Abstract
Proteolytic processing of viral membrane proteins is common among enveloped viruses and facilitates virus entry. The Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) occlusion-derived virus (ODV) protein P74 is part of a complex of essential peroral infectivity factors (PIFs). Here we report that P74 is efficiently cleaved into two fragments of about equal size by an occlusion body (OB) endogenous alkaline protease during ODV release when AcMNPV OBs are derived from larvae. The cleavage is specific for P74, since the other known peroral infectivity factors in the same complex (PIF1, PIF2, and PIF3) were not cleaved under the same conditions. P74 cleavage was not observed in OBs produced in three different insect cell lines, suggesting a larval host origin of the responsible protease. P74 in OBs produced in larvae of two different host species was cleaved into fragments with the same apparent molecular mass, indicating that the virus incorporates a similar alkaline protease from different hosts. Coimmunoprecipitation analysis revealed that the two P74 subunit fragments remain associated with the recently discovered PIF complex. We propose that under in vivo ODV infection conditions, P74 undergoes two sequential cleavage events, the first one being performed by an ODV-associated host alkaline protease and the second carried out by trypsin in the host midgut.
INTRODUCTION
For many enveloped viruses, proteolytic processing of virus membrane proteins is required to facilitate virus entry. In principle, the proteolytic cleavage converts a proprotein into an active conformation and/or exposes the functional domain, e.g., the fusion domain, to mediate virus binding and/or fusion. Cleavage of these proproteins may occur posttranslationally during transportation of the protein through the trans-Golgi network. The F protein of many paramyxoviruses, for instance, is cleaved by the host protein furin (35). Alternatively, the protein is cleaved during virus entry either at the surface of recipient cells, such as influenza virus hemagglutinin (HA), which is cleaved by human airway trypsin-like protease (HAT) (6), or in the endosome, for example, the cleavage of the severe acute respiratory syndrome coronavirus (SARS-CoV) S protein by cathepsin L (31).
Baculoviruses produce two viral phenotypes that follow different routes of infection (reviewed by Slack and Arif [32]). The occlusion-derived virus (ODV) derived from occlusion bodies (OBs) initiates infection in midgut epithelial cells of host larvae, and the budded virus (BV) is involved in systemic spread of the viral infection within the host. Like the F protein of paramyxoviruses, the BV fusion (F) protein of Spodoptera exigua multicapsid nucleopolyhedrovirus (SeMNPV) is posttranslationally cleaved by furin, and this cleavage is essential for the function of F (43).
With baculovirus ODVs, the situation is more complex. ODVs are embedded in a large proteinaceous crystal to form OBs. After being ingested by the insect host per os, the crystal structure of the OBs dissociates under the alkaline conditions in the midgut (pH 9 to 11), thereby releasing the ODVs (reviewed in reference 30). These ODVs then bind and fuse with the microvilli of midgut epithelium cells to start the first round of infection (32), and this process is known as oral infection. So far, six ODV membrane proteins have been found to be essential for oral infectivity, and these were named per os infectivity factors (PIFs; described by Kikhno et al. [19]). They are P74 (PIF0), PIF1, PIF2, PIF3, PIF4, and PIF5 (ODV-E56) (9, 11, 17, 19, 23, 28). Three PIFs, P74, PIF1, and PIF2, have been shown to function in ODV binding (16, 23), while the function(s) of the other three PIFs is still unknown. Recently, Peng et al. (26) reported that at least four of these PIFs, PIF1, -2, and -3 and P74, are present in the ODV membrane in the form of a complex. The P74 protein, representative of a class of highly conserved proteins among baculoviruses, was reported to undergo a proteolytic cleavage mediated by insect midgut trypsins, releasing a ≈20-kDa fragment from the N terminus of P74. This cleavage was shown to be essential for P74-mediated per os infectivity (34).
For a number of baculoviruses, including AcMNPV, an alkaline protease was found to be associated with larvae-derived OBs (L-OBs) (7, 8, 20, 21, 25, 38, 44). This protease was suggested to function in the degradation of the major matrix protein of OBs (polyhedrin) and/or to assist in the release of ODVs (25, 38). However, the functional significance of the presence of this alkaline protease in OBs is not yet fully understood. Recently, Slack and Arif (32) envisioned that this OB-associated alkaline protease could play a synergistic role in infection by proteolytic activation of released ODVs, but experimental data to support this supposition are lacking.
In this study we provide evidence that a potential host-derived alkaline protease associated with AcMNPV L-OBs cleaves P74 efficiently and specifically during ODV release under alkaline conditions. We propose a sequential proteolytic cleavage model of P74 accommodating both the OB endogenous alkaline protease and midgut host trypsin.
MATERIALS AND METHODS
Viruses, cells, and insects.
The AcMNPV E2 strain was used as wild-type (wt) virus in this study. The AcMNPV bacmid is derived from the Bac-to-Bac system (Invitrogen). Spodoptera frugiperda Sf9 cells (Invitrogen) were propagated in Sf-900II medium (Invitrogen) with 5% fetal bovine serum (FBS). BTI-Tn-5B1-4 cells (Tn-High Five; Invitrogen) were grown in Express Five SFM medium (Invitrogen) and Se-UCR (15) in Grace's insect medium (Sigma) supplemented with 5% FBS. All three cell lines were propagated as monolayers at 27°C. Spodoptera exigua and Trichoplusia ni larvae were reared on an artificial diet at 27°C, 40% humidity, with a 16/8 h (light/dark) photoperiod. In this study, OBs produced from cell culture or larvae have been named C-OB or L-OB, respectively. Correspondingly, ODVs purified from C-OB or L-OB are named C-ODV or L-ODV. The origin of OBs and ODVs is also indicated, e.g., Sf9-C-ODV indicates that the ODVs are purified from C-OBs produced in Sf9 cells, while Se-L-OB indicates that the L-OBs are produced in S. exigua larvae.
P74 deletion and repair viruses.
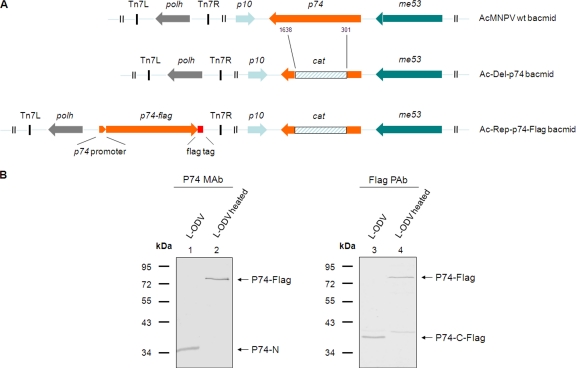
An AcMNPV bacmid with a deletion of the p74 open reading frame (ORF) was constructed as previously described (26). For this, a PCR product with 50-bp overhangs homologous to flanking regions of the p74 gene was generated with primer pair p74-del-F (GCGGCGTCGTGTCCAACACGACGCCGTTCATGTACATGCAGACCTCCGAGACTGCTCGGAT CCACTAGTAACG) and p74-del-R (GCGTATAGCGAGCTAGTGGCTAACGCTTGCCCCACCAAAGTAGATTCGTCAAACCTCTAGATGCATGC TCG) with Phusion polymerase (Finnzymes). Primers were designed to replace a fragment from nucleotide (nt) 301 to 1638 of the p74 ORF with the chloramphenicol resistance gene (cat). Primer sequences for generating the cat gene are underlined. To repair the p74 deletion bacmid, the coding sequence of the p74 gene plus its putative promoter region (from position −150 relative to the ATG start codon) were amplified by PCR using the primer pair p74-rep-F (GCGCCATGGGCACAACGAAATGATTATATATTA) and p74-rep-R (GCGGCATGCTTATTTGTCATCGTCATCCTTATAGTCAAATAACAAATCAATTGTTTTATAAT). NcoI and SphI restriction sites were introduced by the underlined sequences of the primers and used to clone the sequenced PCR product into a modified pFastBacDual vector that lacked the p10 promoter and contains the entire polyhedrin gene (pFBDΔP10-polh) as described before (26). A Flag tag sequence (shown in italics in the sequence) was introduced by the reverse repair primer to generate a P74 protein with a C-terminal Flag tag. The resulting plasmid was used to construct a “p74 repair” bacmid using the Bac-to-Bac transposition protocol (Invitrogen). The bacmids were used to transfect Sf9 cells to generate mutant and repair viruses.
Virus production and purification.
A wt AcMNPV BV stock was generated as previously described (26). To produce and purify C-OBs, Sf9, Tni-High Five, or Se-UCR cells were infected with BVs at a multiplicity of infection of 5 50% tissue culture infectious dose units per cell, and infected cells were collected at 5 days postinfection by low-speed centrifugation. The cell pellet was washed 3 times with deionized water (Millipore) and then resuspended in 0.2% SDS and incubated at 37°C for 2 h or 4°C overnight with gentle rocking. The cell suspension was sonicated and centrifuged at 4,000 × g for 20 min. The pellet was washed and resuspended in phosphate-buffered saline (PBS) and centrifuged over a 35-to-65% (wt/wt) sucrose gradient at 98,000 × g for 60 min at 4°C. The banded OBs were diluted in PBS and collected by centrifugation at 55,000 × g for 60 min. The OB pellet was washed 3 times with PBS and stored at 4°C for later use.
L-OBs were produced in S. exigua or T. ni larvae (third instar) after oral infection with C-OBs produced in Sf9 cells. To purify L-OBs from infected larvae, the liquefied or liquefying larvae were collected and ground in deionized water containing 0.5% SDS and 1% Triton X-100. The suspension was homogenized, sonicated, and passed through two layers of cheesecloth. The L-OB-containing material was then centrifuged at 4,000 × g for 20 min. The pellet was washed twice with 0.5% SDS–1% Triton X-100 in deionized water, resuspended in PBS, and purified through a sucrose gradient in the same way as C-OBs. The resulting L-OB pellet was washed twice with 1 M NaCl and once with PBS. Finally, the L-OBs were stored in PBS.
ODV purification under various conditions.
Prior to ODV purification, the PBS in the OB suspensions was replaced with deionized water. For heat inactivation of the endogenous protease, the L-OB suspension was heated at 80°C for 40 min (heated OB) prior to ODV purification (25). ODVs were either purified at room temperature or at 4°C (low-temperature purification). For low-temperature purification, OBs were incubated on ice water (0°C) for 40 min. OBs were then treated with cold (4°C) alkaline DAS solution (0.1 M Na2CO3, 166 mM NaCl, and 10 mM EDTA; pH 10.5) for 10 min at 4°C, and subsequent steps of purification were also performed at 4°C. In experiments in which the endogenous protease needed to be inhibited, NaOH was added to the ODV-releasing solution at a final concentration of 0.1 M at various time points after the addition of DAS. For time point 0 min, NaOH was mixed with DAS solution, and this mixture was used to dissolve OBs. Nondissolved debris was removed by centrifugation at 1,500 × g for 3 min. The supernatant was collected, and ODVs were pelleted by centrifugation at 20,600 × g for 25 min at 4°C.
In experiments where ODVs needed to be purified over a sucrose gradient, ODVs were released from OBs at 4°C, and the ODV-containing supernatant was layered onto a 25-to-65% (wt/wt) continuous sucrose gradient in 10 mM Tris-HCl, pH 7.5, and centrifuged at 98,000 × g for 90 min at 4°C. The multiple virus bands were collected, diluted in 0.1× TE (1 mM Tris-HCl [pH 7.5], 0.1 mM EDTA), and collected by centrifugation at 55,000 × g for 60 min at 4°C.
Coimmunoprecipitation.
For coimmunoprecipitation (co-IP) analysis, Se-L-OB of the Ac-Rep-P74-Flag virus was used in order to detect both P74-N and P74-C-Flag subunits. For each experiment L-ODVs were liberated from 2 × 109 L-OBs, and the L-ODVs were purified through a sucrose gradient as described above. The ODV pellet was resuspended in 600 μl IP buffer (25 mM Tris-HCl, 150 mM NaCl; pH 7.2) containing 0.5% Triton X-100, sonicated briefly, and incubated at 4°C for 3 h with gentle rotation. Meanwhile, 15 μl of PIF1 antiserum or preimmune serum (26) was incubated with a 20-μl bed volume of protein G-agarose (Pierce) in 500 μl of IP buffer at 4°C for 3 h. Subsequent steps of co-IP were performed as previously described (26). A portion of the supernatant was reserved as input sample. The captured proteins and input sample were treated with Laemmli buffer (125 mM Tris-HCl, 2% sodium dodecyl sulfate, 5% 2-mercaptoethanol, 10% glycerol, 0.001% bromophenol blue; pH 6.8) at 95°C for 10 min. Proteins were then separated by 10% SDS-PAGE.
Polyclonal antibody generation for P74.
A 5′ segment of 1,299 bp of the p74 ORF was selected for making an expression construct for P74 polyclonal antibody (P74-PAb) generation. The DNA fragment was PCR amplified with the primers P74-Ab-F (GCGGGATCCATGGCGGTTTTAACAGCC) and P74-Ab-R (GCGAAGCTTTTAAGCGATTCGAGTTAACGCT). BamHI and HindIII restriction sites (underscored) were introduced by the forward and reverse primers, respectively. The amplified DNA was ligated into the pJet1.2 vector (Fermentas) for sequencing analysis and then cloned into the pET28a vector (Novagen) between the BamHI and HindIII sites for protein expression. The pET28a-P74 construct codes for an N-terminal fusion of a 6×His tag with the AcMNPV P74 fragment. Escherichia coli BL21 cells were transformed with pET28a-P74, and expression was induced with isopropyl-β-d-thiogalactopyranoside (IPTG; 1 mM) for 4 h at 37°C. After washing with PBS three times, the bacteria pellet was resuspended in lysis buffer (0.1% SDS, 1% Triton X-100, PBS [pH 7.4]), incubated at 37°C with gentle rotation for 30 min, and then sonicated. Protein purification from the bacterial inclusion bodies was performed as previously described (26). The purified protein was analyzed with SDS-PAGE, Coomassie brilliant blue staining, and Western blotting with anti-His and P74 monoclonal antibodies (P74 MAb) to determine the specificity, purity, and concentration of the protein. Purified protein was sent to Eurogentec (Seraing, Belgium) to generate polyclonal antibodies in rabbits.
SDS-PAGE, antibodies, and Western blot analysis.
ODV samples were heated in Laemmli buffer at 95°C for 10 min and separated by 10% SDS-PAGE. For nonreducing SDS-PAGE, samples were treated with Laemmli buffer lacking 2-mercaptoethanol. The nonreduced samples were also heated at 95°C for 10 min. Anti-His monoclonal antibody from mouse and anti-Flag polyclonal antibody from rabbit were purchased from Sigma. The P74 MAb (11), anti-PIF1, anti-PIF2, and anti-PIF3 antibodies have previously been described (26). Western blot analysis was performed as previously described (26, 27). Dilutions of the primary antibodies were as follows: His MAb (1:2,000 dilution), Flag PAb (2 μg/ml), PIF1 PAb (1:2,000 dilution), PIF2 PAb (1:2,000 dilution), PIF3 PAb (1:1,000 dilution), P74 MAb (1:50 dilution), and P74 PAb (1:2,000 dilution). Membranes were then probed with appropriate alkaline phosphatase-conjugated secondary antibodies (Sigma). The signal was developed with nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate (Roche).
Computational analysis.
Conserved domain analysis of the P74 amino acid sequence was performed with the NCBI conserved domain server (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Transmembrane domain prediction of P74 was performed with the TMHMM 2.0 server as described before (27). Disulfide bonds were predicted by using the DiANNA server (http://clavius.bc.edu/∼clotelab/DiANNA/) (12).
RESULTS
Cleavage of P74 during ODV release from L-OBs.
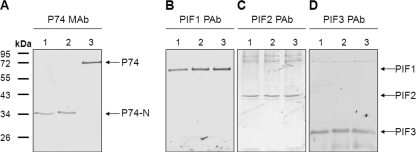
When purified ODV proteins from OBs derived from S. exigua larvae (Se-L-OB; purified at 20°C) were separated by SDS-PAGE and analyzed with P74 MAb, a 35-kDa fragment instead of the 72-kDa full-length P74 protein was detected (Fig. 1A, lane 1). Since the P74 MAb is specific to the N terminus of P74, recognizing an epitope located between S68 and R195 (33), the observed 35-kDa fragment may represent an N-terminal fragment of P74 (P74-N), suggesting proteolytic cleavage of P74. The same ODV protein samples were also analyzed with antibodies against PIF1, PIF2, and PIF3, which are the other three known components of the PIF complex. Under the same purification conditions, these other three PIFs were not cleaved (Fig. 1B to D, lane 1). Cleavage of P74 could be prevented by heating the L-OBs at 80°C for 40 min (Fig. 1A, lane 3). Heating had no effect on the other three PIFs (Fig. 1B to D, lane 3). This indicates that an OB endogenous alkaline protease, which can be inactivated by heating, is responsible for P74 cleavage.
Fig. 1.
P74 is cleaved efficiently and specifically during L-ODV purification. OBs produced from S. exigua larvae were used. L-ODVs were purified under three different conditions. For the samples in lanes 1, L-OBs were dissolved in DAS at room temperature. For the samples in lanes 2, L-OBs were precooled in ice water for 40 min and dissolved with cold DAS (4°C). For the samples in lanes 3, L-OBs were heated at 80°C for 40 min and then dissolved in DAS solution. For all three treatments, L-ODVs were purified at 4°C after DAS treatment. ODV samples were separated by 10% SDS-PAGE and analyzed with P74 MAb (A) or PIF1, PIF2, and PIF3 antibodies (B, C, and D, respectively). Positions of full-length P74, P74-N, PIF1, PIF2, and PIF3 are indicated by arrows. P74-N represents the N-terminal P74 fragment after cleavage.
In previous reports the temperature optimum of the endogenous alkaline protease of L-OBs was reported to be 30 to 40°C (25, 39). Therefore, L-ODVs were purified under low-temperature conditions with the intention to avoid P74 cleavage. L-OBs were incubated at 0°C for 40 min and exposed to cold DAS solution (4°C), and ODVs were purified at 4°C. However, as shown in Fig. 1A, lane 2, even under the low-temperature condition P74 was cleaved completely. Again, the other three PIFs were not affected (Fig. 1B to D, lane 2). These results indicate that the P74 cleavage is not only specific but also very efficient.
No P74 cleavage with OBs produced in cell lines.
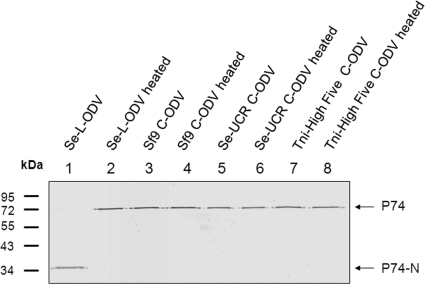
To address the question whether P74 cleavage also occurs in OBs produced in cell cultures (C-OBs), AcMNPV OBs were produced in Sf9, Tni-High Five, and Se-UCR cell lines. ODVs were purified from Se-L-OBs and the different C-OBs with (Fig. 2, lanes 2, 4, 6, and 8) or without (Fig. 2, lanes 1, 3, 5, and 7) heat treatment. P74 in these ODVs was analyzed with P74 MAb. In C-ODVs from three different cell lines, full-length P74 was detected whether or not OBs were treated at 80°C prior to ODV purification (Fig. 2, lanes 3 to 8). Cleavage of P74 did not happen in C-OBs even when the OBs were dissolved at 37°C and ODVs were purified at room temperature (data not shown). In contrast, P74 on L-ODV was cleaved completely (Fig. 2, lanes 1) in nonheated Se-L-OB. As before, heating of Se-L-OB prior to ODV purification prevented P74 cleavage, and full-length P74 was detected (Fig. 2, lane 2). These results suggest that the protease responsible for P74 cleavage is derived from the larval host and not of viral origin.
Fig. 2.
P74 cleavage does not happen in C-OBs produced in three cell lines. LOBs produced in S. exigua larvae and C-OBs produced in Sf9, Tni-High Five, or Se-UCR cells were used in this experiment. OBs were directly used for ODV purification or first heated at 80°C for 40 min before dissolving in DAS at 20°C. Purified ODVs were analyzed by 10% SDS-PAGE, and Western blot analysis was performed with P74 MAb. The full-length P74 and P74-N fragments are indicated by arrows.
P74 cleavage is conserved in AcMNPV L-OBs from two different host larvae.
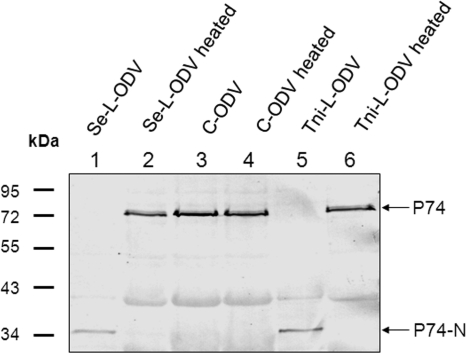
Since Se-L-OBs appear to contain a host protease that cleaves P74, the next question is whether L-OBs derived from different hosts would incorporate similar proteases to cleave P74 in the same way. To address this question, AcMNPV OBs were also produced in T. ni larvae. Like Se-L-OBs, OBs produced from T. ni larvae were named Tni-L-OBs. ODVs were purified from heated or nonheated Se-L-OBs, Tni-L-OBs, and Sf9-C-OBs and analyzed with P74 MAb in immunoblot assays. P74 in nonheated Se- or Tni-L-OBs was cleaved completely (Fig. 3, lanes 1 and 5), and in each case the P74-N fragment migrated with a molecular mass of approximately 35 kDa. Heat treatment blocked P74 cleavage in both Se- and Tni-L-OBs, and full-length P74 was detected (Fig. 3, lanes 2 and 6). In contrast, P74 on C-ODVs was not cleaved and was present as a full-length protein no matter whether the Sf9-C-OBs were heated or not (Fig. 3, lanes 3 and 4). The fact that P74 in L-OBs derived from different hosts was cleaved into similar, if not identical, protein fragments suggests that similar proteases from different hosts are incorporated into AcMNPV OBs. Alternatively, the cleaved region of P74 is sensitive to and can be cleaved by different proteases.
Fig. 3.
P74 cleavage is conserved in L-OBs produced in two different hosts. L-OBs were purified from infected S. exigua and T. ni larvae. Sf9-C-OBs were included as a control. OBs were either treated with DAS to purify ODVs directly or were heated at 80°C for 40 min before dissolving in DAS. Purified ODVs were separated by 10% SDS-PAGE. Western analysis was performed with P74 MAb. Arrows indicate the positions of full-length P74 and the cleaved P74-N fragment.
Cleavage of P74 is highly efficient.
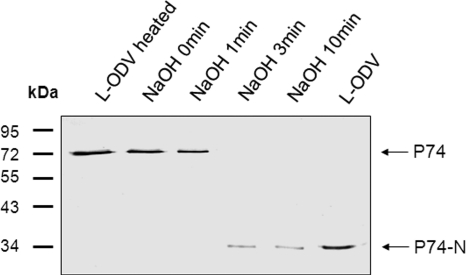
To further estimate the efficiency of P74 cleavage, the endogenous alkaline protease was inactivated with NaOH during ODV release from OBs as previously reported (25). To this aim, L-OBs were exposed to DAS solution supplemented with NaOH to a final concentration of 0.1 M at different times after exposure to DAS. The purified ODVs were then analyzed with P74 MAb. As shown in Fig. 4, if NaOH was added to the DAS solution at 0 or 1 min, only full-length P74 was detected (Fig. 4, lanes 2 and 3), indicating inactivation of the protease. However, if NaOH was added after L-OBs were exposed to DAS solution for 3 or 10 min, only the P74-N cleaved fragment was detected (Fig. 4, lanes 4 and 5). When L-ODVs were purified without prior heat treatment and without NaOH treatment, P74 was also cleaved completely (Fig. 4, lane 6). Like before, when L-OBs were heated prior to ODV purification, full-length P74 was detected (Fig. 4, lane 1). These results showed that the alkaline protease can be inactivated by NaOH, and P74 cleavage was accomplished within 3 min after L-OBs were exposed to alkaline conditions (DAS). It should be noted that in NaOH-treated samples (Fig. 4, lanes 4 and 5), the signal intensity of the P74-N fragment was lower than that of the untreated sample (Fig. 4, lane 6) despite the same amount of OBs being used. The pH of the DAS solution mixed with 0.1 M NaOH was around 12.8, while the pH of DAS solution alone is around 10.5. It is possible that the higher pH in the NaOH-treated solutions weakened the association between the P74-N fragment and the ODV and, therefore, less P74-N fragment was copurified with ODVs. Protein-protein interactions are often sensitive to pH, as has been previously reported (18, 22, 29, 36).
Fig. 4.
P74 cleavage is highly efficient. Se-L-OB was used in this experiment. Heated OBs were incubated at 80°C for 40 min before ODV purification. L-OBs were dissolved in DAS, and NaOH was added to the OB dissolving solution to a final concentration of 0.1 M at the indicated time after adding DAS. For the sample “NaOH 0 min,” NaOH was mixed with DAS and the mixed solution was used to dissolve L-OBs. Purified ODVs were separated by 10% SDS-PAGE, and Western analysis was performed with P74 MAb. Full-length P74 and the cleaved P74-N fragment are indicated by arrows.
P74 is cleaved into two fragments.
Without detecting the P74-C fragment, it is difficult to judge whether P74 is cleaved into two subunits or whether the C-terminal fragment undergoes further processing. A polyclonal antibody of P74 was generated in this study with the intention to cover more epitopes of P74. However, in practice the PAb only efficiently recognized the P74-N fragment, like the P74 MAb, and a potential P74-C fragment(s) was not identified (data not shown). To address this issue, a bacmid with a deletion of the p74 ORF was constructed, and from this a P74 repair bacmid was made that encoded P74 with a C-terminal Flag tag (Fig. 5A). The recombinant virus Ac-Rep-P74-Flag was as infectious as wt AcMNPV, while the p74-deleted virus was not orally infectious (data not shown). The Ac-Rep-P74-Flag virus was then used for analysis of the cleaved P74-C fragment. L-OBs of Ac-Rep-P74-Flag virus were produced in S. exigua larvae and purified. L-ODVs were then prepared from heated or nonheated L-OBs, and ODV samples were separated by SDS-PAGE and analyzed with P74 MAb and Flag PAb to detect P74-N and P74-C subunits, respectively. With the MAb, a 35-kDa P74-N fragment was detected for the nonheated L-OBs (Fig. 5B, lane 1), while a 75-kDa P74-Flag protein was detected with the heated L-OBs (Fig. 5B, lane 2). When the same samples were probed with Flag PAb, a 40-kDa P74-C fragment (P74-C-Flag) and a 75-kDa P74-Flag protein were detected in the nonheated and heated samples, respectively (Fig. 5B, lanes 3 and 4). The molecular masses of P74-N and P74-C-Flag added up approximately to the molecular mass of full-length P74-Flag, about 75 kDa (the predicted size is 74.8 kDa). These results strongly suggest that P74-Flag was cleaved into two fragments: P74-N and P74-C-Flag.
Fig. 5.
P74 is cleaved into two subunits. (A) Schematic of the p74 recombinant bacmids constructed in this study. The orientations of p74 and the adjacent p10 and me53 genes in the bacmids are indicated. All bacmids contain the polh ORF under its own promoter. Part of the p74 ORF, ranging from nt 301 to 1638, was replaced with the chloramphenicol acetyltransferase (cat) gene by homologous recombination to generate the Ac-del-p74 bacmid. In the repaired bacmid, the p74 ORF with its putative promoter region and a Flag tag sequence fused to the 3′ end were included. Del, deletion; Rep, repair. (B) Detection of the P74-N and P74-C-Flag subunits. Ac-Rep-P74-Flag L-OBs were produced and purified from S. exigua larvae. Heating was at 80°C for 40 min for heated OBs. ODVs were purified from nonheated or heated OBs and were separated by 10% SDS-PAGE. Western analysis was performed with P74 MAb or Flag PAb. Positions of full-length P74-Flag and the P74-N and P74-C-Flag fragments are indicated.
Both P74-N and P74-C-Flag fragments are associated with the PIF complex.
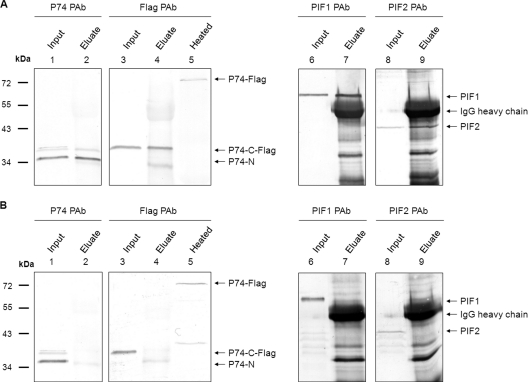
It was noticed that the P74-N subunit could be copurified with ODVs, indicating that the P74-N subunit remains associated with the ODVs after P74 cleavage. Our previous study showed that P74 may associate with a stable complex of PIF1, PIF2, and PIF3 present on the ODV surface (26). To investigate whether in L-ODVs the cleaved P74 subunits still associate with this complex, a co-IP analysis with PIF1 antibody and preimmune serum as a control was performed with L-ODV membrane proteins. Ac-Rep-P74-Flag L-ODVs were used to allow analysis of both P74-N and P74-C-Flag fragments. P74 PAb and Flag PAb were used to detect P74-N and P74-C-Flag fragments, respectively. As shown in Fig. 6A, P74-Flag was cleaved into P74-N and P74-C-Flag subunits (lanes 1 and 3), and both fragments were precipitated by the PIF1 antibody (lanes 2 and 4). In contrast, the two P74 fragments were not precipitated by preimmune serum (Fig. 6B, lanes 2 and 4), although they were present in the input sample (Fig. 6B, lanes 1 and 3). Heated Ac-Rep-P74-Flag L-ODVs were included to show the position of full-length P74-Flag (Fig. 6A and B, lane 5). Similarly, PIF1 and PIF2 were both precipitated by the PIF1 antibody (Fig. 6A, lanes 6 to 9), but not by the preimmune antiserum (Fig. 6B, lanes 6 to 9). These results showed that both the P74-N and P74-C-Flag fragments remain associated with the PIF complex after the cleavage.
Fig. 6.
The cleaved P74-N and P74-C-Flag subunits associate with the PIF complex after cleavage. Ac-Rep-P74-Flag L-ODVs were released from Se-L-OBs and purified through a sucrose gradient. ODV membrane proteins were extracted for co-IP analysis. Co-IP was performed with PIF1 antiserum and with preimmune serum as a negative control. Co-IP input and eluate were separated by 10% SDS-PAGE followed by Western analysis with P74 PAb, Flag PAb, or PIF1 or PIF2 antibodies. To show the position of full-length P74-Flag, L-ODVs were purified from heated L-OBs and detected with Flag PAb. Positions of the full-length P74-Flag, P74-N, P74-C-Flag, PIF1, PIF2, and IgG heavy chain are indicated by arrows. (A) Co-IP results with PIF1 antibody. (B) Co-IP results with PIF1 preimmune serum.
The P74-N and P74-C-Flag fragments do not interact in a covalent way.
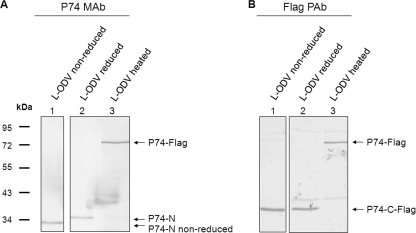
For many viral membrane proteins, the cleaved subunits interact with each other either in a covalent (35, 37, 43) or a noncovalent way (13). The fact that both P74-N and P74-C-Flag subunits were found in association with the PIF complex suggests that these two subunits may also interact with each other. To investigate whether the P74-N and P74-C-Flag subunits interact in a covalent way, nonreducing SDS-PAGE analysis was performed. L-ODVs of Ac-Rep-P74-Flag virus were purified from heated and nonheated L-OBs and treated with protein sample buffers with or without the reducing agent 2-mercaptoethanol. Samples were separated by SDS-PAGE followed by Western analysis with P74 MAb and Flag PAb. In the nonreduced sample the P74-N fragment migrated as a 32-kDa protein (Fig. 7A, lane 1), while in the reduced sample the fragment had a molecular mass of 35 kDa (Fig. 7A, lane 2). The P74-C-Flag subunit migrated with a molecular mass of 40 kDa under both reducing and nonreducing conditions (Fig. 7B, lanes 1 and 2). The position of full-length P74-Flag was shown in heated ODV sample (Fig. 7A and B, lanes 3). These results suggested that the two P74 fragments are not associated covalently through disulfide bonds.
Fig. 7.
P74-N and P74-C-Flag subunits are not associated through disulfide bonding. L-ODVs of Ac-Rep-P74-Flag virus were purified from heated or nonheated L-OBs. The nonheated L-ODV sample was treated with reducing or nonreducing Laemmli buffer, and the heated L-ODVs were treated with reduced Laemmli buffer. Proteins were separated by 10% SDS-PAGE. Western analysis was performed with P74 MAb and Flag PAb. Arrows point to full-length P74-Flag, P74-N, P74-N nonreduced, and P74-C-Flag.
DISCUSSION
Host-derived alkaline proteases have been found in a number of baculovirus L-OBs, but their functional significance remained enigmatic, except for some suggested roles in solubilization of the OB matrix or the release of ODV from polyhedra (25, 38). Recently, Slack and Arif (32) speculated that such proteases could play a synergistic role by ensuring activation of released ODVs for virus infection. Here we show that P74, a conserved ODV protein with midgut epithelium binding properties (16) and a component of the PIF complex, is cleaved by the OB alkaline protease during release of ODVs from OBs and probably prior to infection. This proteolytic cleavage could be part of a mechanism for activation of P74 to function in ODV binding to midgut microvilli and/or further downstream steps in the entry process.
P74 cleavage occurred in a highly efficient way, as evidenced by the fact that cleavage was accomplished within 3 min after L-OBs were exposed to the alkaline solution. Furthermore, P74 was cleaved completely even if L-ODVs were purified at 4°C. It was reported before that ODVs are released from OBs within 12 min after entry into the insect midgut (1, 32). Therefore, it is possible that P74 is already cleaved before ODV is fully released into the midgut lumen and hence before it contacts its host receptor. If this is true the cleaved P74 should represent a functional form. To test this possibility L-ODVs from Tni-L-OBs were purified in the same way as described in the previously reported functional analysis of AcMNPV P74 (16), and we found that P74 was present only in the cleaved form in the ODV (data not shown). Whether it is the cleaved form of P74 that functions in ODV binding needs further investigation.
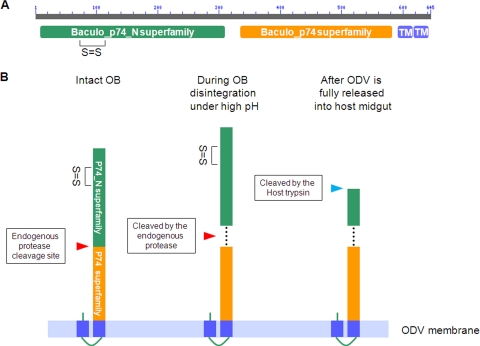
P74 is encoded by a core gene of baculoviruses, and it also has homologues in several other large invertebrate, nuclear-replicating DNA viruses, such as nudiviruses (42), salivary gland hypertrophy viruses (14), and whispoviruses (41), as well as polydnavirus particles (3). Therefore, the function and/or mode of action of P74 in virus entry might be conserved in all these viruses. Analysis of sequence data showed that the P74 protein contains two conserved domains, each belonging to a specific superfamily: the Baculo_p74_N superfamily and the Baculo_p74 superfamily (Fig. 8A). Judging from the size of the P74-N subunit (35 kDa), the cleavage is likely to occur in the region between these two conserved domains. It was previously shown that in ODVs purified from C-OBs, full-length P74 associated with a complex of PIF1, PIF2, and PIF3 (26). Co-IP analysis showed that after cleavage both P74-N and P74-C subunits remained associated with the PIF complex. These results suggest that cleavage of P74 is not a prerequisite for its association with the other three PIFs in the complex. Nonreducing SDS-PAGE showed that the two subunits are not associated covalently, but they may interact with each other in a noncovalent way, like the S1 and S2 fragments of the murine coronavirus S protein or HIV gp120 and gp41 (13, 24). Whether the two subunits interact with the PIF complex separately or whether they interact with each other in a noncovalent way and then associate with the PIF complex is yet unknown. It is tempting to speculate that these two domains form a conserved functional conformation after proteolytic cleavage to facilitate oral infection.
Fig. 8.
Proposed model for sequential cleavage of P74 in L-ODV based on conserved domain predictions and experimental data. (A) Conserved domains in P74. The AcMNPV P74 amino acid sequence was analyzed using the NCBI Conserved Domains server. Baculo_p74_N (green block) and Baculo_p74 superfamilies (orange block) were identified, as indicated. The homology with the Baculo_p74_N superfamily starts with amino acid (aa) 5 and lasts to aa 309 of AcMNPV P74. The Baculo_p74 superfamily domain starts at aa 333 and lasts until aa 583. The only disulfide bond (between cysteine 72 and cysteine 109) predicted with high probability by the DiANNA server is indicated. The score for this prediction was 0.99261, within a range of 0 to 1. Two conserved C-terminal transmembrane domains predicted by the TMHMM 2.0 server are also indicated (TM; blue blocks). (B) Model of P74 sequential cleavage. The two conserved P74 domains and the potential intramolecular disulfide bond in the P74-N subunit are indicated. The first cleavage of P74 is proposed to happen between the two conserved domains and during OB disintegration under alkaline conditions. A potential mode of interaction between the two fragments is indicated by dashed lines. After the first cleavage by the endogenous protease, the P74-N subunit is predicted to undergo a second proteolytic cleavage by a host trypsin present in the insect midgut.
Cleavage of P74 was not observed in OBs produced in cell lines derived from three different insects. Similar results were reported in previous studies (25, 44). This suggests that the protease in the L-OBs is not encoded by the virus but is derived from the host. The fact that P74 was cleaved in the same way in AcMNPV L-OBs produced in different hosts indicates that the virus is able to select similar proteases from different hosts to cleave P74. This conservation further suggests a functional significance for the association of the protease with the OBs and the subsequent P74 cleavage event. If the protease is indeed derived from the host, how the protease is recognized and accommodated into the OB structure and whether certain viral proteins are involved in these processes are highly interesting questions for further research.
Where this alkaline protease is located in the AcMNPV OB structure is also unclear. The protease is unlikely to be present on the surface of L-OBs, as in this study the OBs were routinely washed with 0.5% SDS and 1 M NaCl while P74 cleavage was preserved. Spiking C-OBs with larval homogenate before OB/ODV purification did not lead to P74 cleavage (data not shown), thereby excluding the possibility that cleavage by the protease is due to surface contamination during OB purification. It is possible that the alkaline protease is associated with the polyhedrin matrix. However, this localization may not give the enzyme quick and proper access to P74 upon ODV release. For Spodoptera littoralis NPV, an alkaline protease was found to localize on the ODV membrane (25). Considering the high efficiency of P74 cleavage, it is reasonable to assume that the protease, or at least some of the protease, is also located on the ODV membrane in AcMNPV.
The biological significance of the alkaline protease is not clear, since in a previous study AcMNPV OBs (C-OBs) produced in vitro were quantitatively as infectious to cabbage looper larvae as those produced in vivo (L-OBs) (40). However, a later study showed that mortality occurred significantly more rapidly following infection with L-OBs than with C-OBs (5). The endogenous P74 cleavage, which probably happens before ODVs are fully released and contacts its putative host receptor, could be one of the reasons for this faster mortality. Cleavage of P74 into two associating fragments, as demonstrated by co-IP, may facilitate the protein to switch to an active conformation and/or expose certain functional domains. The P74 on C-ODVs may be proteolytically cleaved by the host alkaline protease(s) present in the midgut after the ODVs are fully released. The delay of C-ODV P74 activation may delay the entry process and hence the time of death. The incorporation of alkaline proteases into the ODVs may therefore have an evolutionary advantage by speeding up the infection process.
Proteolytic cleavage is a strategy employed by many viruses to activate their membrane proteins. Examples include the SARS-CoV S protein (2), paramyxovirus F protein (35), and influenza virus HA (37). In these viruses cleavage occurs during virus entry on the cell surface or in endosomes inside the cell, or posttranslationally during protein translocation. The situation is different in the case of baculovirus P74 cleavage. It seems that the virus is able to specifically capture a host-encoded alkaline protease and incorporate it into the virus structure, which will become activated only under alkaline conditions, e.g., in the midgut of the infected host insect. The P74 cleavage event reported in this study is likely to happen during the release of ODVs from OBs and results in two associating fragments: P74-N and P74-C. After this cleavage, the P74-N fragment probably undergoes a second proteolytic cleavage by a host midgut trypsin (34), which has been shown to be essential for infectivity (at least for C-OBs). The proposed two-step sequential proteolytic cleavage may represent a novel virus membrane protein activation mechanism (Fig. 8B), which in contrast to vertebrate virus membrane protein activation mechanisms occurs under alkaline conditions. Incorporation of an endogenous alkaline protease into the virus has also been reported for other insect viruses, such as Choristoneura biennis entomopoxvirus (4) and invertebrate iridescent virus 6 (10). This common feature suggests that incorporation of a protease into the virus structure has an evolutionary advantage for some invertebrate insect viruses.
The present study reports the efficient and specific cleavage of P74 by an endogenous alkaline protease during the release of ODVs from L-OBs and sheds light on the potential significance of this protease in the process of ODV entry. To further investigate the biological significance of this processing event, the cleavage site in P74 for the alkaline protease will be identified in order to generate mutants defective in this P74 cleavage. Computational analysis of P74 proteins of closely related baculoviruses have not (yet) revealed a common motif for potential alkaline protease cleavage, which complicates the genetic approach somewhat. It would also be interesting to purify and identify the endogenous alkaline protease to address the question of how it is selected and incorporated by the virus.
ACKNOWLEDGMENTS
This work was supported by a grant (07PhD05) in the Joint PhD Training Program provided by the Chinese Academy of Sciences and the Royal Dutch Academy of Sciences.
We are grateful to Gary Blissard from Boyce Thompson Institute for Plant Research, Cornell University, for providing the P74 MAb. We are in debt to Berend Jan Bosch and Peter J. Rottier from Utrecht University, Faculty of Veterinary Medicine, Division of Virology, Utrecht, Netherlands, for insightful discussions.
Footnotes
Published ahead of print on 17 August 2011.
REFERENCES
- 1. Adams J. R., McClintock J. T. 1991. Baculoviridae, nuclear polyhedrosis viruses, part 1. Nuclear polyhedrosis viruses of insects, p. 87–204. In Adams J. R., Bonami J. R. (ed.), Atlas of invertebrate viruses. CRC Press, Boca Raton, FL. [Google Scholar]
- 2. Belouzard S., Chu V. C., Whittaker G. R. 2009. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U. S. A. 106:5871–5876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bezier A., et al. 2009. Polydnaviruses of braconid wasps derive from an ancestral nudivirus. Science 323:926–930 [DOI] [PubMed] [Google Scholar]
- 4. Bilimoria S. L., Arif B. M. 1979. Subunit protein and alkaline protease of entomopoxvirus spheroids. Virology 96:596–603 [DOI] [PubMed] [Google Scholar]
- 5. Bonning B. C., Hoover K., Duffey S., Hammock B. D. 1995. Production of polyhedra of the Autographa californica nuclear polyhedrosis virus using the Sf21 and Tn5B1-4 cell lines and comparison with host-derived polyhedra by bioassay. J. Invertebr. Pathol. 66:224–230 [DOI] [PubMed] [Google Scholar]
- 6. Bottcher-Friebertshauser E., et al. 2010. Cleavage of influenza virus hemagglutinin by airway proteases TMPRSS2 and HAT differs in subcellular localization and susceptibility to protease inhibitors. J. Virol. 84:5605–5614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Crawford A. M., Kalmakoff J. 1977. Effect of alkaline protease on the antigenic nature of wiseana nuclear polyhedrosis virus polyhedron protein. J. Virol. 24:412–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eppstein D. A., Thoma J. A., Scott H. A., Young S. Y., III 1975. Degradation of matrix protein from a nuclear-polyhedrosis virus of Trichoplusia ni by an endogenous protease. Virology 67:591–594 [DOI] [PubMed] [Google Scholar]
- 9. Fang M., Nie Y., Harris S., Erlandson M. A., Theilmann D. A. 2009. Autographa californica multiple nucleopolyhedrovirus core gene ac96 encodes a per os infectivity factor (PIF-4). J. Virol. 83:12569–12578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Farara T., Attias J. 1986. Further characterization of an alkaline protease activity associated with iridescent virus type 6. Brief report. Arch. Virol. 87:307–314 [DOI] [PubMed] [Google Scholar]
- 11. Faulkner P., Kuzio J., Williams G. V., Wilson J. A. 1997. Analysis of P74, a PDV envelope protein of Autographa californica nucleopolyhedrovirus required for occlusion body infectivity in vivo. J. Gen. Virol. 78:3091–3100 [DOI] [PubMed] [Google Scholar]
- 12. Ferre F., Clote P. 2005. DiANNA: a web server for disulfide connectivity prediction. Nucleic Acids Res. 33:W230–W232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gallagher T. M., Buchmeier M. J. 2001. Coronavirus spike proteins in viral entry and pathogenesis. Virology 279:371–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garcia-Maruniak A., et al. 2009. Two viruses that cause salivary gland hypertrophy in Glossina pallidipes and Musca domestica are related and form a distinct phylogenetic clade. J. Gen. Virol. 90:334–346 [DOI] [PubMed] [Google Scholar]
- 15. Gelernter W. D., Federici B. A. 1986. Continuous cell line from Spodoptera exigua (Lepidoptera: Noctuidae) that supports replication of nuclear polyhedrosis viruses from Spodoptera exigua and Autographa californica. J. Invertebr. Pathol. 48:199–207 [Google Scholar]
- 16. Haas-Stapleton E. J., Washburn J. O., Volkman L. E. 2004. P74 mediates specific binding of Autographa californica M nucleopolyhedrovirus occlusion-derived virus to primary cellular targets in the midgut epithelia of Heliothis virescens larvae. J. Virol. 78:6786–6791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harrison R. L., Sparks W. O., Bonning B. C. 2010. Autographa californica multiple nucleopolyhedrovirus ODV-E56 envelope protein is required for oral infectivity and can be substituted functionally by Rachiplusia ou multiple nucleopolyhedrovirus ODV-E56. J. Gen. Virol. 91:1173–1182 [DOI] [PubMed] [Google Scholar]
- 18. Janknecht R., Sander C., Pongs O. 1991. (HX)n repeats: a pH-controlled protein-protein interaction motif of eukaryotic transcription factors? FEBS Lett. 295:1–2 [DOI] [PubMed] [Google Scholar]
- 19. Kikhno I., Gutierrez S., Croizier L., Croizier G., Ferber M. L. 2002. Characterization of pif, a gene required for the per os infectivity of Spodoptera littoralis nucleopolyhedrovirus. J. Gen. Virol. 83:3013–3022 [DOI] [PubMed] [Google Scholar]
- 20. Kozlov E. A., Sidorova N. M., Serebryani S. B. 1975. Proteolytic cleavage of polyhedral protein during dissolution of inclusion bodies of the nuclear polyhedrosis viruses of Bombyx mori and Galleria mellonella under alkaline conditions. J. Invertebr. Pathol. 25:97–101 [DOI] [PubMed] [Google Scholar]
- 21. Langridge W. H., Balter K. 1981. Protease activity associated with the capsule protein of Estigmene acres granulosis virus. Virology 114:595–600 [DOI] [PubMed] [Google Scholar]
- 22. Leach J. L., et al. 1996. Isolation from human placenta of the IgG transporter, FcRn, and localization to the syncytiotrophoblast: implications for maternal-fetal antibody transport. J. Immunol. 157:3317–3322 [PubMed] [Google Scholar]
- 23. Ohkawa T., Washburn J. O., Sitapara R., Sid E., Volkman L. E. 2005. Specific binding of Autographa californica M nucleopolyhedrovirus occlusion-derived virus to midgut cells of Heliothis virescens larvae is mediated by products of pif genes Ac119 and Ac022 but not by Ac115. J. Virol. 79:15258–15264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pancera M., et al. 2010. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc. Natl. Acad. Sci. U. S. A. 107:1166–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Payne C. C., Kalmakoff J. 1978. Alkaline protease associated with virus particles of a nuclear polyhedrosis virus: assay, purification, and properties. J. Virol. 26:84–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peng K., van Oers M. M., Hu Z., van Lent J. W., Vlak J. M. 2010. Baculovirus per os infectivity factors form a complex on the surface of occlusion-derived virus. J. Virol. 84:9497–9504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peng K., et al. 2010. Identification of protein-protein interactions of the occlusion-derived virus-associated proteins of Helicoverpa armigera nucleopolyhedrovirus. J. Gen. Virol. 91:659–670 [DOI] [PubMed] [Google Scholar]
- 28. Pijlman G. P., Pruijssers A. J., Vlak J. M. 2003. Identification of pif-2, a third conserved baculovirus gene required for per os infection of insects. J. Gen. Virol. 84:2041–2049 [DOI] [PubMed] [Google Scholar]
- 29. Qiao S. W., et al. 2008. Dependence of antibody-mediated presentation of antigen on FcRn. Proc. Natl. Acad. Sci. U. S. A. 105:9337–9342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rohrmann G. F. 2010. Baculovirus molecular biology. National Library of Medicine, National Center for Biotechnology Information, Bethesda, MD. [Google Scholar]
- 31. Simmons G., et al. 2005. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U. S. A. 102:11876–11881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Slack J., Arif B. M. 2007. The baculoviruses occlusion-derived virus: virion structure and function. Adv. Virus Res. 69:99–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Slack J. M., Lawrence S. D. 2005. Evidence for proteolytic cleavage of the baculovirus occlusion-derived virion envelope protein P74. J. Gen. Virol. 86:1637–1643 [DOI] [PubMed] [Google Scholar]
- 34. Slack J. M., Lawrence S. D., Krell P. J., Arif B. M. 2008. Trypsin cleavage of the baculovirus occlusion-derived virus attachment protein P74 is prerequisite in per os infection. J. Gen. Virol. 89:2388–2397 [DOI] [PubMed] [Google Scholar]
- 35. Smith E. C., Popa A., Chang A., Masante C., Dutch R. E. 2009. Viral entry mechanisms: the increasing diversity of paramyxovirus entry. FEBS J. 276:7217–7227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sprague E. R., Martin W. L., Bjorkman P. J. 2004. pH dependence and stoichiometry of binding to the Fc region of IgG by the herpes simplex virus Fc receptor gE-gI. J. Biol. Chem. 279:14184–14193 [DOI] [PubMed] [Google Scholar]
- 37. Steinhauer D. A. 1999. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology 258:1–20 [DOI] [PubMed] [Google Scholar]
- 38. Summers M. D., Smith G. E. 1975. Trichoplusia ni granulosis virus granulin: a phenol-soluble, phosphorylated protein. J. Virol. 16:1108–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tweeten K. A., Bulla L. A., Jr., Consigli R. A. 1978. Characterization of an alkaline protease associated with a granulosis virus of Plodia interpunctella. J. Virol. 26:703–711 [PMC free article] [PubMed] [Google Scholar]
- 40. Vail P. V., Jay D. L., Hink W. F. 1973. Replication and infectivity of the nuclear polyhedrosis virus of the alfalfa looper, Autographa californica, produced in cells grown in vitro. J. Invertebr. Pathol. 22:231–237 [Google Scholar]
- 41. Wang Y., Bininda-Emonds O. R., van Oers M. M., Vlak J. M., Jehle J. A. 2011. The genome of Oryctes rhinoceros nudivirus provides novel insight into the evolution of nuclear arthropod-specific large circular double-stranded DNA viruses. Virus Genes 42:444–456 [DOI] [PubMed] [Google Scholar]
- 42. Wang Y., Jehle J. A. 2009. Nudiviruses and other large, double-stranded circular DNA viruses of invertebrates: new insights on an old topic. J. Invertebr. Pathol. 101:187–193 [DOI] [PubMed] [Google Scholar]
- 43. Westenberg M., et al. 2002. Furin is involved in baculovirus envelope fusion protein activation. J. Virol. 76:178–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wood H. A. 1980. Protease degradation of Autographa californica nuclear polyhedrosis virus proteins. Virology 103:392–399 [DOI] [PubMed] [Google Scholar]