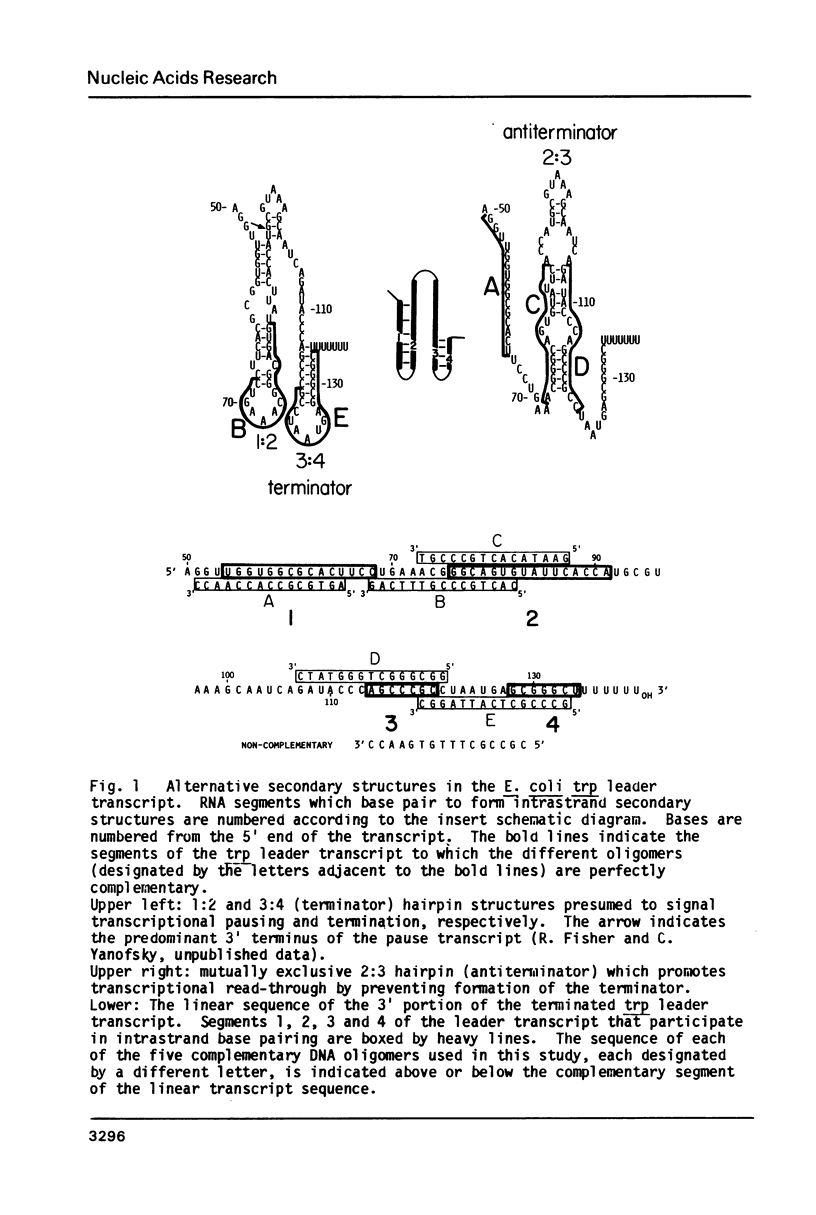
Abstract
DNA oligomers were synthesized that are perfectly complementary to different segments of the tryptophan (trp) operon leader transcript. These 15 nucleotide long oligomers were used as probes of the involvement of transcript secondary structures in two processes: transcription pausing at the pause site located near base pair 90 in the leader region, and transcription termination at the attenuator. The 15-mers were complementary to the four segments of the trp leader transcript which have been shown to form the alternative secondary structures that are believed to be responsible for pausing, termination, and antitermination. Oligomers complementary to RNA segments 1 and 3 relieved termination while the 15-mer complementary to RNA segment 1 relieved pausing. 15-mers complementary to segment 2 had no effect on pausing and the oligomer complementary to segment 4 had virtually no effect on termination.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barnes W. M. DNA sequence from the histidine operon control region: seven histidine codons in a row. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4281–4285. doi: 10.1073/pnas.75.9.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford I. P., Stauffer G. V. Regulation of tryptophan biosynthesis. Annu Rev Biochem. 1980;49:163–195. doi: 10.1146/annurev.bi.49.070180.001115. [DOI] [PubMed] [Google Scholar]
- Das A., Urbanowski J., Weissbach H., Nestor J., Yanofsky C. In vitro synthesis of the tryptophan operon leader peptides of Escherichia coli, Serratia marcescens, and Salmonella typhimurium. Proc Natl Acad Sci U S A. 1983 May;80(10):2879–2883. doi: 10.1073/pnas.80.10.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nocera P. P., Blasi F., Di Lauro R., Frunzio R., Bruni C. B. Nucleotide sequence of the attenuator region of the histidine operon of Escherichia coli K-12. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4276–4280. doi: 10.1073/pnas.75.9.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnham P. J., Platt T. Effects of DNA base analogs on transcription termination at the tryptophan operon attenuator of EScherichia coli. Proc Natl Acad Sci U S A. 1982 Feb;79(4):998–1002. doi: 10.1073/pnas.79.4.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnham P. J., Platt T. Rho-independent termination: dyad symmetry in DNA causes RNA polymerase to pause during transcription in vitro. Nucleic Acids Res. 1981 Feb 11;9(3):563–577. doi: 10.1093/nar/9.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. F., Yanofsky C. Mutations of the beta subunit of RNA polymerase alter both transcription pausing and transcription termination in the trp operon leader region in vitro. J Biol Chem. 1983 Jul 10;258(13):8146–8150. [PubMed] [Google Scholar]
- Fisher R., Yanofsky C. A complementary DNA oligomer releases a transcription pause complex. J Biol Chem. 1983 Aug 10;258(15):9208–9212. [PubMed] [Google Scholar]
- Gardner J. F. Regulation of the threonine operon: tandem threonine and isoleucine codons in the control region and translational control of transcription termination. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1706–1710. doi: 10.1073/pnas.76.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmill R. M., Wessler S. R., Keller E. B., Calvo J. M. leu operon of Salmonella typhimurium is controlled by an attenuation mechanism. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4941–4945. doi: 10.1073/pnas.76.10.4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston H. M., Roth J. R. Genetic analysis of the histidine operon control region of Salmonella typhimurium. J Mol Biol. 1981 Feb 5;145(4):713–734. doi: 10.1016/0022-2836(81)90311-9. [DOI] [PubMed] [Google Scholar]
- Kolter R., Yanofsky C. Attenuation in amino acid biosynthetic operons. Annu Rev Genet. 1982;16:113–134. doi: 10.1146/annurev.ge.16.120182.000553. [DOI] [PubMed] [Google Scholar]
- Lawther R. P., Hatfield G. W. Multivalent translational control of transcription termination at attenuator of ilvGEDA operon of Escherichia coli K-12. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1862–1866. doi: 10.1073/pnas.77.4.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee F., Yanofsky C. Transcription termination at the trp operon attenuators of Escherichia coli and Salmonella typhimurium: RNA secondary structure and regulation of termination. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4365–4369. doi: 10.1073/pnas.74.10.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miozzari G. F., Yanofsky C. Translation of the leader region of the Escherichia coli tryptophan operon. J Bacteriol. 1978 Mar;133(3):1457–1466. doi: 10.1128/jb.133.3.1457-1466.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K., Arentzen R., Huang T., Itakura K. Solid-phase synthesis of polynucleotides. IV. Usage of polystyrene resins for the synthesis of polydeoxyribonucleotides by the phosphostriester method. Nucleic Acids Res. 1980 Nov 25;8(22):5507–5517. doi: 10.1093/nar/8.22.5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K., Huang T., Itakura K. Solid-phase synthesis of polynucleotides. III. Synthesis of polynucleotides with defined sequences by the block coupling phosphotriester method. Nucleic Acids Res. 1980 Nov 25;8(22):5491–5505. doi: 10.1093/nar/8.22.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narang S. A., Brousseau R., Hsiung H. M., Michniewicz J. J. Chemical synthesis of deoxyoligonucleotides by the modified triester method. Methods Enzymol. 1980;65(1):610–620. doi: 10.1016/s0076-6879(80)65063-0. [DOI] [PubMed] [Google Scholar]
- Oxender D. L., Zurawski G., Yanofsky C. Attenuation in the Escherichia coli tryptophan operon: role of RNA secondary structure involving the tryptophan codon region. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5524–5528. doi: 10.1073/pnas.76.11.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T. Termination of transcription and its regulation in the tryptophan operon of E. coli. Cell. 1981 Apr;24(1):10–23. doi: 10.1016/0092-8674(81)90496-7. [DOI] [PubMed] [Google Scholar]
- Ryan T., Chamberlin M. J. Transcription analyses with heteroduplex trp attenuator templates indicate that the transcript stem and loop structure serves as the termination signal. J Biol Chem. 1983 Apr 25;258(8):4690–4693. [PubMed] [Google Scholar]
- Schmeissner U., Ganem D., Miller J. H. Genetic studies of the lac repressor. II. Fine structure deletion map of the lacI gene, and its correlation with the physical map. J Mol Biol. 1977 Jan 15;109(2):303–326. doi: 10.1016/s0022-2836(77)80036-3. [DOI] [PubMed] [Google Scholar]
- Stauffer G. V., Zurawski G., Yanofsky C. Single base-pair alterations in the Escherichia coli trp operon leader region that relieve transcription termination at the trp attenuator. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4833–4837. doi: 10.1073/pnas.75.10.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroynowski I., Kuroda M., Yanofsky C. Transcription termination in vitro at the tryptophan operon attenuator is controlled by secondary structures in the leader transcript. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2206–2210. doi: 10.1073/pnas.80.8.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroynowski I., Yanofsky C. Transcript secondary structures regulate transcription termination at the attenuator of S. marcescens tryptophan operon. Nature. 1982 Jul 1;298(5869):34–38. doi: 10.1038/298034a0. [DOI] [PubMed] [Google Scholar]
- Winkler M. E., Mullis K., Barnett J., Stroynowski I., Yanofsky C. Transcription termination at the tryptophan operon attenuator is decreased in vitro by an oligomer complementary to a segment of the leader transcript. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2181–2185. doi: 10.1073/pnas.79.7.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler M. E., Yanofsky C. Pausing of RNA polymerase during in vitro transcription of the tryptophan operon leader region. Biochemistry. 1981 Jun 23;20(13):3738–3744. doi: 10.1021/bi00516a011. [DOI] [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981 Feb 26;289(5800):751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- Zurawski G., Brown K., Killingly D., Yanofsky C. Nucleotide sequence of the leader region of the phenylalanine operon of Escherichia coli. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4271–4275. doi: 10.1073/pnas.75.9.4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski G., Yanofsky C. Escherichia coli tryptophan operon leader mutations, which relieve transcription termination, are cis-dominant to trp leader mutations, which increase transcription termination. J Mol Biol. 1980 Sep 5;142(1):123–129. doi: 10.1016/0022-2836(80)90210-7. [DOI] [PubMed] [Google Scholar]


