Abstract
Members of the Enterovirus genus of the Picornaviridae family are abundant, with common human pathogens that belong to the rhinovirus (HRV) and enterovirus (EV) species, including diverse echo-, coxsackie- and polioviruses. They cause a wide spectrum of clinical manifestations ranging from asymptomatic to severe diseases with neurological and/or cardiac manifestations. Pandemic outbreaks of EVs may be accompanied by meningitis and/or paralysis and can be fatal. However, no effective prophylaxis or antiviral treatment against most EVs is available. The EV RNA genome directs the synthesis of a single polyprotein that is autocatalytically processed into mature proteins at Gln↓Gly cleavage sites by the 3C protease (3Cpro), which has narrow, conserved substrate specificity. These cleavages are essential for virus replication, making 3Cpro an excellent target for antivirus drug development. In this study, we report the first determination of the crystal structure of 3Cpro from an enterovirus B, EV-93, a recently identified pathogen, alone and in complex with the anti-HRV molecules compound 1 (AG7404) and rupintrivir (AG7088) at resolutions of 1.9, 1.3, and 1.5 Å, respectively. The EV-93 3Cpro adopts a chymotrypsin-like fold with a canonically configured oxyanion hole and a substrate binding pocket similar to that of rhino-, coxsackie- and poliovirus 3C proteases. We show that compound 1 and rupintrivir are both active against EV-93 in infected cells and inhibit the proteolytic activity of EV-93 3Cpro in vitro. These results provide a framework for further structure-guided optimization of the tested compounds to produce antiviral drugs against a broad range of EV species.
INTRODUCTION
Enteroviruses (EVs) are small, nonenveloped, icosahedral, positive-sense, single-stranded RNA viruses classified into a genus of the Picornaviridae family, one of the largest and most important families of viral pathogens of vertebrates, including humans (36). The Enterovirus genus encompasses 234 human pathogens that form 7 species spread worldwide: human enteroviruses A through D (HEV-A, HEV-B, HEV-C, and HEV-D) and human rhinoviruses A through C (HRV-A, HRV-B, and HRV-C) (23). Echoviruses and coxsackievirus B (CV-B) are classified within the HEV-B species, and polioviruses (PVs) are classified within HEV-C. There are also EVs that infect nonhuman primates, cattle, and swine that may play roles in zoonotic spread and the emergence of new human pathogens. In humans, infections range from asymptomatic to more severe illnesses that are manifested as aseptic meningitis, encephalitis, gastroenteritis, myocarditis, paralysis, and poliomyelitis, with high mortality rates in infected newborn infants. Outbreaks of different EVs are frequently reported. For example, EV-71 of HEV-A caused serious complications—encephalitis and myocarditis—and death during epidemics of hand, foot, and mouth disease in Asia in 1997, 1998, 2000, and 2008 (44). The disease manifestation of acute flaccid paralysis is also associated with nonpoliovirus EVs within the HEV-B species, including newly discovered viruses like EV-93, which is the subject of this study (20). Despite the enormous health care impact of EV infections, no antiviral drugs have been approved to control these infections (for a recent review, see reference 11).
The EV genome is a positive-sense, single-stranded RNA of between 7.4 and 7.5 kb with a single open reading frame translated into a large polyprotein of approximately 2,200 amino acids (∼250 kDa) (13, 38). This polyprotein is rapidly processed by co- and posttranslational cleavages into three precursor molecules, P1, P2, and P3, and then into mature viral proteins: the structural proteins VP4 to VP1 from P1 and the nonstructural proteins associated with replication, 2A to 2C and 3A to 3D from P2 and P3, respectively, from the N to the C terminus (22). Most of these cleavages are mediated by the 3C protease (3Cpro) either alone or as a domain of 3CDpro. In addition to its key role in processing the polyprotein, 3Cpro cleaves a number of host proteins to remodel the cellular environment for virus reproduction (12, 43). Due to its central role in the control of genome expression, 3Cpro could be considered the “main” protease (49). Crystal structures have been determined for EV 3Cpro from CV-B3, HRV-2, HRV-14, PV-1, and EV-71 (7, 10, 27, 30, 31, 34). The protease adopts a chymotrypsin-like fold with the Cys-His-Glu catalytic triad present in a shallow groove between two topologically equivalent six-stranded β barrels. It cleaves sites that have predominantly Gln↓Gly at the P1↓P1′ positions and a restricted evolutionary variation at the P4 position (17, 45).
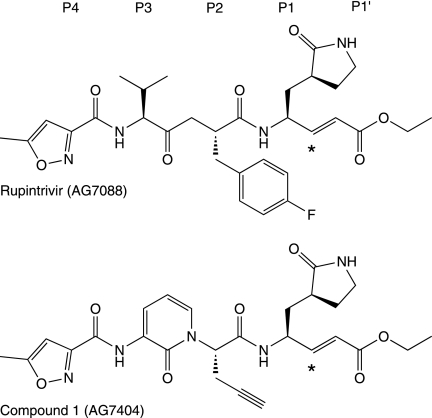
Because of their essential role in virus replication and a narrow substrate specificity, EV 3Cpros are excellent targets for antiviral therapy and have been the focus of extensive structure/activity studies to develop inhibitor compounds, mainly against HRVs (for a recent review, see reference 11). Two compounds against the common cold, rupintrivir (AG7088) and its orally bioavailable analogue compound 1 (AG7404), progressed to phase-II/I clinical trials (19, 41). Both compounds (Fig. 1) are peptidomimetic inhibitors and imitate the P4 to P1 peptide substrate, with an α,β-unsaturated ester at P1′ as a Michael acceptor to form an irreversible covalent bond with the active-site Cys residue. Rupintrivir was shown to have low toxicity and potent antiviral activity against all HRV serotypes tested (48), with a mean 50% effective concentration (EC50) of 23 nM, and also against four related EVs (40). Similar results were later obtained against HRV clinical isolates and four additional EVs, as well as with compound 1 (5, 21, 41).
Fig. 1.
Chemical structure of rupintrivir (AG7088) and its orally bioavailable analogue compound 1 (AG7404). Asterisks indicate carbon atoms that make an irreversible covalent bond with the active-site cysteine residue of 3Cpro.
Here we present the crystal structure of the main protease of human enterovirus B EV-93 (EV-93 3Cpro) alone and in complex with compound 1 and rupintrivir at resolutions of 1.9 Å, 1.3 Å, and 1.5 Å, respectively. This 20.2-kDa cysteine protease adopts a chymotrypsin-like fold with a catalytic triad, Cys147-His40-Glu71, located in the cleft between the two six-stranded β barrels. The residues that form the inhibitor binding pockets are highly conserved among EV 3Cpros, which explains the broad activity of compounds developed initially against a specific virus. To corroborate this finding, we showed that both compound 1 and rupintrivir are active against EV-93 in infected cells and inhibit the proteolytic activity of EV-93 3Cpro in vitro.
MATERIALS AND METHODS
Protein purification.
EV-93 3Cpro was expressed in Escherichia coli cells and purified at high yields (∼25 mg per liter of cell culture). For this process, the 3C protease gene from HEV-B EV-93 strain 38-03 (20) was cloned into a pDEST14 expression vector (Invitrogen) with an N-terminal MK, as the additional Lys can improve protein expression (8), and a C-terminal His6 tag. Four E. coli strains (BL21 pLysS, Rosetta, Origami pLysS, and C41) were tested to identify the optimal conditions for protein overexpression. Soluble protein was detected by dot blotting for three media and temperatures for a total of 36 conditions (4). The best results were obtained using E. coli BL21(DE3)-pLysS grown in Superior Broth medium (Athena) at 17°C. Cells were grown in this medium (3 liters), which contained 100 μg ml−1 ampicillin and 34 μg ml−1 chloramphenicol, at 37°C until an optical density at 600 nm (OD600) of 0.6 was reached. They were then induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside and further incubated overnight at 17°C. Cells were harvested by centrifugation, resuspended in extraction buffer (50 mM Tris-HCl [pH 8], 300 mM NaCl, 10 mM imidazole, 0.1% [vol/vol] Triton X-100, 5% [vol/vol] glycerol, 0.25 mg/ml lysozyme, and Complete EDTA-free antiproteases [Roche]), and then stored at −80°C. Thawed cells were lysed by sonication on ice and centrifuged at 30,000 × g for 1 h at 4°C. The supernatant was loaded into a 5-ml Ni-affinity HisTrap HP column (GE Healthcare), and recombinant EV-93 3Cpro was eluted with 0.5 M imidazole in 50 mM Tris-HCl (pH 8)–300 mM NaCl. EV-93 3Cpro was further purified through a gel filtration column (HiLoad 16/60 Superdex 200; GE Healthcare) in 10 mM bicine (pH 8.5)–300 mM NaCl and concentrated for crystallization to 2 mg/ml, as determined spectrophotometrically using a theoretical extinction coefficient of 0.677 ml mg−1 cm−1 at 280 nm. EV-93 3Cpro was purified without dithiothreitol (DTT) or with 1 mM DTT in all buffers except the extraction buffer (5 mM). The C147A mutant of EV-93 3Cpro was obtained by site-directed mutagenesis (Stratagene), checked by DNA sequencing, and purified as the native protein. The EV-93 3Cpro-compound 1 and EV-93 3Cpro-rupintrivir complexes were purified by gel filtration after overnight incubation at 20°C with a 2-fold molar excess of the inhibitor. The antiviral agents rupintrivir (AG7088) and compound 1 (AG7404) were both generously provided by Pfizer. Selected fractions were concentrated to 3 mg/ml for the EV-93 3Cpro-compound 1 complex and to 5 mg/ml for the EV-93 3Cpro-rupintrivir complex.
Crystallization and structure determination.
Crystallization trials were set up at 20°C with a NanoDrop dispenser in 96-well sitting drop plates using commercial screens. Drops were prepared by mixing 100 nl of protein solution with 100 nl of reservoir solutions. Crystal optimization was performed manually with 1+1 μl sitting drops. Needle-like crystals of EV-93 3Cpro grew in 18% (vol/vol) PEG 8000, 0.2 M Mg acetate, and 0.1 M cacodylate (pH 7). Crystals were briefly soaked in a cryo-protectant solution comprising mother liquor with an additional 20% (vol/vol) glycerol and then flash cooled in liquid nitrogen. EV-93 3Cpro-compound 1 crystals were grown in 1.8 M (NH4)2HPO4–0.1 M Tris (pH 8.3) and were briefly soaked in 2.3 M (NH4)2HPO4–0.1 M Tris (pH 8.3) supplemented with 13.4% ethylene glycol prior to being flash cooled. EV-93 3Cpro-rupintrivir crystals were obtained in 0.25 M MgCl2, 0.1 M Tris (pH 8.5), and 25% PEG 8000 and soaked in mother liquor with an additional 25% PEG 400 before being flash cooled.
Diffraction intensities were recorded on microfocus beamline ID23-2 at the European Synchrotron Radiation Facility (ESRF; Grenoble, France) at a wavelength of 0.8726 Å and a beam diameter of 10 μm. Data for native protein and the compound 1 complex were indexed, integrated, and scaled using the MOSFLM and SCALA programs (9, 46), while the DENZO and SCALEPACK programs (37) were used for the rupintrivir complex data. Statistics for the best-measured data sets are reported in Table 1. EV-93 3Cpro crystals belong to the monoclinic space group P21 and have unit cell parameters that are similar for native protein and the compound 1 and rupintrivir complexes. The calculated Matthews coefficients (VM = 1.9, 2.0, and 1.9 Å3 Da−1) indicate the presence of two molecules of EV-93 3Cpro in the asymmetric units with corresponding solvent contents of 37.5%, 38.9%, and 36.6%, respectively (29).
Table 1.
Crystallographic data and refinement statistics
| Parameter | Values for indicated protease or compounda |
||
|---|---|---|---|
| EV-93 3Cpro | EV-93 3Cpro-compound 1 | EV-93 3Cpro-rupintrivir | |
| Resolution range (Å) | 66.4–1.9 (2.0–1.90) | 39.0–1.32 (1.39–1.32) | 30.0–1.50 (1.55–1.50) |
| Space group | P21 | P21 | P21 |
| Cell dimensions (Å, degrees) | a = 39.07, b = 65.22, c = 66.36, β = 90.67 | a = 39.04, b = 64.45, c = 68.74, β = 90.81 | a = 39.00, b = 63.91, c = 66.36, β = 90.43 |
| No. of observed reflections | 117,117 | 268,892 | 194,865 |
| No. of unique reflections | 26,356 (3,828) | 79,978 (11,666) | 52,513 (5,196) |
| Completeness (%) | 99.9 (100) | 100 (100) | 100 (100) |
| Mean multiplicity | 4.4 (3.1) | 3.4 (3.3) | 3.7 (3.6) |
| Rmergeb | 0.127 (0.412) | 0.085 (0.508) | 0.063 (0.488) |
| <I/σ(I)> | 5.1 (1.6) | 5.2 (1.5) | 17.7 (2.5) |
| Rworkc | 0.148 | 0.125 | 0.166 |
| Rfreec | 0.210 | 0.170 | 0.198 |
| No. of protein atoms (non-H) | 3,037 | 3,322 | 3,011 |
| No. of water molecules | 339 | 543 | 375 |
| No. of hetero compounds | 2 Mg2+ ions, 6 glycerol molecules | 2 compound 1 molecules, 8 ethylene glycol molecules, 3 HPO42− ions, 2 NH4+ ions | 2 rupintrivir molecules, 2 Mg2+ ions, Cl− ions |
| RMSD for bond length (Å); RMSD for bond angles (°) | 0.012; 1.40 | 0.012; 1.60 | 0.008; 1.41 |
| Mean B value (Å2) | 11.89 | 9.29 | 12.21 |
| Mean B value for inhibitor molecules (Å2) | 20.2, 19.7 | 16.8, 22.0 | |
| Ramachandran analysisd: most/additionally/generously allowed region (%) | 88.8/10.2/1.0 | 89.1/9.6/1.3 | 97.26/2.74/0.0 |
Values in parentheses refer to the high-resolution shell.
, where Ii,h is the ith-intensity measurement of reflection h and <Ih> is the average intensity for multiple measurements.
Rfree was calculated for 5% of the reflections not used for refinement.
Ramachandran analysis was done with PROCHECK.
The initial phases were obtained by molecular replacement using the program PHASER (32) with the PV-1 3Cpro structure as a search model (PDB code, 1L1N) (34). The EV-93 3Cpro structure was built initially with ARP/wARP 6.1.1 (42) and then by iterative cycles of restrained refinement with REFMAC5 (9, 35) and model building/solvent addition with COOT (15). The EV-93 3Cpro-compound 1 structure was refined with PHENIX (1), while REFMAC5 was used to refine the EV-93 3Cpro-rupintrivir complex. Geometry restraint information for compound 1 was generated with eLBOW from the SMILES description of the antiviral method and using the semiempirical quantum-mechanical optimization method AM1 (33). Geometry restraint information for rupintrivir was calculated with SKETCHER within CCP4i (9). In the first two structures (EV-93 3Cpro alone and in complex with compound 1), hydrogen atoms were added in their riding positions, but they were not added for the rupintrivir complex structure. Refinement statistics are reported in Table 1. Structural figures were made with PyMOL (www.pymol.org). The sequence alignment was obtained from Muscle-mediated (14), polyprotein-wide EV alignment built using the Viralis software platform (16). The sequence alignment figure was generated with ESPript (18). Amino acid conservation scores were calculated and mapped onto the protein structure with the ConSurf server (25).
Antiviral activity in infected cells.
The antiviral activities of rupintrivir (AG7088) and compound 1 (AG7404) were determined by infecting monolayers of human rhabdomyosarcoma cells (RD) with 100 or 1,000 50% cell culture infective doses (CCID50) of EV-93 in two 96-well plates. After an adsorption period of 2 h at 37°C, the virus was removed and serial dilutions of one compound per plate were added. The cultures were further incubated for 7 days, until the complete cytopathogenic effect (CPE) was observed in the wells with infected and untreated virus controls. The wells were read visually. The compounds were diluted from 1 to 0.0016 μg/ml. After 7 days of culture, the supernatant was collected from wells that exhibited the full CPE in the presence of the lowest concentration of the compounds used. This virus was used for successive rounds of infection, a procedure that was repeated 15 times in order to generate drug-resistant virus.
In vitro proteolytic activity assay.
A peptide representing the predicted 2C↓3A cleavage site of EV-93 (Ac-RHSVGATLEALFQ↓GPPVYREIKIS-NH2; Genepep) was used to test EV-93 3Cpro proteolytic activity in vitro and its inhibition by two antiviral agents, rupintrivir (AG7088) and compound 1 (AG7404). The lyophilized peptide and antiviral agents were dissolved in 100% dimethyl sulfoxide (DMSO) at a concentration of 0.5 mM. Cleavage reaction mixtures containing 30 μM peptide, 3 μM EV-93 3Cpro, 50 mM HEPES (pH 7.2), and 150 mM NaCl in a total volume of 100 μl were incubated at 30°C. The reactions were stopped after 20 h by the addition of 0.5% (final concentration) trifluoroacetic acid (TFA) or by freezing at −20°C. Samples were analyzed by reverse-phase high-performance liquid chromatography (HPLC) on a Source 5RPC ST 4.6/150 column (GE Healthcare) using a 2% to 90% linear gradient of acetonitrile in 0.1% TFA. To keep them independent of the initial amount of the substrate, trans cleavage efficiencies (E) are reported as the fraction of the substrate converted to products, based on the integrated peak areas at 215 nm corresponding to the remaining substrate (r) and the products (pi), as follows: E = Σ pi/(r + Σ pi). The substrate and products were analyzed by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF) and comparison with the reference samples (the synthesized full peptides, Ac-RHSVGATLEALFQ-OH and H-GPPVYREIKIS-NH2). The presence of the protein was confirmed by tandem mass spectrometry (MS-MS) fragmentation and SDS-PAGE.
Protein structure accession numbers.
Atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 3Q3X, 3Q3Y, and 3RUO for EV-93 3Cpro, EV-93 3Cpro-compound 1, and EV-93 3Cpro-rupintrivir, respectively.
RESULTS AND DISCUSSION
Structure determination and model quality.
EV-93 3Cpro was expressed in E. coli cells and purified at a high yield (∼25 mg per liter of cell culture). Needle-like crystals, up to approximately 10 by 10 by 170 μm, were obtained in a few weeks from a solution containing PEG 8000, magnesium acetate, and cacodylate. Further optimizations of the crystallization conditions, including additive screen and macro- or microseeding, did not yield larger crystals of the native protein. Taking advantage of the highly focused beam of the ID23-2 line at the European Synchrotron Radiation Facility (ESRF; Grenoble, France), we collected two data sets from a single crystal, irradiating each time a fresh crystal portion along its long dimension, with a shifted but overlapping φ-angle range. Merging the best parts of these data sets allowed us to obtain a complete data set at high resolution (Table 1). The stability of the crystal during irradiation was most likely due to its low solvent content, 37.5%, corresponding to dense crystal packing with two molecules in the asymmetric unit.
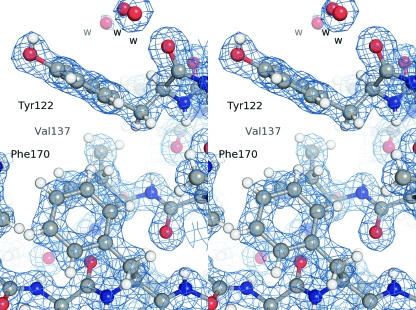
We solved the structure of EV-93 3Cpro by molecular replacement, using the structure of the homologous PV-1 3Cpro as a search model, the two proteins sharing 61% of identical amino acids. The structure of EV-93 3Cpro was refined at 1.9 Å to a final Rwork of 14.8% (Rfree = 21%; see Table 1 for refinement statistics). We modeled all residues except the His6 tag and the last two and three C-terminal residues for chains A and B, respectively, which we did not model because of poor electron density. Figure 2 shows the electron density in a representative region of the structure. The Ramachandran plot shows 88.4% of the residues to be in the most favored regions, 10.3% and 1.3% in the additionally and generously allowed regions, respectively (26). The two molecules of the asymmetric unit are related by a noncrystallographic 2-fold axis. The refined structure contains 339 water molecules, 2 magnesium ions, and 6 glycerol molecules.
Fig. 2.
Typical σA-weighted 2Fo−Fc electron density map of the crystal structure of EV-93 3Cpro-compound 1. The electron density, contoured at 1.3σ, is shown as a blue mesh with the residues depicted in ball-and-stick format (stereo view).
The structure of EV-93 3Cpro in complex with compound 1 was solved by molecular replacement using the native structure and was refined at 1.32 Å to a final Rwork of 12.5% (Rfree = 17%; Table 1). The final model encompasses all EV-93 3Cpro amino acids except the first N-terminal and the last three C-terminal (and His6 tag) residues. One molecule of compound 1 per protein chain was unequivocally and precisely defined (see Fig. 5a), the ethoxycarbonyl group corresponding to the P1′ position (Fig. 1) being more mobile. The structure also contains 543 water, 8 ethylene glycol, 3 HPO42−, and 2 NH4+ molecules. The root mean square deviations (RMSDs) between the two structures are 0.465 Å (for 267 superimposed Cα atoms) for both chains and 0.262 Å (for 137 superimposed Cα atoms) for one protein chain, with an angular shift of about 2° between the second chains of the two structures.
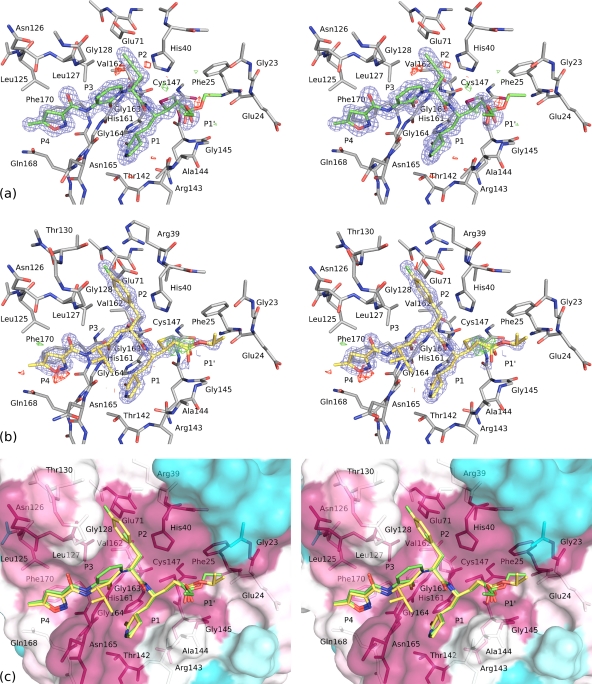
Fig. 5.
Stereo views of compound 1 and rupintrivir bound to the EV-93 3Cpro active-site pocket. (a and b) Refined structures of EV-93 3Cpro in complex with compound 1 (a) and rupintrivir (b). Both compounds are represented as sticks, with their 2Fo−Fc-weighted electron density contoured at 1.5 σ and represented as a blue mesh and their Fo−Fc-weighted-difference electron densities contoured at −3 σ and +3 σ and shown in red and green, respectively. EV-93 3Cpro residues interacting with the compounds are shown as sticks and are labeled. (c) Conservation of the compound 1 and rupintrivir binding pocket. The rupintrivir structure (yellow carbon atoms) is overlaid on the EV-93 3Cpro-compound 1 structure (green carbon atoms). The EV-93 3Cpro molecular surface is colored from cyan to magenta for variable to conserved residues, respectively, based on the multiple-sequence alignment presented in Fig. 6. EV-93 3Cpro residues interacting with the antiviral compounds are shown as sticks and are labeled. Positions P4 to P1′ are labeled in all figures.
The structure of EV-93 3Cpro in complex with rupintrivir was solved by molecular replacement using the native structure as a starting model. One rupintrivir molecule per protein chain, including molecules from the 1-fluorobenzene-4-yl group, which is different from the corresponding compound 1 group (Fig. 1, P2 position), was clearly defined in both 2Fo−Fc and Fo−Fc electron density maps. Given the higher resolution of the EV-93 3Cpro-compound 1 complex structure, the molecular replacement calculation was repeated with protein molecule A of the complex structure (without ligand and solvent atoms) as the initial model. The electron density maps showed a position for the rupintrivir molecule identical to that shown when using the native structure as the starting model, so the molecule was added. The structure was refined at 1.50 Å to a final Rwork of 16.6% (Rfree = 19.8%; Table 1). The structure also contains 375 water molecules, 2 Mg2+ ions, and 1 Cl− ion.
Overall structure and active site of EV-93 3Cpro.
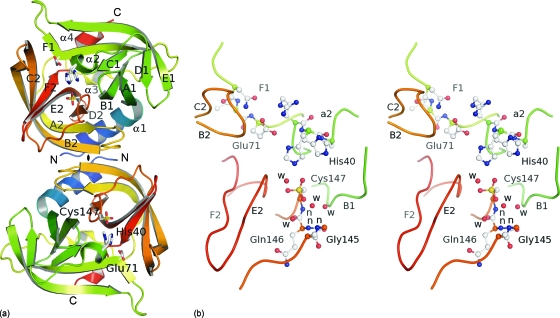
EV-93 3Cpro folds into two antiparallel β barrels (residues 15 to 77 and 97 to 173, respectively) that are oriented 90° apart, linked by a 20-amino-acid loop with a short α-helix in its middle, and flanked by two other α-helixes at the N and C termini, 14 and 6 amino acids long, respectively (Fig. 3 a). The two barrels are topologically equivalent and are formed by six antiparallel β strands with the first four (A to D) organized into a Greek key motif. Our structure confirms that EV-93 3Cpro adopts a chymotrypsin-like fold similar to that of other picornavirus 3Cpros. The RMSDs between 3Cpros from EV-93 and other EVs are 0.28 Å (160 Cα) for CV-B3, 0.70 Å (152 Cα) for PV-1, 0.77 Å (150 Cα) for HRV-2, 1.02 Å (150 Cα) for HRV-14, and 1.16 Å (170 Cα) for EV-71 (7, 10, 27, 30, 34). These figures underline the high conservation of the 3Cpro structure in viruses of the Enterovirus genus. Analysis of EV-93 3Cpro interfaces with the PISA server (24) suggests that the homodimer formed by the two chains of the asymmetric unit (Fig. 3a) might be stable in solution with an interface area of 1,710 Å2 mainly involving parts of the α1-helix and B2 strand (residues 1, 5, 107, 113, and 143 from both chains). However, a gel filtration chromatogram showed only one peak corresponding to a monomer of EV-93 3Cpro, and there is no further evidence that dimerization is required for the proteolytic activity of EV-93 3Cpro.
Fig. 3.
Crystal structure of EV-93 3Cpro. (a) Ribbon representation of the two molecules present in the asymmetric unit with the noncrystallographic 2-fold axis (♦) perpendicular to the plane. The protease folds into two antiparallel β barrels (in green and orange tones from the N to the C terminus), forming the chymotrypsin-like fold. The catalytic triad is highlighted as stick representations. (b) Active site of EV-93 3Cpro. Key residues are highlighted as ball-and-stick representations (stereo view). Main-chain amides forming the oxyanion hole are indicated by “n.”
The active-site residues are located in the cleft between the two barrels with the nucleophilic Cys147 from the C-terminal barrel and the general acid base pair His40-Glu71 from the N-terminal barrel (Fig. 3). Early in the refinement of the native structure of EV-93 3Cpro, residual-difference density close to the sulfur of the active-site cysteine in both copies indicated that Cys147 residues were oxidized at least to the stage of sulfinic acid, −SO2−, and partially to that of sulfonic acid, −SO3−. We therefore modeled both residues as the latter case, with partial occupations for the Oδ atom (Fig. 3b). Since the purified protein is active (see below), such oxidation must have occurred during crystallization or X-ray data collection. Oxidized active-site Cys residues were also observed in other picornaviral 3Cpros (2, 3). As a result, the side chains of His40 adopt two conformations, one corresponding to the canonical orientation, as seen in other EV 3Cpros, and making hydrogen bonds with the two other active-site residues, and the second corresponding to the oxidized-cysteine state, being rotated about 120° out of the active site.
Substrate hydrolysis by cysteine proteases occurs through a covalent tetrahedral intermediate between the active-site nucleophile and the carbonyl carbon of the scissile bond. The resulting oxyanion is stabilized by strong hydrogen bonds with amide groups of the protease, which are collectively called the oxyanion hole. In the EV-93 3Cpro structure (Fig. 3b), the amide groups of Cys147, Gln146, and Gly145 form this oxyanion hole with a conformation similar to that of other EV 3Cpros, which is adequate to stabilize the tetrahedral intermediate.
3C proteases were shown to recognize amino acid residues around the cleavage site, mostly at the P4…P1 ↓ P1′ positions that fit into corresponding specific binding subsites (S4…S1, S1′) of the protease. Based on comparison of the structure of HRV-14 3Cpro covalently bound to a peptide (acetyl-LEALFQ-ethyl propionate) inhibitor, including P6 to P1 substrate residues (7), with the EV-93 3Cpro structure, we propose that the substrate binding pocket of EV-93 3Cpro is formed by residues belonging to the β strands B2, E2, F2 and to the C2 to D2 loop head of the oxyanion hole (Fig. 3b). In particular, mutations in the B2 to C2 loop were shown to have a significant impact on the proteolytic activity of 3Cpro from foot-and-mouth disease virus, which belongs to another genus of the Picornaviridae family (47).
Rupintrivir and compound 1 inhibit EV-93 replication in infected cells.
Since compound 1 and rupintrivir, developed as irreversible inhibitors of HRV 3Cpros, inhibit 3Cpros encoded by viruses of HEV species (40, 41), we reasoned that they could also be active against EV-93. Indeed, rupintrivir inhibited EV-93 replication in infected RD cells, a prototype cell line for enterovirus growth (48), with a mean EC50 of 33 nM (range, 17 to 50 nM). The EC50 for compound 1 was 93 nM (range, 37 to 112 nM). These results are consistent with mean EC50s obtained for related HEVs: 88 nM (range, 7 to 183 nM) and 75 nM (range, 7 to 249 nM) for rupintrivir and compound 1, respectively (40, 41). We were unable to isolate a drug-resistant virus after 15 successive passages of the virus in the presence of either of the compounds at concentrations that allowed observation of the full cytopathogenic effect caused by the virus. For HRVs, most mutations conferring resistance to rupintrivir were obtained after only a few serial passages, 3 for HRV-14, 4 for HRV-2 and HRV-39, and 6 for HRV-Hanks (6), and thus 15 passages were considered sufficient. As mutating residues in HRVs are mostly conserved in sequence and structure compared to those in EV-93, we cannot rule out that the difference between EV-93 and HRVs in obtaining drug-resistant viruses is due to different passaging procedures.
Rupintrivir and compound 1 inhibit EV-93 3Cpro protease in vitro.
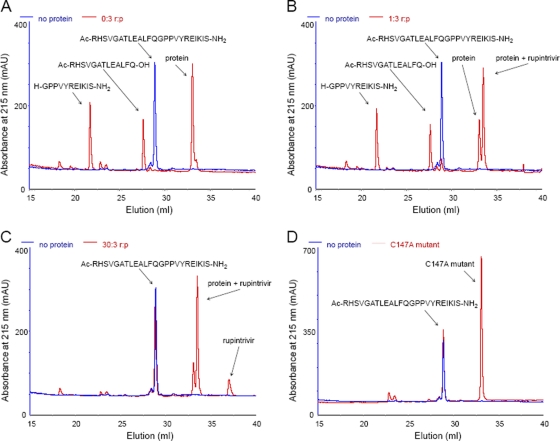
In order to verify that compound 1 and rupintrivir could also target the EV-93 3C protease, we tested the effect of each antiviral agent on the in vitro proteolytic activity of EV-93 3Cpro on a cognate peptide substrate (P13 to P11′) that mimics the 2C↓3A cleavage site. As illustrated in Fig. 4 A, reverse-phase HPLC analyses of overnight incubations of the peptide with EV-93 3Cpro provide clear evidence of the expected proteolytic cleavage. The identities of the substrate and its cleavage products were verified by MALDI-TOF and comparison with the reference samples, while the presence of the protein was confirmed by MS-MS fragmentation and SDS-PAGE. The trans cleavage efficiency (E) of EV-93 3Cpro was estimated to be more than 95% for the native enzyme, regardless of the presence or absence of DTT (Fig. 4A and Table 2). This finding suggests that the oxidation of the active-site Cys147, as seen in the EV-93 3Cpro structure (and thus the inactivation of the enzyme), occurred after its purification, during its crystallization or during X-ray data collection. The proteolytic assay performed with the C147A mutant of EV-93 3Cpro showed no effect on the substrate and did not render detectable cleavage products (Fig. 4D), thereby confirming that Cys147 is the active nucleophilic residue. Proteolytic assays performed in the presence of an inhibitor with three different inhibitor-to-protease molar ratios indicated that both rupintrivir and compound 1 efficiently inhibit native EV-93 3Cpro in vitro (Fig. 4B and C and Table 2). No cleavage product was detected with a 10-fold excess of inhibitors over the enzyme, and cleavage efficiencies of less than 10% were observed with equimolar amounts of the two antivirals relative to the 3Cpro (Table 2). These results are consistent with prior results for potency and irreversible inhibition by these compounds of 3Cpro from HRVs and EVs (40, 41) and corroborate their strong antiviral effect against a broad spectrum of picornaviruses.
Fig. 4.
Inhibition of the in vitro proteolytic activity of EV-93 3Cpro by rupintrivir. Reverse-phase chromatograms show in red the products of the digestion of the EV-93 2C↓3A peptide (Ac-RHSVGATLEALFQ↓GPPVYREIKIS-NH2) by EV-93 3Cpro (p) without rupintrivir (r) (A) or with rupintrivir at two inhibitor-to-protein molar ratios, 1:3 (B) and 30:3 (C). (D) C147A mutant protein. Chromatograms of the substrate peptide alone are shown in blue in all panels.
Table 2.
trans-cleavage efficiencies of 2C↓3A peptide by EV-93 3Cpro
| Protein | Inhibitor | Inhibitor-to-protein molar ratio (μM) | Cleavage efficiency Ea |
|---|---|---|---|
| EV-93 3Cpro | +++ | ||
| EV-93 3Cpro purified without DTT | +++ | ||
| EV-93 3Cpro C147A mutant | 0 | ||
| EV-93 3Cpro | Rupintrivir | 1:3 | +++ |
| 3:3 | + | ||
| 30:3 | 0 | ||
| EV-93 3Cpro | Compound 1 | 1:3 | ++ |
| 3:3 | + | ||
| 30:3 | 0 |
+++, efficiency of >90%; ++, efficiency of <20%; +, efficiency of <10%; 0, no visible product peaks.
Rupintrivir and compound 1 as potential antivirals against all EVs.
To characterize the molecular interactions of compound 1 and rupintrivir with EV-93 3Cpro, we cocrystallized their complexes and solved their crystallographic structures at very high resolutions. The structure of EV-93 3Cpro-compound 1 is the first known structure of a protease in complex with this antiviral agent. The electron density allowed us to unequivocally and precisely build one molecule of compound 1 or rupintrivir per protein (Fig. 5). In both cases, the inhibitor electrophilic β carbon (Fig. 1, asterisk) is covalently bound to the active-site Cys147 after its Michael addition, forming a stable tetrahedral adduct and resulting in the irreversible inactivation of the protease. Compound 1 binds to EV-93 3Cpro in a partially extended conformation with its peptidomimetic backbone making antiparallel β-sheet-type hydrogen bonds with part of the solvent-exposed β strand, E2, of the protein (residues 162 to 164). The inhibitor's P4 part (Fig. 5a) lies in the deep groove formed by the β strands E2, F2, and B2 and interacts with protein residues 125 to 128, 164 to 165, 168, and 170. The P3 part makes two main-chain hydrogen bonds with Gly163–164, and the hetero ring is mainly solvent exposed, interacting with Gly128 only on one side. P2 2-propynyl stacks against His40, and residues 71 and 127 further constrain its conformation. The P1 part is deeply inserted between β strands E2 (residues 162 to 164) and loop 142 to 144, making this pocket wider by about 1 Å than that of the native structure. A P1 glutamine-like side chain makes hydrogen bonds with Thr142 and His161, most probably mimicking the recognition of the natural substrate P1 Gln, which is highly conserved in 3C cleavage sequences. The P1′ carbonyl oxygen of the ethyl ester is positioned above the oxyanion hole formed by the amide groups of Cys147, Gln146, and Gly145 but makes a hydrogen bond only with the latter. The ethoxycarbonyl group is more mobile and is either solvent exposed or interacts with residues 23 to 25.
The binding mode of rupintrivir 1 to EV-93 3Cpro is very similar to that of compound 1. The differences between the two antivirals lie in the least-conserved P3 and P2 positions (Fig. 1 and 5b), where a 2-pyridon-1,3-diyl group that cycles with the following amine is replaced by a valine amino acid (4-methyl-pentan-2-one-1,3-diyl) and the ethynyl group is replaced by a 1-fluorobenzen-4-yl group in rupintrivir. Variations at the P3 position imply the loss of van der Waals contacts with Gly128 in the case of rupintrivir, since its valine side chain is exposed to the solvent and does not interact with the protein. In contrast, compared with that in compound 1 the P2 substitution in rupintrivir results in additional stacking interactions of the 1-flurobenzen-4-yl ring with His40 and Glu71 side chains and new interactions of the fluorine atom with Thr130 and Arg39. The stacking interactions of the inhibitor ring with His40 and Glu71 were also observed in the HRV-2 3Cpro-rupintrivir complex (30), as was the H bond of the fluorine atom with Thr130, which is an Asn in HRV-2 3Cpro. The contact with Arg39 is not present in the HRV-2 3Cpro-rupintrivir complex, since this residue is a Thr in the HRV-2 protein and has a much shorter side chain. The enhanced interactions of rupintrivir with the proteases at site S2 could explain why EC50s for this antiviral are almost systematically lower than those obtained for compound 1 against EV-93 and HRVs (see above and references 40 and 41). Interactions of rupintrivir with EV-93 and HRV-2 3Cpros also changed for the following residues: Leu125 and G128, which correspond to Ile and Ser in the HRV-2 protein, respectively, and Asn165 and Thr142, which have different side chain conformations in the two complexes.
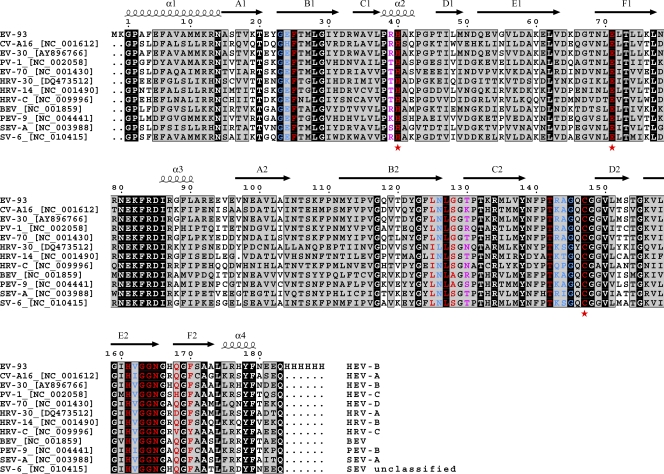
A noticeable feature of the binding pocket is the conservation of EV-93 3Cpro residues interacting with compound 1 within all EVs. For 11 EV species representing the entire genetic diversity of this genus, most of the 3Cpro residues making side chain interactions (71%; in red in Fig. 6) or only main-chain interactions (62%; in blue) with compound 1 are identical or physico-chemically similar. The four not strictly conserved residues involved in side chain interactions (Leu125, Gly128, Thr130, and Phe170) make steric interactions with compound 1 or rupintrivir that are compatible with the amino acid diversity observed. Thr130 interacts only with the fluorine end of the 1-fluorobenzen-4-yl group of rupintrivir (Fig. 1, P2), as was discussed above. A higher level of conservation of the 3C inhibitor binding pocket is observed in HRV serotypes (5).
Fig. 6.
Multiple-sequence alignment of 3Cpros for 11 EVs representing the currently known species diversity of the Enterovirus genus. The sequence names indicate the names and accession IDs of the viruses, and the corresponding species are indicated at the ends of the sequences. Identical and similar residues are boxed in black and gray, respectively. Residues making side chain interactions or only main-chain interactions between EV-93 3Cpro and compound 1 are highlighted in red and blue, respectively. Additional residues of EV-93 3Cpro that interact with rupintrivir are highlighted in magenta. Catalytic residues Cys174, His40, and Glu71 are indicated by red asterisks. Secondary structural elements correspond to EV-93 3Cpro, for which the sequence (top line) has been modified with N- and C-terminal extensions in this study. BEV, bovine enterovirus; PEV, porcine enterovirus; SEV, simian enterovirus.
In studies to control natural rhinovirus infection by 3Cpro inhibitors, compound 1 or rupintrivir showed unsatisfactory performance and was therefore excluded from further clinical development (39). Our results with EV-93 indicate that these compounds could be valuable antivirals against other EV species. The high level of conservation among EVs of the residues forming the 3Cpro binding pockets for compound 1 and rupintrivir and the broad-spectrum antiviral activity of these compounds in vitro reinforce their potential as excellent candidates for developing potent antivirals against all EVs (28). These results also suggest that the level of conservation of the residues forming the substrate binding pocket could be useful in the process of designing antiviral compounds against new, emerging enteroviruses.
Conclusion.
In summary, we report the first determination of the crystallographic structure of the main protease from a human enterovirus B (EV-93 3Cpro) alone and in complex with the HRV antiviral molecules compound 1 and rupintrivir at resolutions of 1.9, 1.3, and 1.5 Å, respectively. The chymotrypsin-like fold of the protease presents the catalytic triad Cys-His-Glu in the cleft between the two six-stranded β barrels, adjacent to a canonically configured oxyanion hole. We showed that compound 1 and rupintrivir inhibit the proteolytic activity of EV-93 3Cpro in vitro and are active against EV-93 in infected cells. The primary and tertiary structures of the 3Cpro binding pockets for these two compounds are highly conserved among EVs, which explains their broad-spectrum antiviral capacity. These results reinforce the structural framework for designing antiviral drugs against the 3Cpro to control enterovirus infections.
ACKNOWLEDGMENTS
This work was supported by the EC (Integrated Project VIZIER, contract LSHG-CT-2004-511960, and Cooperation Project SILVER, GA 260644), the Spanish Ministerio de Ciencia e Innovación (BFU2008-02372/BMC to M.C.), and the Generalitat de Catalunya (2009SGR-1309 to M.C.).
We thank Violaine Lantez and Karen Dalle for technical assistance in protein production. We also thank the personnel of the Automated Crystallography Platform (Barcelona Science Park, Spain) for technical assistance with crystallization and preliminary X-ray data collection; Chris Lauber for help with alignment; and Igor Sidorov, Alexander Kravchenko, and Dmitry Samborskiy for administering the Viralis software platform. We acknowledge the European Synchrotron Radiation Facility for providing access to synchrotron radiation facilities. We thank Pfizer for generously providing rupintrivir and compound 1.
Footnotes
Published ahead of print on 10 August 2011.
REFERENCES
- 1. Adams P. D., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anand K., et al. 2002. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 21:3213–3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bergmann E. M., Mosimann S. C., Chernaia M. M., Malcolm B. A., James M. N. 1997. The refined crystal structure of the 3C gene product from hepatitis A virus: specific proteinase activity and RNA recognition. J. Virol. 71:2436–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berrow N. S., et al. 2006. Recombinant protein expression and solubility screening in Escherichia coli: a comparative study. Acta Crystallogr. D Biol. Crystallogr. 62:1218–1226 [DOI] [PubMed] [Google Scholar]
- 5. Binford S. L., et al. 2005. Conservation of amino acids in human rhinovirus 3C protease correlates with broad-spectrum antiviral activity of rupintrivir, a novel human rhinovirus 3C protease inhibitor. Antimicrob. Agents Chemother. 49:619–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Binford S. L., et al. 2007. In vitro resistance study of rupintrivir, a novel inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 51:4366–4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bjorndahl T. C., Andrew L. C., Semenchenko V., Wishart D. S. 2007. NMR solution structures of the apo and peptide-inhibited human rhinovirus 3C protease (serotype 14): structural and dynamic comparison. Biochemistry 46:12945–12958 [DOI] [PubMed] [Google Scholar]
- 8. Care S., et al. 2008. The translation of recombinant proteins in E. coli can be improved by in silico generating and screening random libraries of a −70/+96 mRNA region with respect to the translation initiation codon. Nucleic Acids Res. 36:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Collaborative Computational Project Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 10. Cui S., et al. 2011. Crystal structure of human enterovirus 71 3C protease. J. Mol. Biol. 408:449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. De Palma A. M., Vliegen I., De Clercq E., Neyts J. 2008. Selective inhibitors of picornavirus replication. Med. Res. Rev. 28:823–884 [DOI] [PubMed] [Google Scholar]
- 12. Dougherty J. D., Park N., Gustin K. E., Lloyd R. E. 2010. Interference with cellular gene expression, p. 165–180In Ehrenfeld E., Domingo E., Roos R. P.(ed.), The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 13. Dougherty W. G., Semler B. L. 1993. Expression of virus-encoded proteinases: functional and structural similarities with cellular enzymes. Microbiol. Rev. 57:781–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Emsley P., Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 16. Gorbalenya A. E., et al. 2010. Practical application of bioinformatics by the multidisciplinary VIZIER consortium. Antiviral Res. 87:95–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gorbalenya A. E., Snijder E. J. 1996. Viral cysteine proteinases. Perspect. Drug Discov. Des. 6:64–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gouet P., Courcelle E., Stuart D. I., Metoz F. 1999. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15:305–308 [DOI] [PubMed] [Google Scholar]
- 19. Hayden F. G., et al. 2003. Phase II, randomized, double-blind, placebo-controlled studies of rupintrivir nasal spray 2-percent suspension for prevention and treatment of experimentally induced rhinovirus colds in healthy volunteers. Antimicrob. Agents Chemother. 47:3907–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Junttila N., et al. 2007. New enteroviruses, EV-93 and EV-94, associated with acute flaccid paralysis in the Democratic Republic of the Congo. J. Med. Virol. 79:393–400 [DOI] [PubMed] [Google Scholar]
- 21. Kaiser L., Crump C. E., Hayden F. G. 2000. In vitro activity of pleconaril and AG7088 against selected serotypes and clinical isolates of human rhinoviruses. Antiviral Res. 47:215–220 [DOI] [PubMed] [Google Scholar]
- 22. Kitamura N., et al. 1981. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature 291:547–553 [DOI] [PubMed] [Google Scholar]
- 23. Knowles N. J., Hovi T., King A. M. Q., Stanway G. 2010. Overview of taxonomy, p. 19–32In Ehrenfeld E., Domingo E., Roos R. P. (ed.), The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 24. Krissinel E., Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774–797 [DOI] [PubMed] [Google Scholar]
- 25. Landau M., et al. 2005. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33:W299–W302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26:283–291 [Google Scholar]
- 27. Lee C.-C., et al. 2009. Structural basis of inhibition specificities of 3C and 3C-like proteases by zinc-coordinating and peptidomimetic compounds. J. Biol. Chem. 284:7646–7655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee E. S., et al. 2007. Development of potent inhibitors of the coxsackievirus 3C protease. Biochem. Biophys. Res. Commun. 358:7–11 [DOI] [PubMed] [Google Scholar]
- 29. Matthews B. W. 1968. Solvent content of protein crystals. J. Mol. Biol. 33:491–497 [DOI] [PubMed] [Google Scholar]
- 30. Matthews D. A., et al. 1999. Structure-assisted design of mechanism-based irreversible inhibitors of human rhinovirus 3C protease with potent antiviral activity against multiple rhinovirus serotypes. Proc. Natl. Acad. Sci. U. S. A. 96:11000–11007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Matthews D. A., et al. 1994. Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA-binding site, and means for cleaving precursor polyprotein. Cell 77:761–771 [DOI] [PubMed] [Google Scholar]
- 32. McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. 2005. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61:458–464 [DOI] [PubMed] [Google Scholar]
- 33. Moriarty N. W., Grosse-Kunstleve R. W., Adams P. D. 2009. Electronic ligand builder and optimization workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 65:1074–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mosimann S. C., Cherney M. M., Sia S., Plotch S., James M. N. 1997. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J. Mol. Biol. 273:1032–1047 [DOI] [PubMed] [Google Scholar]
- 35. Murshudov G. N., Vagin A. A., Dodson E. J. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255 [DOI] [PubMed] [Google Scholar]
- 36. Norder H., et al. 2011. Picornavirus non-structural proteins as targets for new anti-virals with broad activity. Antiviral Res. 89:204–218 [DOI] [PubMed] [Google Scholar]
- 37. Otwinowski Z., Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode, p. 307–326In Carter C. W. J., Sweet R. M. (ed.), Methods in enzymology, vol . 276, part A Academic Press, New York, NY: [DOI] [PubMed] [Google Scholar]
- 38. Palmenberg A. C. 1990. Proteolytic processing of picornaviral polyprotein. Annu. Rev. Microbiol. 44:603–623 [DOI] [PubMed] [Google Scholar]
- 39. Patick A. K. 2006. Rhinovirus chemotherapy. Antiviral Res. 71:391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Patick A. K., et al. 1999. In vitro antiviral activity of AG7088, a potent inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 43:2444–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Patick A. K., et al. 2005. In vitro antiviral activity and single-dose pharmacokinetics in humans of a novel, orally bioavailable inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 49:2267–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Perrakis A., Morris R., Lamzin V. S. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6:458–463 [DOI] [PubMed] [Google Scholar]
- 43. Porter A. G. 1993. Picornavirus nonstructural proteins: emerging roles in virus replication and inhibition of host cell functions. J. Virol. 67:6917–6921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Qiu J. 2008. Enterovirus 71 infection: a new threat to global public health? Lancet Neurol. 7:868–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Skern T., et al. 2002. Structure and function of picornavirus proteinases, p. 199–212In Semler B. L., Wimmer E.(ed.), Molecular biology of picornaviruses. American Society for Microbiology, Washington, DC [Google Scholar]
- 46. Steller I., Bolotovsky R., Rossmann M. G. 1997. An algorithm for automatic indexing of oscillation images using Fourier analysis. J. Appl. Cryst. 30:1036–1040 [Google Scholar]
- 47. Sweeney T. R., Roque-Rosell N., Birtley J. R., Leatherbarrow R. J., Curry S. 2007. Structural and mutagenic analysis of foot-and-mouth disease virus 3C protease reveals the role of the beta-ribbon in proteolysis. J. Virol. 81:115–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wecker I., ter Meulen V. 1977. RD cells in the laboratory diagnosis of enteroviruses. Med. Microbiol. Immunol. 163:233–240 [DOI] [PubMed] [Google Scholar]
- 49. Ziebuhr J., Snijder E. J., Gorbalenya A. E. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853–879 [DOI] [PubMed] [Google Scholar]