Abstract
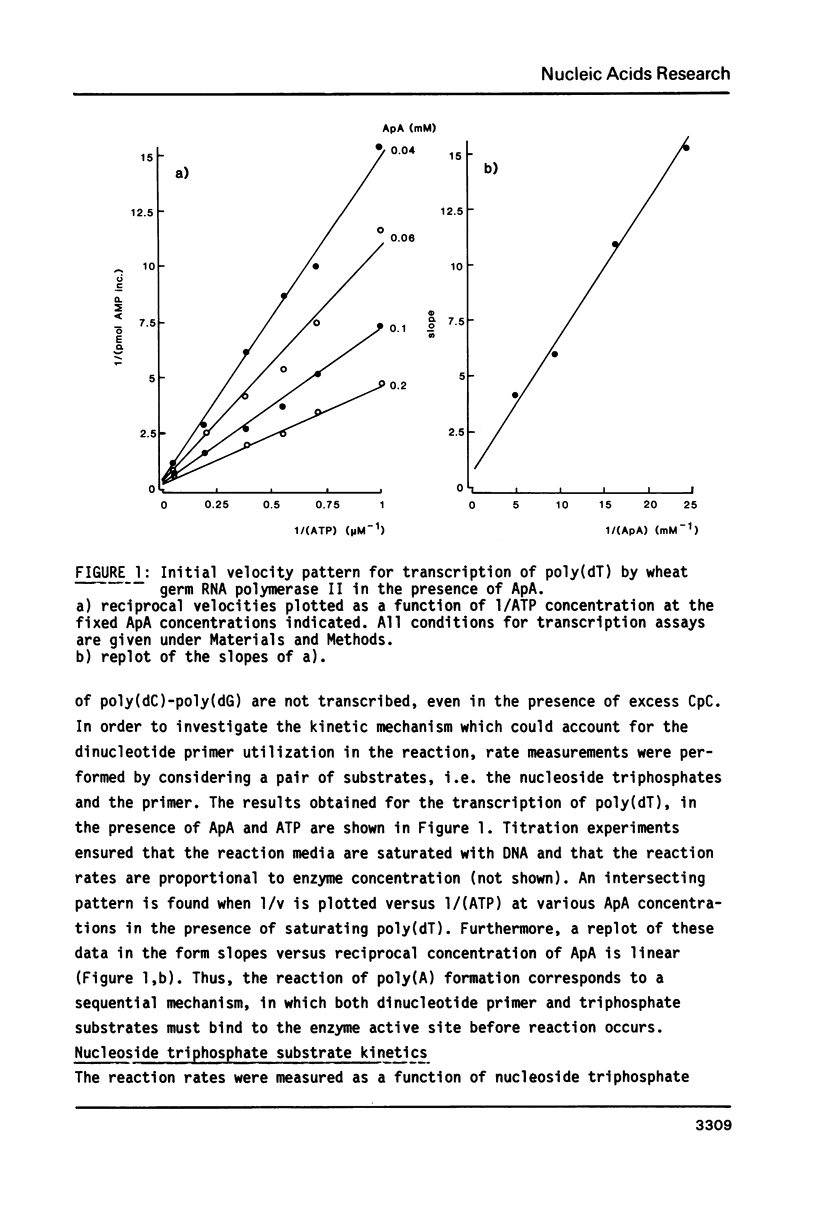
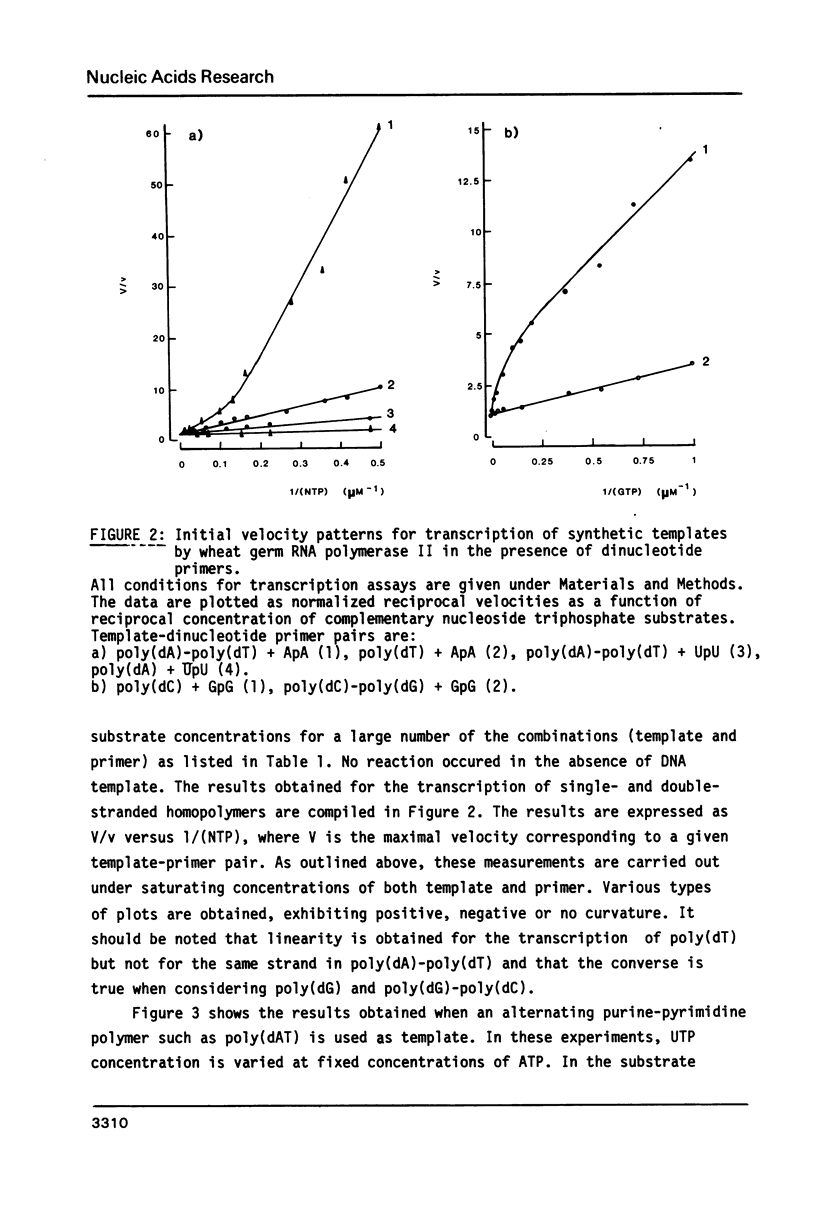
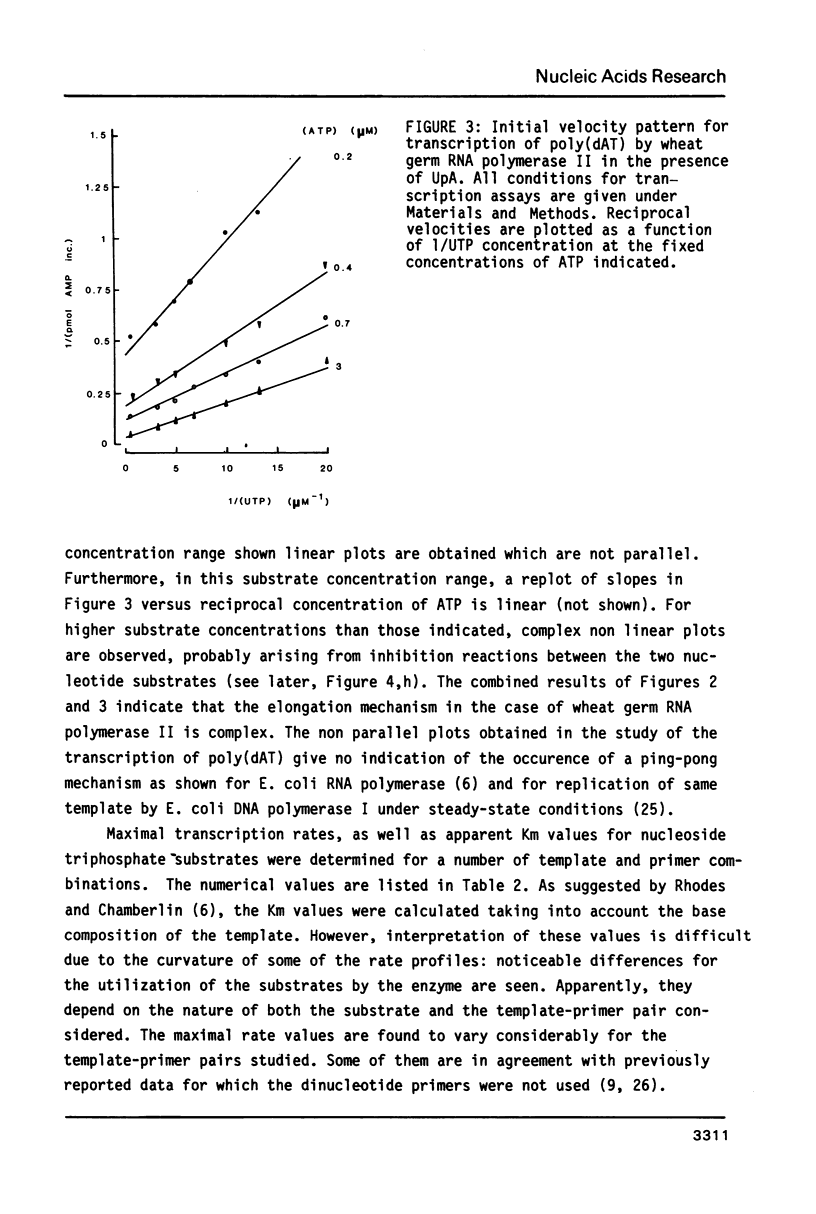
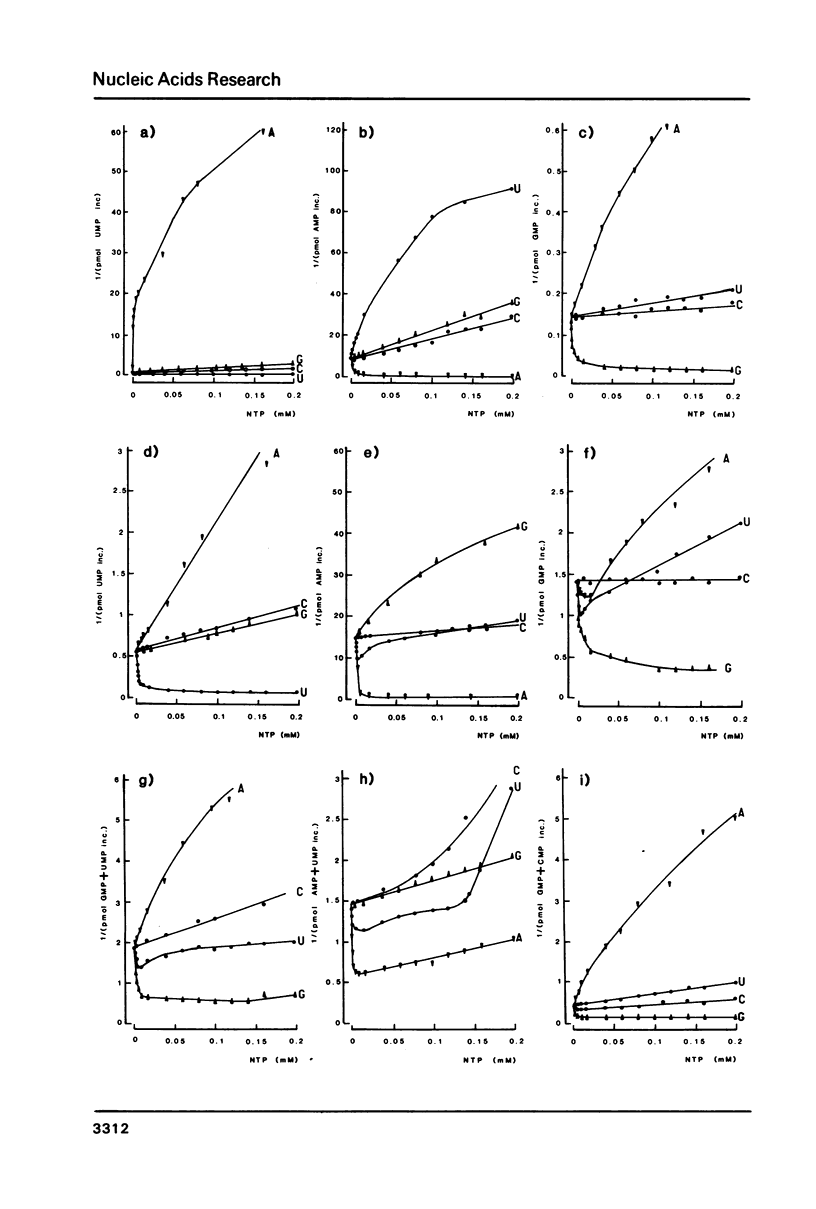
Kinetics of RNA chain elongation catalyzed by wheat germ RNA polymerase II have been studied using various synthetic DNA templates in the presence of excess dinucleotide monophosphate primers. With single- or double-stranded homopolymer templates, the double reciprocal plots 1/(velocity) as a function of 1/(nucleotide substrate) exhibit positive, negative or no curvature. With poly(dAT) as template, the mechanism of nucleoside monophosphate incorporation into RNA is not the ping-pong kinetic mechanism which was derived for E. coli RNA polymerase (6). Noncomplementary nucleoside triphosphates inhibit RNA transcription allosterically. Cordycepin triphosphate behaves as ATP, and not only inhibits AMP incorporation but also that of UMP and GMP on appropriate templates. The reason for this complex kinetic behavior is not yet understood. Possibilities are raised that there are several nucleoside triphosphate binding sites on wheat germ RNA polymerase II, that additional nucleoside triphosphate dependent enzymatic activities are required for reaction to occur or that the Km value for incorporation of a given nucleoside monophosphate into RNA is dependent on the length of the RNA chain and/or the nucleotide sequence surrounding the complementary base on the DNA template.
Full text
PDF










Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackerman S., Bunick D., Zandomeni R., Weinmann R. RNA polymerase II ternary transcription complexes generated in vitro. Nucleic Acids Res. 1983 Sep 10;11(17):6041–6064. doi: 10.1093/nar/11.17.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony D. D., Goldthwait D. A., Wu C. W. Studies with the ribonucleic acid polymerase. II. Kinetic aspects of initiation and polymerization. Biochemistry. 1969 Jan;8(1):246–256. doi: 10.1021/bi00829a035. [DOI] [PubMed] [Google Scholar]
- Badaracco G., Plevani P., Cassani G. Stimulation of poly(dT) transcription by Bacillus subtilis RNA polymerase in the presence of adenosine monophosphate. Biochem Biophys Res Commun. 1981 Mar 16;99(1):23–29. doi: 10.1016/0006-291x(81)91707-1. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bull P., Garrido J. Structure of yeast RNA polymerases determined by electron microscopy. Arch Biochem Biophys. 1982 Nov;219(1):163–166. doi: 10.1016/0003-9861(82)90145-x. [DOI] [PubMed] [Google Scholar]
- Cooke R. M., Penon P., Got C., Miassod R. Selective transcription of a cloned cauliflower mosaic virus DNA fragment in vitro by soybean RNA polymerase II in the presence of dinucleotide primers. Eur J Biochem. 1983 Dec 1;137(1-2):365–371. doi: 10.1111/j.1432-1033.1983.tb07837.x. [DOI] [PubMed] [Google Scholar]
- Dauphinais C. The control of ribosomal RNA transcription in lymphocytes. Evidence that the rate of chain elongation is the limiting factor. Eur J Biochem. 1981 Mar;114(3):487–492. doi: 10.1111/j.1432-1033.1981.tb05171.x. [DOI] [PubMed] [Google Scholar]
- Dennis D., Sylvester J. E. RNA polymerase: a model for rotational translocation. FEBS Lett. 1981 Feb 23;124(2):135–139. doi: 10.1016/0014-5793(81)80121-4. [DOI] [PubMed] [Google Scholar]
- Downey K. M., Jurmark B. S., So A. G. Determination of nucleotide sequences at promoter regions by the use of dinucleotides. Biochemistry. 1971 Dec 21;10(26):4970–4975. doi: 10.1021/bi00802a021. [DOI] [PubMed] [Google Scholar]
- Downey K. M., So A. G. Studies on the kinetics of ribonucleic acid chain initiation and elongation. Biochemistry. 1970 Jun 9;9(12):2520–2525. doi: 10.1021/bi00814a019. [DOI] [PubMed] [Google Scholar]
- Durand R., Job C., Teissère M., Job D. Non-processive transcription of poly[d(A-T)] by wheat germ RNA polymerase II. FEBS Lett. 1982 Dec 27;150(2):477–481. doi: 10.1016/0014-5793(82)80793-x. [DOI] [PubMed] [Google Scholar]
- Durand R., Job C., Zarling D. A., Teissère M., Jovin T. M., Job D. Comparative transcription of right- and left-handed poly[d(G-C)] by wheat germ RNA polymerase II. EMBO J. 1983;2(10):1707–1714. doi: 10.1002/j.1460-2075.1983.tb01646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann K., Seitz H. U. Cooperative effects of RNA polymerase from higher plant cells and Escherichia coli: a comparison. FEBS Lett. 1980 Jul 28;116(2):193–195. doi: 10.1016/0014-5793(80)80641-7. [DOI] [PubMed] [Google Scholar]
- Grossmann K., Seitz U. RNA polymerase I from higher plants. Evidence for allosteric regulation and interaction with a nuclear phosphatase activity controlled NTP pool. Nucleic Acids Res. 1979 Dec 11;7(7):2015–2029. doi: 10.1093/nar/7.7.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C. Y., Dennis D. RNA polymerase: linear competitive inhibition by bis-(3' to 5')-cyclic dinucleotides,NpNp. Nucleic Acids Res. 1982 Sep 25;10(18):5637–5647. doi: 10.1093/nar/10.18.5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendrisak J. J., Burgess R. R. A new method for the large-scale purification of wheat germ DNA-dependent RNA polymerase II. Biochemistry. 1975 Oct 21;14(21):4639–4645. doi: 10.1021/bi00692a012. [DOI] [PubMed] [Google Scholar]
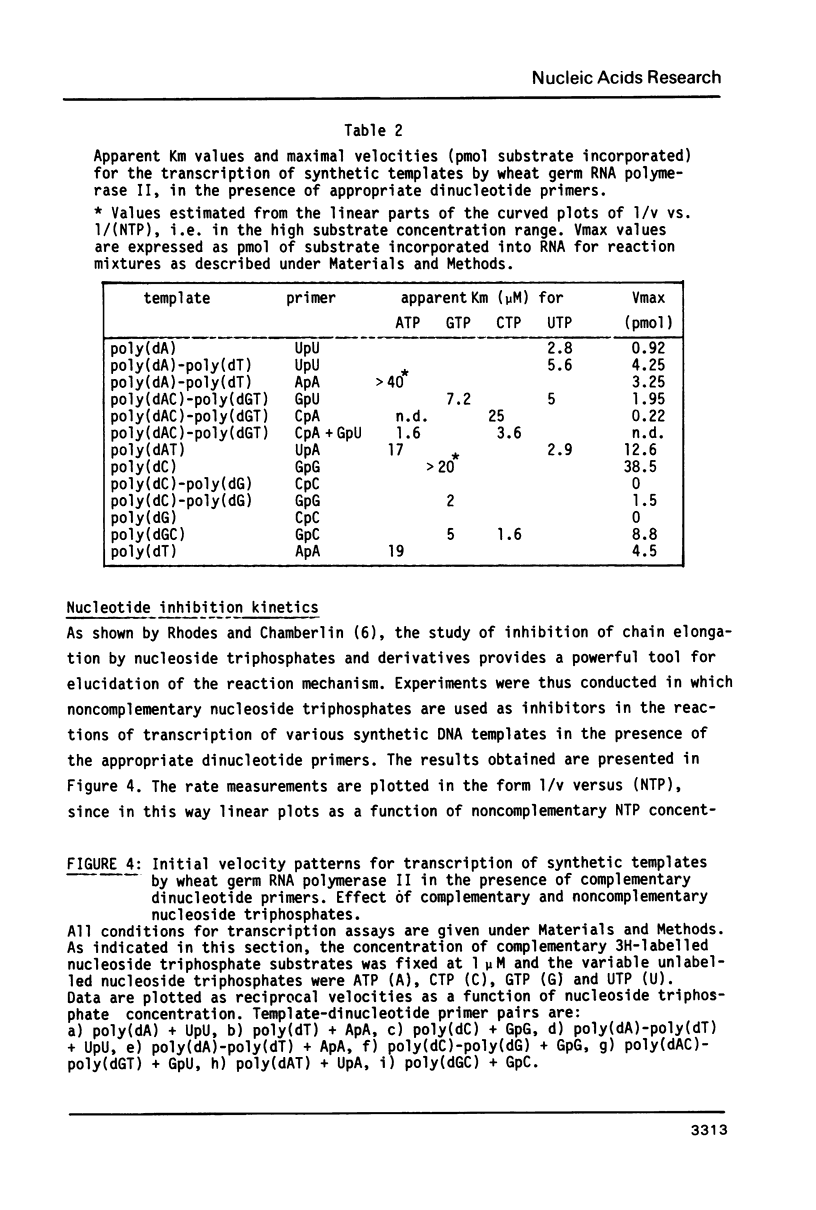
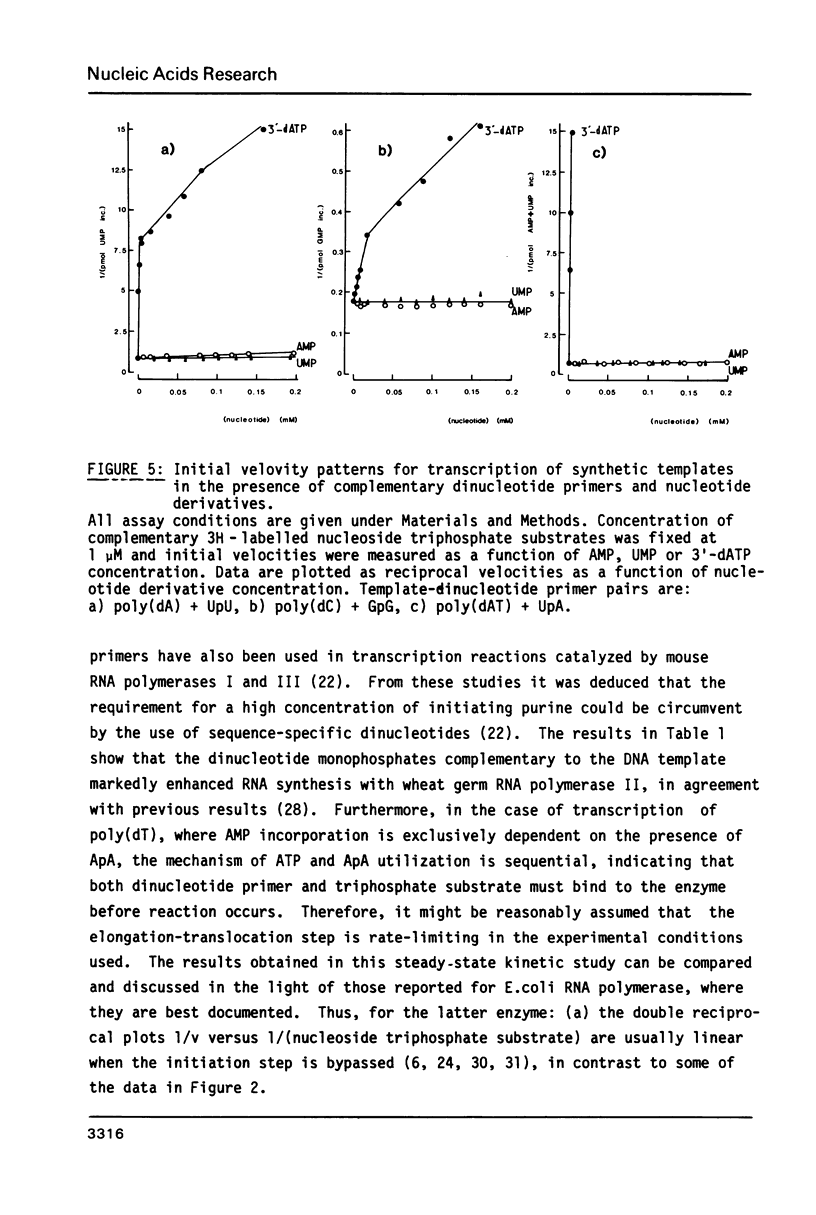
- Job D., Durand R., Teissere M. Enzymatic properties and cooperative effects in the kinetics of wheat-germ RNA polymerases. A comparative study of the three nuclear enzyme classes. Eur J Biochem. 1982 Nov;128(1):35–39. doi: 10.1111/j.1432-1033.1982.tb06927.x. [DOI] [PubMed] [Google Scholar]
- Kumar S. A. The structure and mechanism of action of bacterial DNA-dependent RNA polymerase. Prog Biophys Mol Biol. 1981;38(3):165–210. doi: 10.1016/0079-6107(81)90013-4. [DOI] [PubMed] [Google Scholar]
- Lescure B., Bennetzen J., Sentenac A. In vitro transcription of the yeast alcohol dehydrogenase I gene by homologous RNA polymerase B (II). Selective initiation and discontinuous elongation on a supercoiled template. J Biol Chem. 1981 Nov 10;256(21):11018–11024. [PubMed] [Google Scholar]
- Lescure B., Williamson V., Sentenac A. Efficient and selective initiation by yeast RNA polymerase B in a dinucleotide-primed reaction. Nucleic Acids Res. 1981 Jan 10;9(1):31–45. doi: 10.1093/nar/9.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure W. R., Jovin T. M. The steady state kinetic parameters and non-processivity of Escherichia coli deoxyribonucleic acid polymerase I. J Biol Chem. 1975 Jun 10;250(11):4073–4080. [PubMed] [Google Scholar]
- NIYOGI S. K., STEVENS A. STUDIES OF THE RIBONUCLEIC ACID POLYMERASE FROM ESCHERICHIA COLI. 3. STUDIES WITH SYNTHETIC POLYRIBONUCLEOTIDES AS TEMPLATES. J Biol Chem. 1965 Jun;240:2587–2592. [PubMed] [Google Scholar]
- Nierman W. C., Chamberlin M. J. Studies of RNA chain initiation by Escherichia coli RNA polymerase bound to T7 DNA. Direct analysis of the kinetics and extent of RNA chain initiation at T7 promoter A1. J Biol Chem. 1979 Aug 25;254(16):7921–7926. [PubMed] [Google Scholar]
- Nierman W. C., Chamberlin M. J. The effect of low substrate concentrations on the extent of productive RNA chain initiation from T7 promoters A1 and A2 by Escherichia coli RNA polymerase. J Biol Chem. 1980 May 25;255(10):4495–4500. [PubMed] [Google Scholar]
- Ninio J., Bernardi F., Brun G., Assairi L., Lauber M., Chapeville F. On the mechanism of nucleotide incorporation into DNA and RNA. FEBS Lett. 1975 Sep 15;57(2):139–144. doi: 10.1016/0014-5793(75)80702-2. [DOI] [PubMed] [Google Scholar]
- Rackwitz H. R., Rohde W., Sänger H. L. DNA-dependent RNA polymerase II of plant origin transcribes viroid RNA into full-length copies. Nature. 1981 May 28;291(5813):297–301. doi: 10.1038/291297a0. [DOI] [PubMed] [Google Scholar]
- Rhodes G., Chamberlin M. J. Ribonucleic acid chain elongation by Escherichia coli ribonucleic acid polymerase. I. Isolation of ternary complexes and the kinetics of elongation. J Biol Chem. 1974 Oct 25;249(20):6675–6683. [PubMed] [Google Scholar]
- Rose K. M., Stetler D. A., Jacob S. T. Protein kinase activity of RNA polymerase I purified from a rat hepatoma: probable function of Mr 42,000 and 24,600 polypeptides. Proc Natl Acad Sci U S A. 1981 May;78(5):2833–2837. doi: 10.1073/pnas.78.5.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saneyoshi M., Tohyama J., Nakayama C., Takiya S., Iwabuchi M. Inhibitory effects of 3'deoxycytidine 5'-triphosphate and 3'-deoxyuridine 5'-triphosphate on DNA-dependent RNA polymerases I and II purified from Dictyostelium discoideum cells. Nucleic Acids Res. 1981 Jul 10;9(13):3129–3138. doi: 10.1093/nar/9.13.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y., Goto H., Ohta H., Kamikubo T. Template activity of synthetic deoxyribonucleotide polymers in the eukaryotic DNA-dependent RNA polymerase reaction. Eur J Biochem. 1976 Nov 15;70(2):369–375. doi: 10.1111/j.1432-1033.1976.tb11026.x. [DOI] [PubMed] [Google Scholar]
- Shaw P. A., Marshall M. V., Saunders G. F. Dinucleotide priming of RNA synthesis. Cytogenet Cell Genet. 1980;26(2-4):211–222. doi: 10.1159/000131442. [DOI] [PubMed] [Google Scholar]
- Shaw P. A., Saunders G. F. Stimulation of RNA synthesis by dinucleotides with eukaryotic RNA polymerase. FEBS Lett. 1979 Oct 1;106(1):104–110. doi: 10.1016/0014-5793(79)80704-8. [DOI] [PubMed] [Google Scholar]
- Shimamoto N., Wu C. W. Mechanism of ribonucleic acid chain initiation. 2. A real time analysis of initiation by the rapid kinetic technique. Biochemistry. 1980 Mar 4;19(5):849–856. doi: 10.1021/bi00546a004. [DOI] [PubMed] [Google Scholar]
- Vaisius A. C., Wieland T. Formation of a single phosphodiester bond by RNA polymerase B from calf thymus is not inhibited by alpha-amanitin. Biochemistry. 1982 Jun 22;21(13):3097–3101. doi: 10.1021/bi00256a010. [DOI] [PubMed] [Google Scholar]
- Volloch V. Z., Rits S., Tumerman L. A possible mechanism responsible for the correction of transcription errors. Nucleic Acids Res. 1979 Apr;6(4):1535–1546. doi: 10.1093/nar/6.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson J. A., Miller K. G., Sollner-Webb B. Dinucleotide primers facilitate convenient identification of the mouse ribosomal DNA transcription initiation site. A general method for analysis of transcription by RNA polymerases I and III. J Biol Chem. 1983 Nov 25;258(22):13919–13928. [PubMed] [Google Scholar]
- Yarbrough L. R. Utilization of primers and primer-templates by wheat germ RNA polymerase II. J Biol Chem. 1982 Jun 10;257(11):6171–6177. [PubMed] [Google Scholar]


