Abstract
YidC has an essential but poorly defined function in membrane protein insertion and folding in bacteria. The yidC gene is located in a gene cluster that is highly conserved in Gram-negative bacteria, the gene order being rpmH, rnpA, yidD, yidC, and trmE. Here, we show that Escherichia coli yidD, which overlaps with rnpA and is only 2 bp upstream of yidC, is expressed and localizes to the inner membrane, probably through an amphipathic helix. Inactivation of yidD had no discernible effect on cell growth and viability. However, compared to control cells, ΔyidD cells were affected in the insertion and processing of three YidC-dependent inner membrane proteins. Furthermore, in vitro cross-linking showed that YidD is in proximity of a nascent inner membrane protein during its localization in the Sec-YidC translocon, suggesting that YidD might be involved in the insertion process.
INTRODUCTION
Membrane proteins function in critical cellular processes such as energy transduction, transport of molecules, cell communication, and cell division. Upon membrane targeting and insertion, the membrane proteins need to fold and often to assemble into oligomeric structures for proper functioning (31, 39). The Sec translocon plays an important role in the insertion of most membrane proteins into the inner membrane (IM) of Escherichia coli (12). It consists of the conserved heterotrimeric channel complex SecYEG and the accessory components SecDF-YajC, whose function is not entirely clear although a recent study suggested that SecDF functions as a proton motive force (PMF)-dependent chaperone for preproteins (41). Cross-linking and pulldown experiments have identified YidC as another Sec-associated factor (36). For a subset of inner membrane proteins (IMPs) such as Lep, FtsQ, and MtlA, YidC has been shown to associate with the transmembrane (TM) segments of nascent protein chains upon their lateral exit from the Sec translocon (7, 50). However, depletion of YidC in vivo only slightly affects the levels of these Sec-dependent IMPs, indicating that it is not essential for insertion. YidC can also act upstream of the Sec translocon, as demonstrated for the lipoprotein CyoA (subunit II of the cytochrome bo3 oxidase) (4, 13, 45). YidC is sufficient to catalyze insertion of the N-terminal domain of CyoA while translocation of the large C-terminal domain requires the Sec translocon. For another subset of IMPs, YidC alone is sufficient for insertion and assembly of the complete protein into the IM. Substrates of this YidC-only pathway include small coat proteins of the M13 and Pf3 phages, the subunits a and c of the F1Fo ATPase (Foa and Foc), and the subunit K of the NADH dehydrogenase complex (34, 56). YidC has also been copurified with the membrane protease FtsH and its modulator proteins HflK/HflC, suggesting an early, linked role in the quality control of membrane proteins (44).
The coding sequence for the essential yidC gene is located in a cluster of genes involved in protein synthesis and membrane targeting that is highly conserved in Gram-negative bacteria, with the gene order being rpmH, rnpA, yidD, yidC, and trmE (see Fig. S1 in the supplemental material). In general, such a conserved genetic organization suggests coordinated gene expression and a related function. Indeed, three promoters have been located upstream of rpmH, one of which generates a polycistronic mRNA (21). rpmH codes for the ribosomal protein L34, a small basic protein of the large ribosomal subunit. L34 has been cross-linked to L23 that participates in a docking site (48) for the signal recognition particle (SRP), SecA, and the chaperone trigger factor near the nascent chain exit region. rnpA encodes the protein component of RNase P, an endoribonuclease that processes tRNA precursor molecules but also the 4.5S RNA component of the SRP (2). trmE encodes an Era-type GTPase that functions in a tRNA modification pathway and has been implicated in the control of the glutamate-dependent acid resistance system (20). Interestingly, the growth defect of YidC-depleted cells can be alleviated by overexpression of GadX and GadY, transcriptional activators that are also involved in regulation of this acid resistance mechanism (54).
In comparison to YidC, very little is known about the expression and function of yidD, but its presence and location in the gene cluster are highly conserved. We set out to characterize yidD because of our interest in inner membrane protein biogenesis and the functional relation of the remainder of the gene cluster in that process. The yidD gene is sandwiched between rnpA (37-bp overlap) and yidC (2-bp spacing) and is likely to contain an internal promoter for yidC (3). In the present study, we have verified the expression of yidD in E. coli. YidD was found to be associated with the IM via a putative amphipathic α-helix in its N-terminal region. Although YidD is not essential for growth, our data indicate that YidD is required for efficient insertion and maturation of YidC-dependent IMPs. Furthermore, using an in vitro sulfhydryl cross-linking approach, YidD was found in close proximity to a short nascent IMP, suggesting a direct role for YidD during IMP biogenesis.
MATERIALS AND METHODS
Enzymes and material.
Restriction endonucleases and other DNA-modifying enzymes were obtained from Roche and Invitrogen. Bis-maleimidoethane (BMOE) was from Pierce. All other chemicals were supplied by Sigma. Antisera against the His tag and the hemagglutinin (HA) tag were purchased from Roche and Sigma, respectively. Antisera against YidC, PspA, Lep, Foc, and TolC have been described previously (27, 47).
Strains, plasmids, and primers.
All strains, plasmids, and primers are listed in Table 1. All strains were grown in Luria-Bertani (LB) medium with appropriate antibiotics, unless stated otherwise. E. coli strain Top10F′ (Invitrogen) was used for cloning and maintenance of plasmid constructs. Strain MC4100-A was routinely used for expression and preparation of inner membrane vesicles (IMVs) and translation extracts. The MC4100-A strain (23) was a generous gift of Frank Sargent (University of East Anglia, Norwich, United Kingdom). The ΔyidD strain was constructed according to Datsenko and Wanner (8). The primers YidD_Ecoli_swap_H1 and YidD_Ecoli_swap_H2 were used to amplify the kanamycin cassette from pKD13 (8). The PCR product was electroporated into MC4100(DE3) (53) containing the red-mediated recombination system, after which kanamycin-resistant colonies were selected. Proper inactivation of yidD was verified by PCR.
Table 1.
Strains, plasmids, and primers
| Strain, plasmid, or primer | Description or sequence | Reference or source |
|---|---|---|
| E. coli strains | ||
| Top10F′ | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL(Strr) endA1 λ− | Invitrogen |
| MC4100-A | F− ΔlacU169 araD139 rpsL150 relA1 ptsF rbs flbB5301; arabinose resistant | 23 |
| MC4100(DE3) | MC4100 with λ DE3 | 53 |
| ΔyidD strain | yidD knockout in MC4100 (DE3) | This study |
| Plasmids | ||
| pKD13 | Template containing kanamycin cassette | 8 |
| pTL61T | lacZ ORF | 30 |
| pTL61T-Nco | NcoI site at the start of the lacZ ORF | This study |
| pTL61T-Nco YidDprom | yidD promoter sequence fused to lacZ ORF | This study |
| pEH3 | IPTG inducible expression plasmid | 22 |
| pEH3His-YidD | yidD ORF with 5′ His tag | This study |
| pEGFP | gfp ORF | Clontech |
| pEH3GFP-YidD | gfp ORF fused to 5′ of yidD ORF | This study |
| pEH3GFP-YidDH1 | gfp ORF fused to 5′ of yidD helix 1 ORF | This study |
| pEH3GFP | gfp ORF | This study |
| pCL-M13P2 | m13p2 ORF | 46 |
| pCL-CyoA-HA | cyoA ORF with 3′ HA tag | 45 |
| pC4Meth108FtsQCys15 | Sequence coding for 108 residues of FtsQ with a cysteine at position 15 | 55 |
| pC4Meth108FtsQCys36 | Sequence coding for 108 residues of FtsQ with a cysteine at position 36 | 55 |
| pC4Meth108FtsQCys61 | Sequence coding for 108 residues of FtsQ with a cysteine at position 61 | 55 |
| Primers | ||
| RT1 | TAGCCCTCATTCGGGTCTATC | |
| RT2 | TCTCTGGTATCAAATGGTC | |
| RT3 | ACCTCAGGCCCAACAGACCAC | |
| RT4 | ACGCCGTCGCGAAATAC | |
| RT5 | AGCCGGTACTGGTTCAGCCTG | |
| RT6 | TCTTCAGCTGCATCGGGTC | |
| pTL61T-NcoFw | CACACAGGAAACAGCCATGGCCATGATTACGGATTC | |
| pTL61T-NcoRv | GAATCCGTAATCATGGCCATGGCTGTTTCCTGTGTG | |
| YidDpromFw | GGCTATGTCGACCCTAAA CTGCGCAAGAGATC | |
| YidDpromRv | TTAGGCGCCATGGTTTTTCCAACGCTTCCG | |
| YidDpromminNcoFw | TTAATCACCCTTGGCTTCGG | |
| YidDpromminNcoRv | CCGAAGCCAAGGGTGATTAAAG | |
| YidDNterm-Xba-SD-His6Fw | TATCTAGATAGGAGGAGTTTTTACCATGCACCACCACCACCACCACGGATCTATGGCGCCGCCACTGTCG | |
| YidDCterm-SacIRv | TATAGAGCTCATTAGTGTTCTCTGGTATCAAATG | |
| GFP5′XbaFw | CCGGGTCTAGAAGAAGGAGATATACATATGAGTAAAGGAGAAGAAC | |
| YidD3′stopRv | TTCCGAGCTCTTATTAGTGTTCTCTGGTATC | |
| YidD3′NtermstopRv | TTCCGAGCTCTTATTAGAGTAGCGGACTAATCAGGCG | |
| GFP3′SacIRv | TTCCGAGCTCTTATTATTTGTATAGTTCATCCATG | |
| YidD_Ecoli_swap_H1 | GACTAAAAGATTGCGTTGCGAATCCATCGTTAGTGTTCTCTGGTATCAAAATTCCGGGGATCCGTCGACC | |
| YidD_Ecoli_swap_H2 | TTGGAAAAATTATGGCGCCGCCACTGTCGCCTGGCTCGCGGGTCCTGATATGTAGGCTGGAGCTGCTTCG |
Plasmid pTL61T (30) served as a template to construct the lacZ transcriptional fusion pTL61T-NcoI-yidDprom. First, an NcoI site was introduced with the primers pTL61T-NcoFw and pTL61T-NcoRv at the start of the lacZ open reading frame (ORF) in pTL61T, resulting in pTL61T-NcoI, using a QuikChange Site-Directed Mutagenesis Kit (Stratagene). Then, a −924/+3 region 5′ of the yidD start codon was mutated to remove an endogenous NcoI site using the primers YidDpromminNcoFw and YidDpromminNcoRv. Then, this region was amplified using the primers YidDpromFw and YidDpromRv to introduce an NcoI site at the start codon of YidD and cloned into the NcoI site of pTL61T-Nco, resulting in pTL61T-NcoI-YidDprom.
Plasmid pEH3 (22) was used for the expression of His-YidD. The gene encoding yidD was amplified using E. coli MC4100 genomic DNA as a template with the primers YidDNterm-Xba-SD-His6Fw and YidDCterm-SacIRv and cloned into XbaI/SacI-digested pEH3 to construct pEH3His-YidD. pEH3GFP-YidD (where GFP is green fluorescent protein), pEH3GFP-YidDH1, and pEH3GFP were constructed as follows. The coding sequence of GFP was amplified from the pEGFP plasmid (Clontech) using the primers GFP5′XbaFw and GFP3′GGSRv while the coding sequence for yidD was amplified from MC4100 genomic DNA with the primers YidD5′GGSFw and YidD3′stopRv. These two PCR products were used as templates for the subsequent overlapping PCR with the primers GFP5′XbaFw and YidD3′stopRv to yield the amplification products encoding the GFP-YidD fusion flanked by the XbaI and SacI sites. This product was cloned into XbaI/SacI-digested pEH3 to yield pEH3GFP-YidD. Similarly, to construct pEH3GFP-YidDH1 containing gfp fused 3′ to the first 81 bases of yidD, the product of the PCR using the primers GFP5′XbaFw and GFP3′GGSRv was combined with the PCR product using the primers YidD5′GGSFw and YidD3′NtermstopRv and used as a template in an overlapping PCR with the primers GFP5′XbaFw and YidD3′NtermstopRv. To construct pEH3GFP, the coding sequence of GFP was amplified with the primers GFP5′XbaFw and GFP3′SacIRv and cloned into XbaI/SacI digested pEH3.
yidD transcription and translation.
For reverse transcription-PCR (RT-PCR), total RNA from strain MC4100 grown to mid-log phase was isolated using an RNeasy minikit (Qiagen) and treated with RNase-free DNase (Qiagen) according to the manufacturer's protocol. cDNA was synthesized from total RNA using avian myeloblastosis virus (AMV) reverse transcriptase (Promega) with the sequence-specific primer RT6 and used as a template for the subsequent PCR with the yidD and yidC sequence-specific primers RT1 to RT6 (Table 1; see also Fig. 2A).
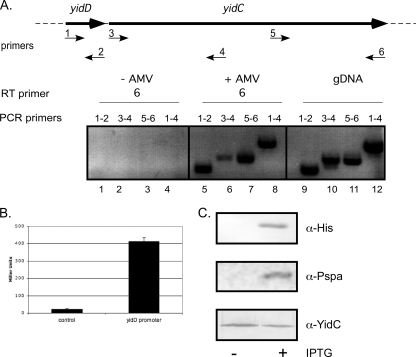
Fig. 2.
Transcription and translation of yidD. (A) RT-PCR analysis of E. coli RNA using AMV RT polymerase with the indicated primers showed that yidD gene is cotranscribed with yidC. gDNA, genomic DNA. (B) β-Galactosidase assay using MC4100-A harboring the reporter plasmid pTL61T-Nco-YidDprom showed that yidD mRNA is translated. As a negative control, MC4100-A harboring the empty pTL61T-Nco was used. The results are an average of three independent measurements. (C) MC4100-A cells containing pEH3His-YidD were grown in LB medium to an OD660 of 0.3 and induced with 1 mM IPTG. Whole-cell samples were taken 1 h later and analyzed by SDS-PAGE and Western blotting using antisera against the His tag, YidC, and PspA. α, anti.
To examine translation E. coli MC4100 carrying plasmids pTL61T-Nco or pTL61T-Nco-YidDprom were grown for 3 h in LB medium. β-Galactosidase activity was determined as described by Miller (32) and quantified as follows: 1,000 × (OD420 × 1.75 × OD550)/(OD600 × reaction time × volume), where the optical density (OD) is measured at 420, 550, and 600 nm, respectively.
Membrane separation by sucrose gradient centrifugation.
E. coli MC4100-A cells harboring pEH3His-YidD or control plasmid pEH3 were induced with 100 μM isopropyl β-d-1-thiogalactopyranoside (IPTG) when cultures reached an OD660 of ∼0.3 and harvested 1 h later. Membranes were isolated and separated into IM and outer membrane (OM) fractions by isopycnic sucrose gradient centrifugation essentially as described previously (11) with the following modification: sucrose gradients were centrifuged for 20 h at 80,220 × g using a TST41.14 rotor. Fractions, withdrawn from the top of the gradients, were analyzed by SDS-PAGE and Western blotting. In addition, the IM fractions were pooled and used as IMVs for further experiments.
Differential solubilization of IM with sarcosyl.
Crude membranes were isolated and subjected to differential solubilization with sodium lauryl sarcosinate (sarcosyl) as described previously (6). Essentially, crude membranes were incubated with 0.5% Sarkosyl for 20 min at room temperature and centrifuged for 1 h at 271,667 × g. Samples of both the supernatant (enriched IMs) and pellet (enriched OMs) were analyzed by SDS-PAGE and Western blotting.
GFP fluorescence microscopy.
MC4100-A cells harboring pEH3GFP-YidD, pEH3GFP-YidDH1, or pEH3GFP were grown in M9 medium and induced with 50 μM IPTG when an OD660 of ∼0.3 was reached. After 1 h of induction, the cells were collected, fixed, and subjected to fluorescence imaging as described previously (10).
In vitro translation, translation, and cross-linking.
Truncated mRNA was prepared from HindIII-linearized plasmid pC4Meth108FtsQCys15, -36, and -61, which introduces a Cys residue in FtsQ at positions 15, 36, and 61, respectively, as described previously (36). In vitro translation, insertion in IMVs, cross-linking with BMOE, and sodium carbonate extraction were carried out as described previously (55). Immunoprecipitations (IPs) on sodium carbonate-extracted samples were carried out as described previously (45). The samples were analyzed by SDS-PAGE and phosphorimaging using a Molecular Dynamics PhsophorImager 473 and quantified using Imagequant software from Molecular Dynamics.
Protein sample analysis.
Protein samples were routinely analyzed by SDS-PAGE and Western blotting. Protein quantifications were performed using ChemiDoc XRS (Bio-Rad) with Quantity One (Bio-Rad) software.
RESULTS
Genomic organization of yidD.
The E. coli yidD is located in an interesting gene cluster comprising the following genes: rpmH, rnpA, yidD, yidC, and trmE (Fig. 1A). This region has been strikingly conserved among the Gram-negative bacteria (see Fig. S1 in the supplemental material) (16). Homologs of yidD are also widely spread in Gram-positive bacteria, but the genomic context in these cases is less conserved. Interestingly, yidD homologs, defined by the presence of the conserved domain of unknown function 37 (DUF37) (5), are found in all plants sequenced so far but not in yeast, Caenorhabditis elegans, or mammals. In addition, yidD is found in the genome of Haemophilus influenzae phage HP1 (17). The yidD homologs in plants contained neither a clear mitochondrial nor a chloroplast localization signal as predicted by the subcellular localization programs PSORTII (24) and TargetP (15). Nor were the yidD homologs found to colocalize with alb3 and oxa1 in the genome.
Fig. 1.
In silico analysis of E. coli yidD. (A) Genomic context of the yidD gene in E. coli. ORFs are represented by arrows in 5′ to 3′ direction. (B) ClustalW alignment of selected yidD gene family members containing the DUF31 conserved sequence. (C) Schematic representation of the predicted secondary structure of E. coli YidD. The boxes represent putative α-helices, and the lines represent coiled regions of YidD. (D) Helical representation of the first putative helix of E. coli YidD as predicted by http://cti.itc.virginia.edu/∼cmg/Demo/wheel/wheelApp.html. M. tuberculosis, Mycobacterium tuberculosis; B. subtilis, Bacillus subtilis; S. aureus, Staphylococcus aureus; P. aeruginosa, Pseudomonas aeruginosa; N. meningitidis, Neisseria meningitidis; A. thaliana, Arabidopsis thaliana; H. pylori, Helicobacter pylori.
E. coli YidD has a predicted size of 9.3 kDa and a pI of 10.2. The DUF37 domain in the yidD sequence is characterized by three conserved cysteines (Fig. 1B and C). Some members of the yidD family have been annotated as hemolysins, which resulted from the unpublished observation reported in GenBank L36462 that the Aeromonas hydrophila YidD homolog HlyA possesses alpha-hemolysin activity (35). However, the Aeromonas HlyA does not appear to be related to any of the known hemolysins. Secondary structure prediction indicated the presence of three α-helices in YidD (Fig. 1C). None of the three α-helices appeared sufficiently hydrophobic to serve as a TM, suggesting a cytoplasmic localization for YidD. However, a closer examination of the α-helical wheel projection of the predicted first α-helix in YidD suggested an amphipathic structure (Fig. 1D).
Transcription and translation of yidD.
The Proteus mirabilis yidD open reading frame (ORF) has been shown to be expressed in E. coli by using a translational LacZ fusion (38). However, there are no reports available on the putative E. coli yidD, and we therefore verified its expression. First, we determined whether mRNA transcript coding for YidD is produced in E. coli MC4100 cells by performing reverse transcription-PCR (RT-PCR) on total RNA isolated from logarithmically growing MC4100 cells (Fig. 2A). Using primers that were specific for the yidD sequence, we could detect yidD mRNA (lane 5). A reaction with primers for the adjacent yidC gene (which is known to be expressed) served as a positive control and yielded amplification products of the expected sizes (lanes 6 and 7). To ensure that the observed RT-PCR products were not due to DNA contamination, negative controls were performed in which the reverse transcriptase was omitted from the cDNA synthesis reaction. As shown (Fig. 2A, lanes 1 to 4), no amplification products were detected under these conditions, verifying that the products generated by RT-PCR were due to the presence of transcript and not due to genomic DNA contamination. As a further positive control for the PCR amplification, genomic DNA of the strain MC4100 was used as a template, which resulted in PCR products of the same size as the products in the RT-PCR amplification (Fig. 2A, lanes 9 to 12). Furthermore, a PCR product was formed (lane 8) when primers were used that corresponded to the 5′ end of yidD and the center of yidC, indicating that yidD is, at least in part, cotranscribed with yidC. Thus, RT-PCR analysis confirmed that yidD transcript is synthesized and cotranscribed with yidC mRNA.
The transcriptional activity of the putative promoter upstream of yidD was analyzed using a β-galactosidase reporter system. The ∼1-kb region upstream of the coding sequence of yidD was cloned into pTL61-Nco, resulting in a lacZ reporter gene that is directly fused to the start codon of yidD. Using a β-galactosidase assay, significant activity of the yidD-lacZ fusion was detected in E. coli MC4100 compared to activity in the promoterless lacZ control (Fig. 2B). Together, the expression data indicate that yidD is transcribed and translated in E. coli.
Expression and subcellular localization of YidD.
To monitor the expression of yidD, the gene was cloned under lac promoter control in the expression vector pEH3 (22). A sequence encoding a His6 tag was fused to the 5′ end of yidD, allowing the detection of the expressed protein. As shown in Fig. 2C (upper panel), a 14-kDa product was detected in IPTG-induced cells upon Western blotting using antiserum directed against the His6 tag. Apparently, His-YidD migrates more slowly than the predicted 9.4 kDa, which may be caused by the addition of the His tag. His-YidD expression did not affect growth, and the protein could not be detected by total Coomassie protein staining, indicating that his-yidD is not expressed at a high level (not shown). Fusion of the His6 tag-encoding sequence to the 3′ end of yidD yielded only a faintly detectable product of 14 kDa upon Western blotting, suggesting that the C-terminal tag destabilized the protein (data not shown).
IPTG induced expression of His-YidD resulted in a slight increase of PspA expression (Fig. 2C, middle panel). PspA is a peripheral IM-associated stress protein that responds to impaired membrane function (26, 52). Although the biological function of PspA is far from clear, it appears to play a role in the maintenance of the PMF upon cell envelope stress. It was shown previously that YidC-depleted cells have increased levels of PspA to counteract a loss of PMF due to the impaired insertion of respiratory chain complexes (47). However, the level of YidC was not detectably changed upon overexpression of His-YidD (Fig. 2C, lower panel). Therefore, the PspA response—albeit modest—caused by His-YidD overexpression suggests an effect on membrane functioning, independent of the YidC level. The level of the cytoplasmic heat shock protein GroEL was not altered upon His-YidD expression, arguing against a more general stress phenomenon (data not shown).
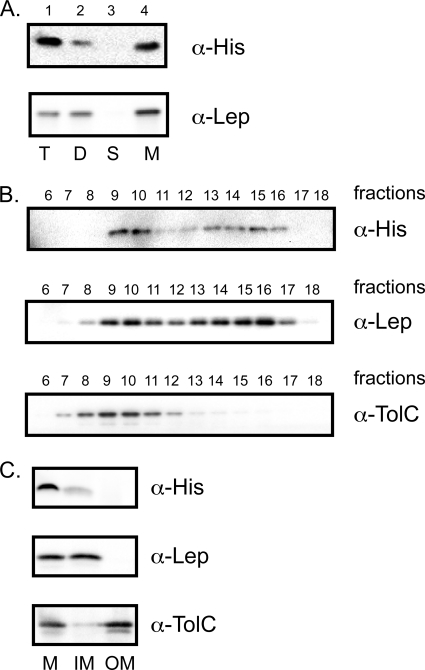
The link with membrane stress was surprising because YidD is predicted to be located in the cytoplasm, considering the absence of a putative signal sequence and of hydrophobic TMs. We therefore experimentally verified the subcellular localization of YidD. Cells in logarithmic growth phase that produced His-YidD were harvested and lysed, after which total membranes were isolated by centrifugation. Interestingly, Western blot analysis of the various fractions taken during the isolation showed that His-YidD cofractionated with membranes in the high-speed pellet (Fig. 3A, lane 4). As a control for the fractionation, the samples were analyzed by Western blotting using antibodies against leader peptidase (Lep), a bona fide IMP. YidD and Lep were found in the same fraction. In these experiments, the His-YidD protein level appeared to be decreased during the fractionation procedure. For the cell debris (D) (Fig. 3A, lane 2) and soluble (S) (lane 3) and membrane (M) (lane 4) fractions, the material loaded was equivalent to four times the amount of the total cell lysate (T) (lane 1) sample taken before fractionation. Nevertheless, the His-YidD signal was lower in the combined fractions than in the total cell sample. This indicates either that the His tag is proteolytically removed during the fractionation or that YidD itself is unstable, consistent with its low expression level. Nonetheless, the data indicate that at least a subpopulation of YidD is membrane associated since the membrane fractions contained considerable amounts of YidD compared to the other fractionation samples.
Fig. 3.
His-YidD is localized at the IM. (A) Subcellular fractionation of MC4100-A containing pEH3His-YidC. Cells were grown in LB medium and induced with 100 μM IPTG for 1 h and fractionated, and the fractions were analyzed by Western blotting using antisera against the His tag and the control IMP Lep. T, total cell sample; D, cellular debris (pellet from low-speed centrifugation); S, soluble fraction (supernatant of high-speed centrifugation); M, membrane fraction (pellet of high-speed centrifugation). Four times the amount of material of the total cell sample was loaded for fractions D, S, and M. (B) Membranes from MC4100-A expressing His-YidD were separated by sucrose gradient centrifugation. Fractions were analyzed by Western blotting using antisera against the His tag, the control IMP Lep, and the control OMP TolC. (C) Membranes from MC4100-A expressing His-YidD were treated with 0.5% Sarkosyl to solubilize the IMs and centrifuged to collect the OMs. Samples of the fractions, derived from equal amounts of cell material, were analyzed by SDS-PAGE and Western blotting using antisera against the His tag, the control IMP Lep, and the control OMP TolC. M, total membranes; IM, inner membrane (supernatant of Sarkosyl extraction); OM, outer membrane (pellet of Sarkosyl extraction).
The absence of an obvious signal sequence would suggest that His-YidD is associated with the IM rather than the OM. To confirm this, crude membranes from His-YidD-expressing cells were subjected to isopycnic sucrose gradient centrifugation for separation of IMs and OMs (Fig. 3B). Analysis of the fractions by Western blotting revealed a similar distribution for His-YidD and the control IMP Lep, peaking in fractions 9 and 10 and 13 to 16, while TolC, a control outer membrane protein (OMP), as almost exclusively localized in fractions 8 to 11. Despite the observation that the more dense OM fractions seemed slightly contaminated with IMPs (but not vice versa), the data clearly indicated that His-YidD is located in or at the IM. Of note, the same distributions of Lep and TolC were observed in membranes derived from cells harboring the empty plasmid pEH3, indicating that His-YidD expression did not interfere with the membrane separation (data not shown). To rule out that His-YidD forms aggregates that cofractionated with E. coli membranes, crude membranes were also subjected to sucrose floatation gradient analysis, showing that His-YidD, similar to Lep, floated with the IMs (data not shown).
To independently corroborate the IM localization of His-YidD, crude membranes were subjected to differential solubilization using the detergent sodium lauryl sarcosinate (Sarkosyl) that preferentially solubilizes IMs (6). The results shown in Fig. 3C indicate that His-YidD, like the control IMP Lep, was solubilized by Sarkosyl in contrast to the control OMP TolC, which was primarily found in the nonextracted pellet fraction, as expected. Only part of His-YidD was recovered after the extraction procedure, which is probably due to the inherent instability of this tagged protein, as discussed above. Together, the data argue that His-YidD is associated with the IM.
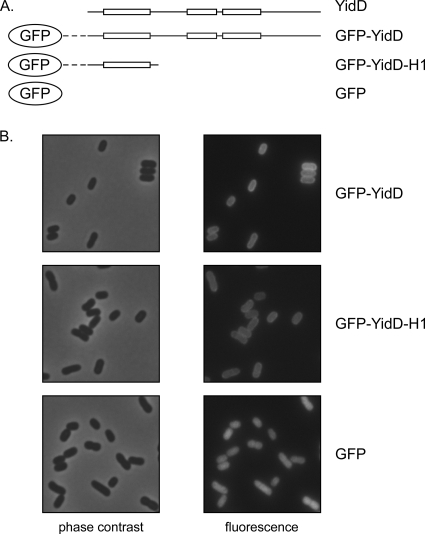
To exclude the possibility that the membrane localization of His-YidD was influenced by the His tag, we replaced the His tag by green fluorescent protein (GFP). The latter fusion protein allowed a more direct evaluation of the location by fluorescence microscopy (Fig. 4B). MC4100-A cells producing GFP-YidD clearly showed a halo-type fluorescence, indicating that YidD localized in the cell envelope and most likely at the IM. In contrast, expression of nonfused GFP resulted in a diffuse signal throughout the cells, consistent with its expected cytoplasmic localization.
Fig. 4.
GFP-YidD and GFP-YidDH1 localize to the membrane. (A) Schematic representation of the constructs used. The boxes represent predicted α-helices while the lines represent coiled regions of YidD (Fig. 1C). The dashed line represents the linker region between GFP and YidD. (B) MC4100-A cells expressing GFP and GFP fusion proteins were grown to the early log phase of growth. Protein expression was induced with 50 μM IPTG for 1 h. Subsequently, the cells were collected, fixed, and imaged by fluorescence microscopy while micrographs of corresponding fields are shown by phase-contrast microscopy.
As mentioned before, YidD does not contain any hydrophobic domains to account for its membrane targeting and insertion. Secondary structure prediction did, however, reveal the presence of two to three potential α-helical regions (Fig. 1C). The most N-terminal of these is relatively conserved and displays a predicted amphipathic character (Fig. 1D). To investigate whether this helix acts as a membrane targeting sequence for YidD, we fused it directly to GFP (Fig. 4A). Indeed, this construct appeared to localize at the cell envelope (Fig. 4B), suggesting that the N-terminal amphipathic helix is responsible for the membrane localization of YidD. The membrane association appeared to withstand extraction with 4 M urea, suggesting that it represents a direct interaction with lipids since this treatment would interfere with protein-protein interactions (data not shown).
Function of YidD in IMP biogenesis.
To examine the function of YidD, a chromosomal knockout mutant was constructed by deleting almost the entire coding sequence according to the procedure described by Datsenko and Wanner (8). Deletion of the yidD gene was confirmed by PCR (data not shown). The deletion was made such that the rnpA ORF remained intact. The ΔyidD cells were viable with no apparent phenotype, in accordance with the results from the Keio project, in which ΔyidD cells were reported to be viable (1). YidC levels in the ΔyidD strain remained virtually identical to the wild-type (WT) level as observed by Western blot analysis (Fig. 5A). Furthermore, we studied the growth in different culture media and the cellular morphology by fluorescence-activated cell sorting (FACS) and microscopy, but we did not observe a difference between WT and ΔyidD strains (data not shown), indicating that YidD is not essential for growth, cell division, and overall morphology.
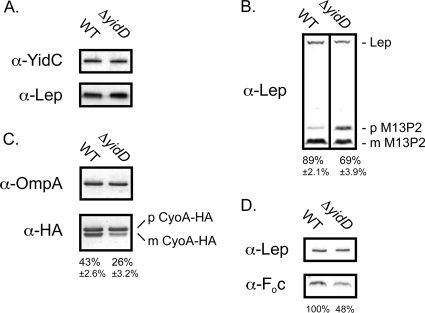
Fig. 5.
YidD is required for optimal processing of YidC substrates. (A) Western blot analysis with antisera against YidC and Lep of samples taken from logarithmically growing MC4100(DE3) (WT) and ΔyidD cells. WT and ΔyidD cells harboring pCL-M13P2 (B) or pCL-CyoA-HA (C) were grown to the early log phase of growth and induced for 1 h with 100 μM IPTG. Samples were analyzed by Western blotting using antisera against the HA tag, Lep, and OmpA (loading control). The precursor and mature forms are indicated by p and m, respectively. The percentage of processed preprotein (amount of mature form/[amount of mature form + amount of precursor]) is given below the lanes and was calculated from four biological replicates. (D) Western blot analysis with antisera against Foc and Lep of IMVs derived from the ΔyidD strain and its isogenic parental strain. The percentage of Foc in the ΔyidD IMVs is given below the lane compared to the level in WT IMVs, which is set at 100%.
To study a possible functional relationship with YidC, we studied the membrane integration of three YidC-dependent IMPs: the chimeric protein M13P2 that consists of phage M13 procoat extended at its C terminus with the periplasmic P2 domain of Lep, the CyoA subunit of the cytochrome o oxidase complex, and the Foc subunit of the F1Fo-ATPase complex. Membrane insertion of M13P2 is exclusively dependent on YidC and can be conveniently monitored by processing of its signal peptide by signal peptidase I (19). Similarly, YidC is sufficient to mediate translocation of the N-terminal periplasmic loop of CyoA and processing of its signal peptide by the lipoprotein-specific signal peptidase II (45). Using Western blot analysis under steady-state conditions, we monitored the processing of both pre-M13P2 and HA-tagged pre-CyoA expressed from expression plasmids introduced in the WT and ΔyidD strains (Fig. 5B and C). Importantly, the HA tag has been shown not to interfere with insertion and maturation of CyoA (45).
In WT cells, most pre-M13P2 was processed to mature form (89% ± 2.1%), whereas pre-M13P2 significantly accumulated in ΔyidD cells resulting in only 69% ± 3.9% mature M13P2 of the total M13P2 produced (Fig. 5B). Endogenous Lep was probed on the same blot to serve as a loading control. Pre-CyoA-HA was also less efficiently processed in the ΔyidD strain (26% ± 3.2%) than in the WT background (43% ± 2.6%) (Fig. 5C). As a control, signal peptide processing of the Sec-dependent but YidC-independent outer membrane protein A (OmpA) appeared unaffected, demonstrating that indirect inactivation of the Sec translocon or signal peptidase had not occurred under these conditions (Fig. 5C). In addition, OmpA served as a loading control in this experiment.
Insertion of the Foc subunit of the F1Fo-ATPase complex is exclusively dependent on YidC which results in a severely reduced level of Foc in YidC-depleted IMVs (47). To study the effect of YidD, the level of Foc was analyzed by Western blotting in IMVs derived from the ΔyidD strain and its isogenic parental strain (Fig. 5D). Compared to the control protein Lep, a clear reduction of Foc was apparent upon inactivation of YidD.
Together, the data demonstrate that YidC-dependent membrane insertion is significantly affected but not completely abolished in the absence of YidD suggesting a functional link between the two proteins.
YidD cross-linking to nascent FtsQ.
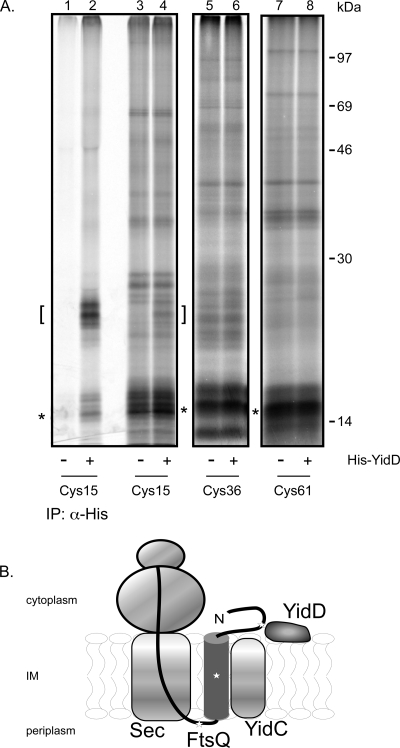
Previous in vitro site-specific cross-linking experiments have shown interactions of nascent IMPs with the translocon components SecY, SecA, and YidC that depended on the length of the nascent chain (36). In this approach, membrane insertion intermediates are generated by the translation of truncated mRNAs that lack a stop codon, thus preventing the release of the polypeptide chain from the ribosome. Considering the localization of YidD at the IM and the apparent role of YidD in YidC-dependent membrane insertion, we hypothesized that YidD is adjacent to a nascent model IMP during membrane insertion. Therefore, we chose to analyze nascent chains of FtsQ with a length of 108 amino acids (108FtsQ). FtsQ is a type II IMP that spans the IM once and plays a critical role in cell division. The TM of the well-characterized 108FtsQ intermediate is located between residues 24 and 49, exposed outside the ribosome, and has been shown to interact with YidC (36, 55). Because YidD contains three conserved cysteine residues, we decided to use sulfhydryl-specific cross-linking upon introduction of single cysteine residues in 108FtsQ to probe a putative interaction. The cysteines were placed at positions 15 (located in the cytoplasmic domain), 36 (in the TM), and 61 (in the periplasmic domain) (Fig. 6B), all expected to be exposed outside the ribosome exit tunnel that covers approximately 35 residues.
Fig. 6.
His-YidD is in close proximity to the cytoplasmic N terminus of nascent FtsQ. (A) Nascent 108FtsQ with a unique cysteine at position 15, 36, or 61 was synthesized in vitro, inserted in IMVs derived from cells induced or not for expression of His-YidD, and cross-linked using the cysteine-specific cross-linker BMOE. Carbonate-resistant pellet fractions are shown. IPs were performed using antiserum against the His tag. The positions of the nascent chains are indicated by asterisks, and the position of His-YidD cross-linking adducts are indicated by brackets. (B) Model for the molecular environment of membrane-inserted nascent 108FtsQ. The stars present the positions of the unique cysteines in 108FtsQ.
Nascent chains of the three single cysteine FtsQ constructs were prepared by translating truncated mRNA in the presence of [35S]methionine in a cell- and membrane-free E. coli lysate. Purified inner membrane vesicles (IMVs) from WT cells and cells that produce His-YidD were added to the translation reaction mixture to allow membrane targeting and insertion of the translation intermediates. Subsequently, the samples were incubated with the homo-bifunctional cysteine-specific reagent BMOE to induce cross-linking followed by carbonate extraction to enrich for membrane-integral proteins. Adducts of ∼25 kDa were specifically detected upon cross-linking of 108FtsQCys15 in His-YidD IMVs (Fig. 6A, lane 4). IP experiments using antibodies directed against the His tag confirmed that the detected adduct, indeed, contained His-YidD cross-linked to 108FtsQCys15 (Fig. 6A, lanes 1 to 2; the adduct is indicated by a bracket). The doublet of adducts might be caused by the observed heterogeneity in nascent chain size. Alternatively, it might reflect the involvement of different cysteine residues in YidD in cross-linking to 108FtsQCys15, which could result in distinct adducts of slightly different gel mobilities. In contrast, 108FtsQCys36 and 108FtsQCys61 did not generate any specific cross-linked adducts in His-YidD compared to WT IMVs (Fig. 6A, lanes 5 to 8). These data suggest that His-YidD is in close vicinity of the nascent chain of FtsQ, presumably near the cytoplasmic N-terminal part of the membrane-inserted nascent chain and possibly to assist YidC with the insertion of the growing polypeptide.
DISCUSSION
In our effort to understand the role of YidC in membrane protein insertion and assembly, we have analyzed the small yidD ORF that is located directly upstream of yidC. YidD belongs to a family of small (∼80 amino acids) hypothetical proteins with a domain of unknown function 37 (DUF37) comprising three conserved cysteine residues (49). The yidD gene is present in all phyla of Bacteria. We showed that E. coli yidD is expressed giving rise to a membrane-associated protein. Although yidD is not essential, we obtained evidence that its gene product is functionally related to YidC: in the absence of YidD the insertion and processing of three YidC substrates are affected, and YidD was found cross-linked to a nascent IMP that is also in vicinity of YidC (55).
The rpmH-rnpA-yidD-yidC-trmE gene cluster belongs to the most conserved clusters in Eubacteria and even Archaea (51). Such a strong conservation of gene proximity and order could point to a coordinated expression and a related function, which may be essential. We propose that this cluster functions in the cotranslational insertion of membrane proteins via YidC, which is considered to be the most ancient protein insertase (33). Interestingly, in several cases there is a partial overlap in the genes, the most extreme example being the complete overlap of the rpmH and rnpA genes in the genus Thermus (14). Although small deviations exist in the presence and order of genes in this cluster within various microorganisms, yidC is usually preceded by yidD, and it is also cotranscribed with yidC (this study). Interestingly, in the genome of Blochmannia pennsylvanicus the yidD gene is fused to the 5′ end of the yidC gene. B. pennsylvanicus is an obligate intracellular symbiotic bacterium of the ant Camponotus pennsylvanicus (9). The B. pennsylvanicus genome consists of only 792 kb that resulted from a massive reduction of genome size compared to its free-living ancestors. The presence of the yidDC gene fusion among the only 610 identified ORFs in this genome strongly suggests an evolutionary pressure to maintain a linked function of the two proteins.
Despite the absence of a putative transmembrane anchor, all our analyses, including cell fractionation and differential detergent solubilization, indicated that YidD localized to the E. coli IM. Most likely, the predicted amphipathic α-helix is responsible for membrane targeting as it is sufficient to target GFP to the membrane. In contrast to YidC, which accumulates at the cell poles (42), YidD fused to GFP showed a circumferential distribution at the cell periphery that is consistent with a dispersed localization in the cell envelope. It should be noted that this chimeric construct is probably present in amounts that exceed the endogenous YidD level. Interestingly, YidD from the obligate intracellular Gram-negative pathogen Chlamydia trachomatis has been shown to associate with lipid droplets when expressed in yeast (29, 37), suggesting that affinity for lipids may be a general feature of YidD orthologs.
What is the molecular environment of YidD in the membrane, and what is its function? His-tagged YidD was cross-linked to the cytosolic domain of the nascent chain of 108 residues of the IMP FtsQ, indicating a location of YidD at the cytosolic side of the IM. In fact, the molecular environment of the 108FtsQ insertion intermediate used is well characterized in cross-linking and structural studies (18, 36, 43, 55) and also includes YidC and parts of the Sec translocon. The combined data indicate that at this stage in translation and insertion of FtsQ, the TM is close to YidC and membrane lipids, whereas the flanking regions interact with SecY and YidD (36, 43) (Fig. 6B). Assuming that the FtsQ intermediate has adopted a stable homogeneous location, this would imply that YidD is also close to SecA, SecY, and perhaps YidC and the ribosomal nascent chain exit region. However, pulldown experiments using cross-linked and non-cross-linked cells and membranes have failed to indicate a physical connection between YidD and components of the ribosome and Sec-YidC translocon (data not shown). Although these negative data are difficult to interpret, they would indicate that any interaction of these components with YidD, if existing, is transient and/or of low affinity.
At present, we can only speculate about the function of YidD. The cross-linking of YidD to the N terminus of nascent 108FtsQ, concomitant with its proximity to YidC, SecA, and SecY, might suggest that YidD functions as an interaction partner, perhaps as a chaperone for the cytosolic part of nascent IMPs during membrane insertion. The basic YidD, positively charged at physiological pH, might also assist in tethering the ribosome to the membrane. It has been shown that the positively charged C-terminal domain of YidC contributes to ribosome binding (28), but this tail is much shorter than the corresponding C-terminal extension of the homologous mitochondrial OxaI protein that is both required and sufficient for ribosome binding during cotranslational membrane insertion in mitochondria (25, 40).
Currently, the role of YidD is being analyzed in more detail by studying proteome-wide effects of YidD deletion and by extensive in vitro cross-linking analyses.
Supplementary Material
ACKNOWLEDGMENTS
We thank W. Jong and A. Sauri for helpful discussions and critical reading of the manuscript. We also thank Gregory Koningstein for technical assistance.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 29 July 2011.
REFERENCES
- 1. Baba T., et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bothwell A. L., Garber R. L., Altman S. 1976. Nucleotide sequence and in vitro processing of a precursor molecule to Escherichia coli 4.5 S RNA. J. Biol. Chem. 251:7709–7716 [PubMed] [Google Scholar]
- 3. Burland V., Plunkett G., III, Daniels D. L., Blattner F. R. 1993. DNA sequence and analysis of 136 kilobases of the Escherichia coli genome: organizational symmetry around the origin of replication. Genomics 16:551–561 [DOI] [PubMed] [Google Scholar]
- 4. Celebi N., Yi L., Facey S. J., Kuhn A., Dalbey R. E. 2006. Membrane biogenesis of subunit II of cytochrome bo oxidase: contrasting requirements for insertion of N-terminal and C-terminal domains. J. Mol. Biol. 357:1428–1436 [DOI] [PubMed] [Google Scholar]
- 5. Chenna R., et al. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chopra I., Shales S. W. 1980. Comparison of the polypeptide composition of Escherichia coli outer membranes prepared by two methods. J. Bacteriol. 144:425–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dalbey R. E., Wang P., Kuhn A. 2011. Assembly of bacterial inner membrane proteins. Annu. Rev. Biochem. 80:161–187 [DOI] [PubMed] [Google Scholar]
- 8. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Degnan P. H., Lazarus A. B., Wernegreen J. J. 2005. Genome sequence of Blochmannia pennsylvanicus indicates parallel evolutionary trends among bacterial mutualists of insects. Genome Res. 15:1023–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Den Blaauwen T., Aarsman M. E., Vischer N. O., Nanninga N. 2003. Penicillin-binding protein PBP2 of Escherichia coli localizes preferentially in the lateral wall and at mid-cell in comparison with the old cell pole. Mol. Microbiol. 47:539–547 [DOI] [PubMed] [Google Scholar]
- 11. De Vrije T., Tommassen J., De Kruijff B. 1987. Optimal posttranslational translocation of the precursor of PhoE protein across Escherichia coli membrane vesicles requires both ATP and the protonmotive force. Biochim. Biophys. Acta 900:63–72 [DOI] [PubMed] [Google Scholar]
- 12. du Plessis D. J., Nouwen N., Driessen A. J. 2011. The Sec translocase. Biochim. Biophys. Acta 1808:851–865 [DOI] [PubMed] [Google Scholar]
- 13. du Plessis D. J., Nouwen N., Driessen A. J. 2006. Subunit a of cytochrome o oxidase requires both YidC and SecYEG for membrane insertion. J. Biol. Chem. 281:12248–12252 [DOI] [PubMed] [Google Scholar]
- 14. Ellis J. C., Brown J. W. 2003. Genes within genes within bacteria. Trends Biochem. Sci. 28:521–523 [DOI] [PubMed] [Google Scholar]
- 15. Emanuelsson O., Nielsen H., Brunak S., von Heijne G. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300:1005–1016 [DOI] [PubMed] [Google Scholar]
- 16. Enault F., Suhre K., Poirot O., Abergel C., Claverie J. M. 2004. Phydbac2: improved inference of gene function using interactive phylogenomic profiling and chromosomal location analysis. Nucleic Acids Res. 32:W336–W339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Esposito D., et al. 1996. The complete nucleotide sequence of bacteriophage HP1 DNA. Nucleic Acids Res. 24:2360–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Frauenfeld J., et al. 2011. Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat. Struct. Mol. Biol. 18:614–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fröderberg L., et al. 2003. Versatility of inner membrane protein biogenesis in Escherichia coli. Mol. Microbiol. 47:1015–1027 [DOI] [PubMed] [Google Scholar]
- 20. Gong S., Ma Z., Foster J. W. 2004. The Era-like GTPase TrmE conditionally activates gadE and glutamate-dependent acid resistance in Escherichia coli. Mol. Microbiol. 54:948–961 [DOI] [PubMed] [Google Scholar]
- 21. Hansen F. G., Hansen E. B., Atlung T. 1982. The nucleotide sequence of the dnaA gene promoter and of the adjacent rpmH gene, coding for the ribosomal protein L34, of Escherichia coli. EMBO J. 1:1043–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hashemzadeh-Bonehi L., et al. 1998. Importance of using lac rather than ara promoter vectors for modulating the levels of toxic gene products in Escherichia coli. Mol. Microbiol. 30:676–678 [DOI] [PubMed] [Google Scholar]
- 23. Hatzixanthis K., Palmer T., Sargent F. 2003. A subset of bacterial inner membrane proteins integrated by the twin-arginine translocase. Mol. Microbiol. 49:1377–1390 [DOI] [PubMed] [Google Scholar]
- 24. Horton P., Nakai K. 1997. Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc. Int. Conf. Intell. Syst. Mol. Biol. 5:147–152 [PubMed] [Google Scholar]
- 25. Jia L., et al. 2003. Yeast OxaI interacts with mitochondrial ribosomes: the importance of the C-terminal region of OxaI. EMBO J. 22:6438–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Joly N., et al. 2010. Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol. Rev. 34:797–827 [DOI] [PubMed] [Google Scholar]
- 27. Jong W. S., et al. 2007. Limited tolerance towards folded elements during secretion of the autotransporter Hbp. Mol. Microbiol. 63:1524–1536 [DOI] [PubMed] [Google Scholar]
- 28. Kohler R., et al. 2009. YidC and OxaI form dimeric insertion pores on the translating ribosome. Mol. Cell 34:344–353 [DOI] [PubMed] [Google Scholar]
- 29. Kumar Y., Cocchiaro J., Valdivia R. H. 2006. The obligate intracellular pathogen Chlamydia trachomatis targets host lipid droplets. Curr. Biol. 16:1646–1651 [DOI] [PubMed] [Google Scholar]
- 30. Linn T., St Pierre R. 1990. Improved vector system for constructing transcriptional fusions that ensures independent translation of lacZ. J. Bacteriol. 172:1077–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maddalo G., et al. 2011. Systematic analysis of native membrane protein complexes in Escherichia coli. J. Proteome Res. 10:1848–1859 [DOI] [PubMed] [Google Scholar]
- 32. Miller J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- 33. Pohlschroder M., Hartmann E., Hand N. J., Dilks K., Haddad A. 2005. Diversity and evolution of protein translocation. Annu. Rev. Microbiol. 59:91–111 [DOI] [PubMed] [Google Scholar]
- 34. Price C. E., Driessen A. J. 2010. Biogenesis of membrane bound respiratory complexes in Escherichia coli. Biochim. Biophys. Acta 1803:748–766 [DOI] [PubMed] [Google Scholar]
- 35. Rudd K. E., Humphery-Smith I., Wasinger V. C., Bairoch A. 1998. Low molecular weight proteins: a challenge for post-genomic research. Electrophoresis 19:536–544 [DOI] [PubMed] [Google Scholar]
- 36. Scotti P. A., et al. 2000. YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 19:542–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sisko J. L., Spaeth K., Kumar Y., Valdivia R. H. 2006. Multifunctional analysis of Chlamydia-specific genes in a yeast expression system. Mol. Microbiol. 60:51–66 [DOI] [PubMed] [Google Scholar]
- 38. Skovgaard O. 1990. Nucleotide sequence of a Proteus mirabilis DNA fragment homologous to the 60K-rnpA-rpmH-dnaA-dnaN-recF-gyrB region of Escherichia coli. Gene 93:27–34 [DOI] [PubMed] [Google Scholar]
- 39. Stenberg F., et al. 2005. Protein complexes of the Escherichia coli cell envelope. J. Biol. Chem. 280:34409–34419 [DOI] [PubMed] [Google Scholar]
- 40. Szyrach G., Ott M., Bonnefoy N., Neupert W., Herrmann J. M. 2003. Ribosome binding to the OxaI complex facilitates co-translational protein insertion in mitochondria. EMBO J. 22:6448–6457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tsukazaki T., et al. 2011. Structure and function of a membrane component SecDF that enhances protein export. Nature 474:235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Urbanus M. L., et al. 2002. Targeting, insertion, and localization of Escherichia coli YidC. J. Biol. Chem. 277:12718–12723 [DOI] [PubMed] [Google Scholar]
- 43. Urbanus M. L., et al. 2001. Sec-dependent membrane protein insertion: sequential interaction of nascent FtsQ with SecY and YidC. EMBO Rep. 2:524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. van Bloois E., et al. 2008. Detection of cross-links between FtsH, YidC, HflK/C suggests a linked role for these proteins in quality control upon insertion of bacterial inner membrane proteins. FEBS Lett. 582:1419–1424 [DOI] [PubMed] [Google Scholar]
- 45. van Bloois E., Haan G. J., de Gier J. W., Oudega B., Luirink J. 2006. Distinct requirements for translocation of the N-tail and C-tail of the Escherichia coli inner membrane protein CyoA. J. Biol. Chem. 281:10002–10009 [DOI] [PubMed] [Google Scholar]
- 46. van Bloois E., Koningstein G., Bauerschmitt H., Herrmann J. M., Luirink J. 2007. Saccharomyces cerevisiae Cox18 complements the essential Sec-independent function of Escherichia coli YidC. FEBS J. 274:5704–5713 [DOI] [PubMed] [Google Scholar]
- 47. van der Laan M., et al. 2003. A conserved function of YidC in the biogenesis of respiratory chain complexes. Proc. Natl. Acad. Sci. U. S. A. 100:5801–5806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Walleczek J., Redl B., Stoffler-Meilicke M., Stoffler G. 1989. Protein-protein cross-linking of the 50 S ribosomal subunit of Escherichia coli using 2-iminothiolane. Identification of cross-links by immunoblotting techniques. J. Biol. Chem. 264:4231–4237 [PubMed] [Google Scholar]
- 49. Wang F., et al. 2008. A systematic survey of mini-proteins in bacteria and archaea. PLoS One 3:e4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang P., Dalbey R. E. 2011. Inserting membrane proteins: the YidC/OxaI/Alb3 machinery in bacteria, mitochondria, and chloroplasts. Biochim. Biophys. Acta 1808:866–875 [DOI] [PubMed] [Google Scholar]
- 51. White T. A., Kell D. B. 2004. Comparative genomic assessment of novel broad-spectrum targets for antibacterial drugs. Comp. Funct. Genomics 5:304–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wickström D., et al. 2011. Characterization of the consequences of YidC depletion on the inner membrane proteome of E. coli using 2D blue native/SDS-PAGE. J. Mol. Biol. 409:124–135 [DOI] [PubMed] [Google Scholar]
- 53. Yahr T. L., Wickner W. T. 2001. Functional reconstitution of bacterial Tat translocation in vitro. EMBO J. 20:2472–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yu Z., et al. 2011. Activators of the glutamate-dependent acid resistance system alleviate deleterious effects of YidC depletion in Escherichia coli. J. Bacteriol. 193:1308–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yu Z., Koningstein G., Pop A., Luirink J. 2008. The conserved third transmembrane segment of YidC contacts nascent Escherichia coli inner membrane proteins. J. Biol. Chem. 283:34635–34642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yuan J., Dalbey R. E., Kuhn A. 2010. Membrane protein insertion in E. coli. Methods Mol. Biol. 619:63–77 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.