for Cwp8430-518 and Cwp84C116A and at 6 h postinduction for Cwp8492-518. The recombinant forms of the protease were then purified by affinity chromatography using the BD Talon cobalt affinity resin (Clontech Laboratories, Inc.) in accordance with the standard protocol provided by the manufacturer. Elution was performed with a gradient of imidazole (10 mM to 1 M), and elution fractions were dialyzed overnight against phosphate-buffered saline (PBS; pH 7.4). Elution fractions were then divided into aliquots and frozen at −80°C for storage until use. These fractions were analyzed using SDS–12% PAGE.
Proteolytic activity of recombinant Cwp84s.
The proteolytic activities of Cwp8430-803, Cwp8430-518, Cwp8492-518, and Cwp84C116A were determined by using azocasein (Sigma) as a substrate, as previously described (20). The proteolytic activity of active forms of the protease was further assayed on fibronectin (Sigma) at an enzyme/substrate ratio of 1:1 or 1:10 at 37°C for 16 h in 25 mM Tris buffer (pH 7.5). Experiments were performed with 2 mM dithiothreitol (DTT), a reducing agent known as cysteine protease activator, and the samples were analyzed using SDS–8% PAGE. Moreover, the proteolytic activity of Cwp8430-803 on Cwp84C116A was analyzed at an enzyme/substrate ratio of 1:100 at 37°C for 0, 2, 4, 6, and 24 h in 200 μl of PBS with 2 mM DTT; Cwp84C116A was incubated alone under the same conditions.
Reassociation of recombinant Cwp84 with the C. difficile and B subtilis cell surface.
The cwp84 mutant and B. subtilis were grown to stationary growth phase in TYG and BHI, respectively. In a first set of experiments, the cwp84 mutant was washed once in PBS and then incubated 1 h at 37°C in anaerobiosis with 20 μg of Cwp8492-518. Bacteria were washed five times with PBS–0.05% Tween in order to eliminate the recombinant protease that had not been associated tightly with the bacterial surface. As a negative control, C. difficile 630fbpA640a::erm and purified recombinant FbpA protein were incubated under the same conditions. To analyze whether the reassociation was specific, we performed a second set of experiments in which we varied the number of bacteria while keeping the concentration of protein constant, as previously described (22). Thus, different dilutions of bacteria were incubated in 100 μl of PBS with 2 μg of Cwp8492-518 for 1 h at 37°C. The samples were centrifuged at 10,000 × g for 5 min, and the pellet was resuspended in 100 μl of PBS. The potential reassociation of Cwp8492-518 with C. difficile and B. subtilis cell surface was tested in the same conditions, except that B. subtilis was grown in an aerobic environment. Analysis was performed by dot blotting with specific anti-Cwp84 antibodies (1:5,000 dilution) to detect Cwp84 that had reassociated with bacterial cells.
Immunofluorescence microscopy.
The localization of Cwp84 was examined by immunofluorescence microscopy. The strains 630 and 630Δerm cwp84347a::erm were cultured in TY for 15 h at 37°C. The cells were washed with PBS, 108 CFU ml−1 was fixed on a slide using ethanol, and 5% bovine serum albumin in PBS was used as a blocking agent. Incubation with a first antibody (specific anti-Cwp84, 1:200) was performed for 1 h at room temperature in blocking buffer. The samples were then washed with PBS and incubated with 1:500 dilution of fluorescein-conjugated Alexa Fluor 488 goat anti-rabbit IgG as the secondary antibody (Molecular Probes/Invitrogen) for 1 h at room temperature. The cells were washed again and stained with the nuclear stain DAPI (4′,6′-diamidino-2-phenylindole; Molecular Probes). Control experiments were carried out similarly. Rabbit anti-FliC (1:200 dilutions) was used as the control for another surface protein, and bacteria were incubated without primary or secondary antibody as negative controls. Fixed samples were analyzed by epifluorescence using a Nikon Eclipse 80i microscope, and the images were analyzed using ImageJ software (31).
MALDI/TOF-MS, LC-MS/MS, and N-terminal protein sequencing.
Purified Cwp8430–803 was incubated with 2 mM DTT for 4 h, in order to collect the mature cleaved form of the protease (mCwp84). This form was subjected to matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF-MS) for the determination of the molecular weight using a Perceptive Biosystems/Applied Biosystems/ABI Voyager DE-STR MALDI-TOF mass spectrometer. Automated Edman sequence analysis of mCwp84 was performed on the proteins transferred onto PVDF membrane (Amersham/GE Healthcare). The blots were stained in 0.1% Coomassie blue and 1% acetic acid and then destained with 50% methanol. Dry membranes were stored at −20°C. N-terminal sequencing was carried out at the Institute of Biochemistry and Molecular and Cellular Biophysics (IBBMC-UMR CNRS 8619; Team Chemistry of Proteins, University Paris-Sud 11) on an Applied Biosystems Procise Sequenator using established protocols. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses were performed using a 1200 Series Nano-LC system (Agilent Technologies), coupled with an Agilent 6330 XCT ion trap equipped with a nanospray Chip Cube ion source, at the proteomic platform of IFR 141 (University Paris-Sud 11).
RESULTS
Localization of Cwp84. (i) In vitro experiments.
In a previous work, we showed by immunoblotting that Cwp84 was surface associated and was recovered mainly in the glycine extract associated with the S layer (20). To confirm that the protease was located on the cell surface, immunofluorescence analysis was performed in the present study on C. difficile strain 630 after growth in TY medium. As shown in Fig. 1A, Cwp84 was localized on the cell surface. In the same conditions, flagella were observed at the bacterial surface of C. difficile (36; data not shown). As expected, the 630Δerm cwp84347a::erm strain (Fig. 1A) and the controls omitting the first or second antibodies were not labeled (data not shown), indicating that the fluorescent signal was Cwp84 specific.
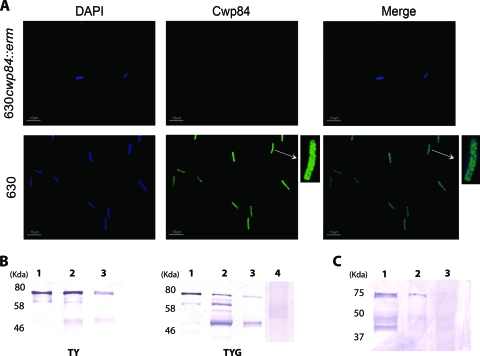
Fig. 1.
Localization of Cwp84 in C. difficile. (A) Immunofluorescence microscopy of Cwp84 (green) by Alexa Fluor 488 in whole bacteria, comparing the 630 and the 630Δerm cwp84347a::erm strains; the nucleus was stained by DAPI (blue). (B and C) Immunoblot analysis using Cwp84 antibodies was carried out on protein extracts collected from bacteria grown in TY and TYG media (B) or from bacteria isolated from monoxenic C. difficile-associated mice (C). Lane 1, glycine extract; lane 2, cell surface-associated proteins; lane 3, extracellular proteins; lane 4, cell surface-associated proteins from the cwp84 mutant strain. The various fractions were subjected to SDS–12% PAGE. The positions of molecular mass marker proteins (kDa) are indicated (Biolabs for the in vitro experiments and Bio-Rad for the in vivo experiment).
To further analyze the exact localization of Cwp84, different fractions of C. difficile proteins, extracted after growth in TY or TYG media, were studied by immunoblot analysis. In the glycine extract, regardless of growth medium, Cwp84 was recovered mainly as a protein of ∼80 kDa, as previously observed (Fig. 1B, lane 1) (20). The glycine extraction method exploits the ability of low pH to remove the S layer and associated proteins from the bacterial surface without producing changes in the SLP; proteins that are extracted by this method should be noncovalently associated with the underlying cell wall.
In the cell surface-associated protein extract (Fig. 1B, lane 2), anti-Cwp84 antibodies mainly revealed three different forms of the protease: the 80-kDa form, an intermediate form of ∼60 kDa, and a mature form of ∼50 kDa as determined by electrophoretic migration. However, some differences were observed in the relative amounts of these three protease forms according to the growth medium. For example, the mature 50-kDa Cwp84 was recovered especially in the TYG medium. The method used to collect these extracts combines the use of mutanolysin to gently disorganize the cell wall with an abrasive method and consequently promotes the release of tightly cell wall-associated proteins. To ensure that these cleaved (intermediate and mature) forms of Cwp84 were not the result of a proteolytic process of the 80-kDa protein during the extraction steps, experiments were performed with E64 in the extraction solutions. No difference was observed (data not shown), suggesting that these different forms are truly associated with the bacteria. Moreover, no reaction was observed with the surface-associated proteins from the cwp84 mutant, indicating that the detection of lower-molecular-weight proteins was not due to a cross-reaction. These results suggest that Cwp84 could exist at the C. difficile surface under different forms anchored to the bacterial cell surface in different ways. In the extracellular fraction, we observed both the 80-kDa form and the mature 50 kDa-form (Fig. 1B, lanes 3).
(ii) In vivo experiments.
After C. difficile challenge in mice, bacteria expressed Cwp84 at the cell surface mainly associated with the S-layer proteins in the glycine extract (Fig. 1C, lanes 1 and 2). The cysteine protease was detected as the 80-kDa form, processed intermediates forms, and the mature cleaved 50-kDa form. Cwp84 was not detected in the extracellular fraction.
Proteolytic processing of recombinant Cwp8430-803.
In our first Cwp84 study, we observed that Cwp8430-803 was processed into a cleaved form, called mCwp84 (20). The mCwp84 migrates on SDS-PAGE at ∼50 kDa similarly to the cleaved mature form found mainly in the cell surface extract. So we hypothesized that the cleavage was due to an autoproteolytic process and we observed that this process began during the metal affinity purification and the dialysis steps.
Therefore, to be able to observe the intermediate forms, we slowed down the maturation process, and we then performed purification of Cwp8430-803 under conditions of partial proteolytic inhibition. SDS-PAGE analysis showed that at 0, 2, and 4 h postincubation with 2 mM DTT intermediate forms of Cwp8430-803 were detected, and the majority of Cwp8430-803 had been converted in the mature form (mCwp84) at 6 h (Fig. 2A, lane 5). Whereas for Cwp8430-803 purified without the cysteine protease inhibitor the protease was quite fully matured after the dialysis (0 h postincubation), intermediate forms were detected only at very low levels (Fig. 2A, lane 6).
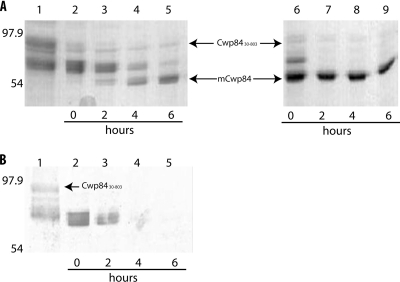
Fig. 2.
Proteolytic process of Cwp8430-803. The recombinant protease Cwp8430-803 was purified in the presence or absence of 50 μM E64 and then incubated under reducing conditions with 2 mM DTT. (A) SDS-PAGE analysis. Purification of Cwp8430-803 in the presence of the cysteine protease inhibitor (lanes 2 to 5) resulted in the detection of intermediate forms at 0, 2, and 4 h and the mature form at 6 h, whereas the purification in the absence of E64 (lanes 6 to 9) resulted in the detection of the mature form from 0 h. Lane 1 corresponds to the protease before dialysis. (B) Immunoblot analysis with anti-His tag antibody. In the presence of E64, the histidine tag fused to the N-terminal end was detected in the total and some of the processed intermediate forms of Cwp84; however, the mature form was not detected, suggesting that proteolytic process of Cwp84 begins at the C-terminal end, and follows at the N-terminal end. The Bio-Rad molecular mass standard (in kilodaltons) was used.
Immunoblot analysis of protease fractions purified with E64 revealed that anti-His tag antibodies reacted with distinct protein bands that were considered to correspond to the whole recombinant Cwp8430-803 and some of its processed intermediate forms (Fig. 2B). The histidine tag at the N-terminal end was still present in Cwp84 cleaved forms after 2 h of incubation with 2 mM DTT (Fig. 2B, lane 3), suggesting that the maturation process of recombinant Cwp8430-803 begins at the C-terminal end. Since no His tag was detected after 4 h (Fig. 2B, lane 4), the maturation of Cwp8430-803 seems to proceed via cleavage at the N-terminal end. These results suggested that the maturation process of Cwp8430-803 occurred via sequential cleavage beginning at the C-terminal end, followed by one or more cleavages at the N-terminal end.
To determine the cleavage sites, the band corresponding to the mature form mCwp84 (∼50 kDa as determined by SDS-PAGE) was analyzed using MALDI-TOF mass spectrometry, MS/MS, and N-terminal Edman sequencing. The molecular mass obtained by MALDI-TOF was 47,136 Da. N-terminal sequencing revealed that a major cleavage site at the N-terminal part of the Cwp8430-803 occurred between the Lys-91 and Ser-92, with an N-terminal amino acid sequence being SSVAYN (Fig. 3). This cleavage seemed to be heterogeneous, with a minor site occurring between the Ser-93 and the Val-94 residues. By MS/MS, we identified a cleavage at the C-terminal end at Lys-491, and we confirmed the cleavage at the N-terminal end at Ser-92. Therefore, the predicted molecular mass of this cleaved form (Ser-92–Lys-491) was 44,500 Da calculated using ExPaSy software, which differed from the predicted molecular mass of 47,136 Da by more than 2,600 Da. This discrepancy indicated that the cleavage at the C-terminal end might take place downstream from this site, most likely between Lys-518 and Val-519 (Fig. 3), and that it has been hidden by trypsin digestion performed for MALDI analysis.
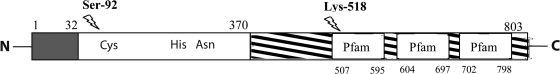
Fig. 3.
Schematic representation of Cwp84 proteolytic processing. cwp84 encodes a protein of 803 amino acid residues. The peptide signal ( ), the catalytic domain (□), and the domain corresponding to the anchoring domain (
), the catalytic domain (□), and the domain corresponding to the anchoring domain ( ) are indicated. The scheme shows the three Pfam 04122 motifs and the catalytic triad (Cys-116, His-262, and Asn-287). The amino acids involved in the proteolytic cleavage are indicated.
) are indicated. The scheme shows the three Pfam 04122 motifs and the catalytic triad (Cys-116, His-262, and Asn-287). The amino acids involved in the proteolytic cleavage are indicated.
To assess whether maturation of Cwp84 was an autoproteolytic process, we constructed an active-site substitution, Cwp84C116A. As expected, Cwp84C116A displayed no proteolytic activity against azocasein. Surprisingly, after purification we did not observe, as expected, only the total protease form but instead four bands corresponding to the total and intermediate forms, suggesting that these forms could ultimately be due to degradation during the purification steps. Incubation of Cwp84C116A with 2 mM DTT for 24 h did not lead to additional cleavage (Fig. 4A), suggesting that the mutated protease was not able to complete its own cleavage process. In contrast, incubation with Cwp8430-803 led to the conversion to intermediate and mature forms at 2, 4, 6, and 24 h (Fig. 4B), similar to the autocatalytic process of Cwp8430-803. This, together with the previous results, suggested that the complete maturation process of Cwp84 proceeds, at least partially, by an autocatalytic mechanism involving a transmaturation.
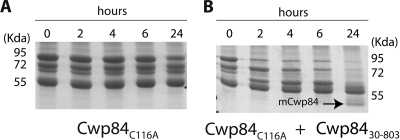
Fig. 4.
Purification of Cwp84C116A and transmaturation by Cwp8430-803. The mutated protease Cwp84C116A was incubated at 37°C alone or with Cwp8430-803 under reducing conditions for 0, 2, 4, 6, and 24 h. Cwp84C116A alone with 2 mM DTT does not show the maturation process (A), whereas incubation of Cwp84C116A with Cwp8430-803 at an enzyme/substrate ratio of 1:100 resulted in the increase of intermediate forms at 4 and 6 h and detection of mature form at 24 h (B). The molecular mass standard used was purchased from Fermentas.
Purification and characterization of the proteolytic activities of Cwp8430-803, Cwp8492-518, and Cwp8430-518.
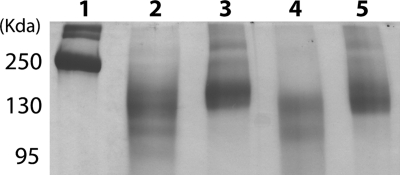
In our previous study, we could not be certain which protein form was responsible for the proteolytic activity (20). Therefore, we cloned the cwp84 gene region corresponding to the mature form (Cwp8492-518), deduced from previous analysis, into a pET expression vector. The purified recombinant Cwp8492-518 was tested for proteolytic activity on azocasein and fibronectin. However, Cwp8492-518 did not show any proteolytic activity on these substrates, suggesting that the mature form was expressed as an inactive enzyme. Thus, a larger DNA fragment corresponding to the mature form, including the propeptide of Cwp84, was cloned into the pET expression vector. This form, named Cwp8430-518, was expressed and purified by using an affinity column. However, after SDS-PAGE analysis, two forms of the protease, Cwp8430-518 and another form with a lower molecular weight, corresponding presumably to mature Cwp8492-518, were detected (data not shown). This purified fraction displayed proteolytic activity on azocasein. As shown in Fig. 5, the recombinant proteases Cwp8430-518 (lanes 4 and 5) and Cwp8430-803 (lanes 2 and 3) were able to degrade fibronectin.
Fig. 5.
Proteolytic activities of Cwp8430-803 and Cwp8430-518 on fibronectin. The recombinant proteases were incubated for 16 h with fibronectin at enzyme/substrate ratios of 1:1 and 1:10. All experiments were performed under reducing conditions (2 mM DTT). Lane 1, fibronectin control (5 μg); lane 2, Cwp8430-803 and fibronectin (1:1); lane 3, Cwp8430-803 and fibronectin (1:10); lane 4, Cwp8430-518 and fibronectin (1:1); lane 5, Cwp8430-518 and fibronectin (1:10). The molecular mass standard used was from Fermentas.
Reassociation of Cwp84 at the C. difficile surface.
The existence of the mature form of the protease both as a surface-associated protein and as an extracellular protein suggests a putative reassociation of this form to the bacterial surface. As shown in Fig. 6A, incubation of 20 μg of recombinant Cwp8492-518 with the cwp84 mutant bacteria resulted in the detection of labeled bacterial cells after immunodetection with specific anti-Cwp84 antibodies. No labeling was observed after incubation in the same conditions of another surface protein FbpA with the corresponding knockout strain 630Δerm fbpA640a::erm. This suggests that the protease is able to reassociate with the C. difficile surface.
Fig. 6.
Reassociation of Cwp84 protein. Bacteria were loaded onto a nitrocellulose membrane and treated with specific anti-Cwp84 or anti-Fbp68 antibodies. (A) C. difficile strains 630Δerm cwp84347a::erm and 630Δerm fbpA640a::erm were incubated with 20 μg of Cwp8492-518 or 20 μg of FbpA (lane 3). Lane 1, positive control Cwp8492-518; lane 2, negative control 630Δerm cwp84347a::erm and 630Δerm fbpA640a::erm not incubated with the recombinant proteins. (B) Different numbers of C. difficile 630Δerm cwp84347a::erm and B. subtilis bacteria. Samples—5 × 108 (lane 1), 108 (lane 2), 107 (lane 3), 106 (lane 4), 105 (lane 5), or 104 (lane 6)—were incubated with 2 μg of Cwp8492-518. The samples were centrifuged, and the reassociation of recombinant Cwp84 was analyzed by dot blotting.
To address whether Cwp84 is able to reassociate specifically with the bacterial surface of C. difficile, or whether it may associate with any bacterial surface, different concentrations of bacteria were incubated with 2 μg of Cwp8492-518. As shown in Fig. 6B, an efficient association of recombinant protein with the surface of C. difficile was detected in a concentration-dependent manner; the level of reassociation of recombinant protein decreased in parallel with the decrease of the bacterial number. When B. subtilis was incubated with Cwp8492-518 under identical conditions, a labeling in a nonuniform manner was observed (Fig. 6B), suggesting that the association at the surface of B. subtilis is nonspecific. These results argued for the presence of a binding site for Cwp8492-518 at the bacterial surface of C. difficile.
DISCUSSION
Cysteine proteases represent a large group of proteolytic enzymes that have been shown to contribute to a large variety of complex functions. Bacterial proteases play a critical role in the pathogenesis of several human and animal diseases by direct damage to host tissues and by activating zymogens and often also participate in the turnover of bacterial proteins (25, 33). Cwp84 is the first characterized protease of C. difficile; this cysteine protease plays a key role in the maturation of S-layer proteins (10, 23). Cwp84 has also been shown to possess in vitro degrading activity against human ECM proteins (20), but the physiological relevance of this property is not yet known. In addition, it was hypothesized that Cwp84 could undergo autocatalytic maturation, as previously observed for numerous papain family cysteine proteases (32). The aim of the present study was to examine the localization of Cwp84 in the bacterial cell and its maturation process.
We have shown that the maturation process of recombinant Cwp8430-803 proceeds in sequential cleavage reactions, beginning at the C-terminal domain. The cleavage at the N-terminal domain is heterogeneous, with the major cleavage site identified at Lys-91, in accordance with the recent results from de la Riva et al. (11). It is noteworthy that the consensus cleavage site of SlpA into the mature S-layer proteins is also located after a serine residue (13, 14), but no other consensus motif has been recovered in Cwp84. The C-terminal cleavage site has been presumably identified at Lys-518. The molecular mass of the mature recombinant Cwp84 has been determined to be 47 kDa. Some members of the M4 family of metalloproteases require processing at the N-terminal end, and at the C-terminal end to become fully active (12). Since we observed a maturation process on purified recombinant protease, it is likely that these cleavages occurred via an autocatalytic mechanism. However, purification of the inactive mutated Cwp84C116A showed that some of the Cwp84 cleavages, leading to intermediary forms of the protease, could be due to a degradation process. Nevertheless, conclusive proof that ultimate conversion to the 47-kDa form proceeds via an autocatalytic mechanism was obtained from our processing experiments, where the different forms of the mutated Cwp84C116A protease were cleaved by active Cwp8430-803, leading to accumulation of the mature form (Fig. 4). Autocatalytic maturation of cysteine proteases could be due to cis or trans processing (7). It is therefore possible that the first cleavages could be due to the activity of an exogenous protease, as recently suggested by de la Riva et al. for the removal of the propeptide leading to the 77-kDa form associated with the S layer; the following cleavages could then result from an autoproteolytic mechanism. Maturation of protease involving both exogenous and autocatalytic processing has already been described (28).
In the mouse model, we found that Cwp84 is associated with the bacterial surface in different forms, suggesting that the proteolytic maturation of Cwp84 occurs in the gut. The absence of Cwp84 in the extracellular fraction could be due to the dilution of the cecal contents.
In the present study, we showed that Cwp84 was expressed as an extracellular protein and mostly localized at the bacterial surface. Surface localization is not usual for bacterial cysteine protease but has been already reported for the SpeB protease of Streptococcus pyogenes (19). We confirmed here that Cwp84 exists at the bacterial cell surface associated with the S-layer proteins mainly as an ∼80-kDa protein, and this is consistent with the recent results from de la Riva et al. (11). Since we had not precisely determined the molecular weight of this form in the bacterial cell, it could correspond either to the proprotein (amino acids 33 to 803) or to a form without the propeptide (amino acids 92 to 803). The association of the protease with the S-layer proteins is not unexpected, since the latter are substrates for Cwp84 (23). Moreover, the C-terminal part of Cwp84 contains three PF04122 Pfam “cell wall binding” motifs, which could mediate anchoring to the cell wall (6, 21). These motifs are also found in three copies in the HMW-SLP (15), suggesting that these two proteins may share a binding mechanism to the C. difficile surface. This mechanism is not yet characterized but is likely to involve a noncovalent attachment mode.
Furthermore, we also recovered the protease as a lower-molecular-weight form, in both the extracellular and the cell wall-associated protein fractions, especially when C. difficile was grown in the TYG medium. This form shared the same molecular weight as the cleaved one, which resulted from maturation of the recombinant Cwp8430-803, and we assume that it is the same mature form of the protease. The mature protease could be secreted into the extracellular fraction after cleavage at the C-terminal part. However, it is surprising to recover this mature form tightly associated with the underlying cell wall. It should be noted that this last result is consistent with a previous proteomic study, where Cwp84 was recovered in the extracts collected after enzymatic degradation of the peptidoglycan (42). One hypothesis is that the secreted cleaved protease could be reassociated with the bacterial surface, since it has been shown for some surface proteins in other bacteria (2, 9), and previously proposed as an anchoring mechanism of GroEL at the C. difficile surface (18). To further investigate this hypothesis, the ability of recombinant Cwp8492-518 to reassociate at the surface of C. difficile was explored. This recombinant protein was able to specifically reassociate at the surface of the 630Δerm cwp84347a::erm mutant; this specific reassociation was not observed with B. subtilis. The recombinant protein Cwp8492-518 does not possess the three complete PF04122 Pfam motifs but only 11 amino acids of the first motif. Therefore, the Pfam motifs may not be necessary for the reassociation of the mature protease. Neither GW nor LysM conserved anchoring motifs have been recovered (4). Thus, the Cwp84 anchoring domain to the underlying cell surface is still unknown but could be mediated by interactions either with the peptidoglycan or lipoteichoic acids.
Cwp84 is located at the bacterial surface and also released extracellularly and may contribute to the pathogenicity of C. difficile by several mechanisms. Cwp84 participates in the turnover of S-layer proteins (23). It is therefore possible that the different forms of Cwp84 may promote either the cleavage of SlpA or additional functions in the development of the infection. We did not investigate in the present study which form of the protease could be responsible for the maturation of the S-layer protein, but it is likely to be the form Cwp8430-803 associated with the S layer. In contrast, degrading activity against human tissues could be expressed by the mature extracellular Cwp84. Indeed, in a previous study, we showed that Cwp84 has a degrading activity on some ECM proteins (20), but we could not assign this role to one defined form of the protease. Here, purification of the cleaved forms helped us to answer this question. We observed that the purified recombinant mature Cwp8492-518 had no proteolytic activity on tested substrates, whereas the purified Cwp8430-518 demonstrated catalytic activity against azocasein and fibronectin. The inactivity of mature Cwp8492-518 may be due to a defective folding of the active site of this recombinant protein. Other recombinant cysteine proteases have been purified directly under the mature form and have also been shown to be inactive, suggesting that the propeptide may play a role in the maturation and correct folding of the catalytically mature protein (3). In fact, the recombinant Cwp8430-518 was also cleaved during the purification in the mature protease; therefore, we could not exclude that the proteolytically active form is the Cwp8492-518 and that presence of propeptide is essential for correct folding before maturation of the active mature protease.
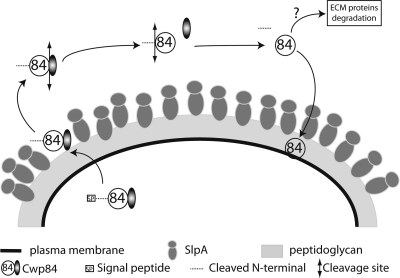
In which subcellular compartment Cwp84 maturation occurs remains to be determined. However, according to our results, we could hypothesize that, after removal of the peptide signal from the preproprotein and translocation across the cytoplasmic membrane, the 84-kDa proprotease or the 77-kDa form (without propeptide) is associated with the S layer. Cleavage at the C-terminal domain leads ultimately to the secretion of mature Cwp84, where it can cleave host proteins and also be reassociated with the bacterial surface (Fig. 7). These different phases may require stringent regulation by environmental conditions.
Fig. 7.
Cwp84 localization and maturation process model. The Cwp84 native form is composed of a signal peptide, an N-terminal propeptide (dashed line), an N-terminal part (empty circle), and a C-terminal part (partially filled circle). The native protein is exported to the S layer probably via the SecA secretion system, where the signal peptide is removed. Cwp8433-803 is then associated with the S layer where the N-terminal peptide (dash line) is potentially removed. At this subcellular position, it is likely that Cwp8430-803 cleaves the SlpA protein and participates in the S-layer turnover. When Cwp8430-803 is released from the bacterial surface, it is first cleaved at the C-terminal part, leading to Cwp8430-518, and then after the N-terminal propeptide, leading to Cwp8492-518. According to our observations in E. coli, at least the final cleavage is probably due to an automaturation mechanism. The mature Cwp8492-518 is thereafter reassociated with the underlying cell wall. When the mature form is not surface bound, it potentially degrades the ECM host proteins.
ACKNOWLEDGMENTS
Diana Chapetón Montes is supported by a doctoral fellowship from the French Ministry of Higher Education and Research.
We thank Celine Boursier for assistance in the proteomic analyses (proteomic platform, IFR 141, Université Paris-Sud) and Séverine Péchiné for valuable advice. We especially thank Neil Fairweather for the gift of the cwp84 mutant strain and assistance with the manuscript.
Footnotes
Published ahead of print on 22 July 2011.
REFERENCES
- 1. Ausiello C. M., et al. 2006. Surface layer proteins from Clostridium difficile induce inflammatory and regulatory cytokines in human monocytes and dendritic cells. Microbes Infect. 8:2640–2646 [DOI] [PubMed] [Google Scholar]
- 2. Bergmann S., Rohde M., Chhatwal G. S., Hammerschmidt S. 2001. α-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 40:1273–1287 [DOI] [PubMed] [Google Scholar]
- 3. Bromme D., Nallaseth F. S., Turk B. 2004. Production and activation of recombinant papain-like cysteine proteases. Methods 32:199–206 [DOI] [PubMed] [Google Scholar]
- 4. Buist G., Steen A., Kok J., Kuipers O. P. 2008. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 68:838–847 [DOI] [PubMed] [Google Scholar]
- 5. Calabi E., Calabi F., Phillips A. D., Fairweather N. F. 2002. Binding of Clostridium difficile surface layer proteins to gastrointestinal tissues. Infect. Immun. 70:5770–5778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Calabi E., et al. 2001. Molecular characterization of the surface layer proteins from Clostridium difficile. Mol. Microbiol. 40:1187–1199 [DOI] [PubMed] [Google Scholar]
- 7. Chen C. Y., et al. 2003. Maturation processing and characterization of streptopain. J. Biol. Chem. 278:17336–17343 [DOI] [PubMed] [Google Scholar]
- 8. Cloud J., Kelly C. P. 2007. Update on Clostridium difficile associated disease. Curr. Opin. Gastroenterol. 23:4–9 [DOI] [PubMed] [Google Scholar]
- 9. Coutte L., et al. 2003. Role of adhesin release for mucosal colonization by a bacterial pathogen. J. Exp. Med. 197:735–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dang T. H., et al. Chemical probes of surface layer biogenesis in Clostridium difficile. ACS Chem. Biol. 5:279–285 [DOI] [PubMed] [Google Scholar]
- 11. de la Riva L., Willing S. E., Tate E. W., Fairweather N. F. 2011. Roles of cysteine proteases Cwp84 and Cwp13 in biogenesis of the cell wall of Clostridium difficile. J. Bacteriol. 193:3276–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Del Papa M. F., Hancock L. E., Thomas V. C., Perego M. 2007. Full activation of Enterococcus faecalis gelatinase by a C-terminal proteolytic cleavage. J. Bacteriol. 189:8835–8843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eidhin D. N., Ryan A. W., Doyle R. M., Walsh J. B., Kelleher D. 2006. Sequence and phylogenetic analysis of the gene for surface layer protein, slpA, from 14 PCR ribotypes of Clostridium difficile. J. Med. Microbiol. 55:69–83 [DOI] [PubMed] [Google Scholar]
- 14. Fagan R. P., et al. 2009. Structural insights into the molecular organization of the S-layer from Clostridium difficile. Mol. Microbiol. 71:1308–1322 [DOI] [PubMed] [Google Scholar]
- 15. Fagan R. P., et al. 2011. A proposed nomenclature for cell wall proteins of Clostridium difficile. J. Med. Microbiol. 60:1225–1228 [DOI] [PubMed] [Google Scholar]
- 16. Heinlen L., Ballard J. D. 2010. Clostridium difficile infection. Am. J. Med. Sci. 340:247–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hennequin C., Janoir C., Barc M. C., Collignon A., Karjalainen T. 2003. Identification and characterization of a fibronectin-binding protein from Clostridium difficile. Microbiology 149:2779–2787 [DOI] [PubMed] [Google Scholar]
- 18. Hennequin C., et al. 2001. GroEL (Hsp60) of Clostridium difficile is involved in cell adherence. Microbiology 147:87–96 [DOI] [PubMed] [Google Scholar]
- 19. Hytonen J., Haataja S., Gerlach D., Podbielski A., Finne J. 2001. The SpeB virulence factor of Streptococcus pyogenes, a multifunctional secreted and cell surface molecule with strepadhesin, laminin-binding and cysteine protease activity. Mol. Microbiol. 39:512–519 [DOI] [PubMed] [Google Scholar]
- 20. Janoir C., Péchiné S., Grosdidier C., Collignon A. 2007. Cwp84, a surface-associated protein of Clostridium difficile, is a cysteine protease with degrading activity on extracellular matrix proteins. J. Bacteriol. 189:7174–7180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karjalainen T., et al. 2001. Molecular and genomic analysis of genes encoding surface-anchored proteins from Clostridium difficile. Infect. Immun. 69:3442–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kern T., et al. 2008. Toward the characterization of peptidoglycan structure and protein-peptidoglycan interactions by solid-state NMR spectroscopy. J. Am. Chem. Soc. 130:5618–5619 [DOI] [PubMed] [Google Scholar]
- 23. Kirby J. M., et al. 2009. Cwp84, a surface-associated cysteine protease, plays a role in the maturation of the surface layer of Clostridium difficile. J. Biol. Chem. 284:34666–34673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuehne S. A., et al. The role of toxin A and toxin B in Clostridium difficile infection. Nature 467:711–713 [DOI] [PubMed] [Google Scholar]
- 25. Lantz M. S. 1997. Are bacterial proteases important virulence factors? J. Periodontal Res. 32:126–132 [DOI] [PubMed] [Google Scholar]
- 26. Lyras D., et al. 2009. Toxin B is essential for virulence of Clostridium difficile. Nature 458:1176–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matsushita O., Okabe A. 2001. Clostridial hydrolytic enzymes degrading extracellular components. Toxicon 39:1769–1780 [DOI] [PubMed] [Google Scholar]
- 28. Nickerson N. N., Prasad L., Jacob L., Delbaere L. T., McGavin M. J. 2007. Activation of the SspA serine protease zymogen of Staphylococcus aureus proceeds through unique variations of a trypsinogen-like mechanism and is dependent on both autocatalytic and metalloprotease-specific processing. J. Biol. Chem. 282:34129–34138 [DOI] [PubMed] [Google Scholar]
- 29. Péchiné S., Janoir C., Collignon A. 2005. Variability of Clostridium difficile surface proteins and specific serum antibody response in patients with Clostridium difficile-associated disease. J. Clin. Microbiol. 43:5018–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pothoulakis C. 2000. Effects of Clostridium difficile toxins on epithelial cell barrier. Ann. N. Y. Acad. Sci. 915:347–356 [DOI] [PubMed] [Google Scholar]
- 31. Rasband W. S. ImageJ. National Institutes of Health, Bethesda, MD: http://rsb.info.nih.gov/ij/.1997–2005 [Google Scholar]
- 32. Rasmussen M., Bjorck L. 2002. Proteolysis and its regulation at the surface of Streptococcus pyogenes. Mol. Microbiol. 43:537–544 [DOI] [PubMed] [Google Scholar]
- 33. Rudenskaya G. N., Pupov D. V. 2008. Cysteine proteinases of microorganisms and viruses. Biochemistry (Moscow) 73:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Savariau-Lacomme M. P., Lebarbier C., Karjalainen T., Collignon A., Janoir C. 2003. Transcription and analysis of polymorphism in a cluster of genes encoding surface-associated proteins of Clostridium difficile. J. Bacteriol. 185:4461–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tasteyre A., Barc M. C., Collignon A., Boureau H., Karjalainen T. 2001. Role of FliC and FliD flagellar proteins of Clostridium difficile in adherence and gut colonization. Infect. Immun. 69:7937–7940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tasteyre A., et al. 2000. A Clostridium difficile gene encoding flagellin. Microbiology 146(Pt. 4):957–966 [DOI] [PubMed] [Google Scholar]
- 37. Voth D. E., Ballard J. D. 2005. Clostridium difficile toxins: mechanism of action and role in disease. Clin. Microbiol. Rev. 18:247–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Waligora A. J., et al. 2001. Characterization of a cell surface protein of Clostridium difficile with adhesive properties. Infect. Immun. 69:2144–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Warny M., et al. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079–1084 [DOI] [PubMed] [Google Scholar]
- 40. Wexler H., Mulligan M. E., Finegold S. M. 1984. Polyacrylamide gel electrophoresis patterns by Clostridium difficile. Rev. Infect. Dis. 6:S229–S234 [DOI] [PubMed] [Google Scholar]
- 41. Wright A., Drudy D., Kyne L., Brown K., Fairweather N. F. 2008. Immunoreactive cell wall proteins of Clostridium difficile identified by human sera. J. Med. Microbiol. 57:750–756 [DOI] [PubMed] [Google Scholar]
- 42. Wright A., et al. 2005. Proteomic analysis of cell surface proteins from Clostridium difficile. Proteomics 5:2443–2452 [DOI] [PubMed] [Google Scholar]