Abstract
Human cytomegalovirus (HCMV) remains the leading viral cause of birth defects and life-threatening disease in transplant recipients. All approved antiviral drugs target the viral DNA polymerase and are associated with severe toxicity issues and the emergence of drug resistance. Attempts to discover improved anti-HCMV drugs led to the identification of the small-molecular-weight compound AIC246 (Letermovir). AIC246 exhibits outstanding anti-HCMV activity in vitro and in vivo and currently is undergoing a clinical phase IIb trial. The initial mode-of-action studies suggested that the drug acts late in the HCMV replication cycle via a mechanism distinct from that of polymerase inhibitors. Here, we extend our mode-of-action analyses and report that AIC246 blocks viral replication without inhibiting the synthesis of progeny HCMV DNA or viral proteins. The genotyping of mutant viruses that escaped AIC246 inhibition uncovered distinct point mutations in the UL56 subunit of the viral terminase complex. Marker transfer analyses confirmed that these mutations were sufficient to mediate AIC246 resistance. The mapping of drug resistance to open reading frame UL56 suggests that viral DNA processing and/or packaging is targeted by AIC246. In line with this, we demonstrate that AIC246 affects the formation of proper unit-length genomes from viral DNA concatemers and interferes with virion maturation. However, since AIC246-resistant viruses do not exhibit cross-resistance to previously published terminase inhibitors, our data suggest that AIC246 interferes with HCMV DNA cleavage/packaging via a molecular mechanism that is distinct from that of other compound classes known to target the viral terminase.
INTRODUCTION
Despite modern prevention and treatment strategies, human cytomegalovirus (HCMV) continues to represent a common opportunistic pathogen associated with serious morbidity and mortality in immunocompromised patients, such as recipients of bone marrow and solid-organ transplants. In addition, HCMV remains one of the most common congenitally acquired infections in the developed world, leading to birth defects, including deafness, mental retardation, and mortality (16, 18, 20, 33).
Current standard therapy for HCMV relies on oral or intravenous ganciclovir (GCV) or its oral prodrug, valganciclovir (VGCV). Although efficacious, GCV treatment suffers from dose-related toxicities, including bone marrow suppression, which interferes with the recovery of the patient's bone marrow and immune system. Foscarnet (FOS) and cidofovir (CDV), the two commonly used second-line treatments for HCMV, also are associated with significant toxicities, including renal toxicity. Moreover, results from animal experiments indicate that all currently available systemic HCMV agents are teratogenic, mutagenic, and potential carcinogens (1, 24, 27, 32). During prolonged and/or repeated application, HCMV can become resistant to GCV and lead to treatment failure in both solid-organ and bone marrow transplant recipients. Depending on the transplanted organ, the incidence of GCV-resistant virus in organ transplant recipients is 5 to 10% (35). Worryingly, GCV resistance in HCMV infections appears to have been increasing during recent years, and the emergence of cross-resistance to either or both second-line agents (FOS and CDV) is encountered in all forms of transplantation (42). Mechanistically, the latter is explained by the fact that all licensed drugs used for the systemic treatment of HCMV share a common target molecule, the viral DNA polymerase pUL54 (15, 35, 42–44). In light of these facts, there is an urgent medical need for new anti-HCMV drugs that do not have the limitations associated with the existing therapeutics and that combine efficacy with a novel mode of action, thus excluding cross-resistance to the available anti-HCMV agents.
A promising new drug candidate with potential to improve HCMV therapy is the small-molecule compound AIC246, which is a representative of a new class of nonnucleoside HCMV inhibitors, the 3,4 dihydro-quinazolines (28). We have shown previously that AIC246 exhibits an outstanding anti-HCMV activity in vitro and demonstrates an antiviral profile superior to that of the commonly used polymerase inhibitors (28). AIC246 is one of the most potent anti-HCMV agents reported to date, with a cell culture EC50 in the one-digit nanomolar range (∼5 nM) and a selectivity index exceeding 15,000 (28). Besides its excellent in vitro inhibitory activity against HCMV laboratory strains, AIC246 also (i) exhibits potent antiviral activity against clinical isolates, (ii) retains activity against virus strains resistant to currently approved antivirals, and (iii) exerts a potent in vivo efficacy in a mouse xenograft model (28). Phase I trials demonstrated that AIC246 was generally well tolerated and showed a high and long-lasting exposure in human subjects, allowing once-daily dosing (25). Moreover, a proof of concept was shown in a recent phase IIa trial and in a patient infected with a multidrug-resistant virus causing multiorgan cytomegalovirus disease (22, 50). Currently, a phase IIb multicenter randomized double-blind placebo-controlled dose escalation trial of AIC246 on the prevention of HCMV infection/reactivation in hematopoietic stem cell transplant recipients is in progress (clinicaltrials.gov).
As outlined above, a critical need exists for new HCMV antivirals with targets other than the viral DNA polymerase. Our previous observations suggested that AIC246 can fulfill this criterion (28). Here, we extend our mode of action studies and report on experiments aiming to more precisely identify the steps during viral life cycle compromised by AIC246 and to elucidate the molecular target for this novel antiviral compound. We demonstrate that neither HCMV DNA replication nor HCMV protein expression is affected by the drug. Instead, our data suggest that AIC246 inhibits HCMV replication through a specific antiviral mechanism that involves the viral terminase subunit pUL56 but that is distinct from that of other compound classes also known to target this enzyme complex.
MATERIALS AND METHODS
Antiviral compounds and drug susceptibility assays.
AIC246, BAY 38-4766, and BDCRB were synthesized at the Medical Chemistry Department of Bayer Schering Pharma AG, Wuppertal, Germany, and stored as a 50 mM stock solution in dimethylsulfoxide (DMSO) for in vitro use. The intravenous formulations of ganciclovir (Cymevene; Roche), foscarnet (Foscavir; AstraZeneca), and cidofovir (Vistide; Gilead) were used as 50 mM solutions in 0.9% saline.
HCMV cytopathic effect reduction assays (CPE-RAs) and green fluorescent protein (GFP)-based fluorescence reduction assays (GFP-RAs) were performed as described previously (28). All assays were done at least three times with duplicate samples, and standard deviations were calculated.
Cells, cell culture, and viruses.
Normal human dermal fibroblast cells (NHDF; no. CC-2511), human lung fibroblast cells (MRC5; no. CCL-171), and HELF human embryonic lung fibroblast cells (HEL299; no. 87042207) were purchased from Clonetics, the American Type Culture Collection (ATCC), and the European Collection of Cell Cultures (ECACC), respectively, and were cultured as described previously (28, 40). HFF cells were prepared from human foreskin tissue as previously described (21). The HCMV laboratory strain AD169 was purchased from the ATCC (ATCC VR 538 and VR-807), and the AD169-derived recombinant virus RV-HG was reconstituted from the HCMV BAC pHG, kindly provided by E. Borst and M. Messerle (9). Inserted in the unique short region, HCMV BAC pHG contains an enhanced green fluorescent protein (EGFP) reporter gene expressed under the control of the major immediate-early (IE) promoter (8). HCMV virus stocks were propagated using NHDF cells and titrated by means of IE1p72 fluorescence as described previously (2, 34). For single-step growth curves, 1.5 × 105 NHDF cells seeded in 12-well dishes were infected in triplicate at a multiplicity of infection (MOI) of 0.1 PFU/cell. Cell supernatants were collected at 24-h intervals for 8 days and stored at −80°C until the end of the experiment. The virus titer was determined by the immediate-early antigen staining of infected cell nuclei.
Selection of resistant mutants.
Drug-resistant HCMV AD169 mutants were selected by the single-step selection method (5, 12). Briefly, 5 × 103 AD169-infected NHDF cells/well (MOI, 0.03 PFU/cell) were seeded into the wells of 30 96-well microtiter plates. The infection was allowed to proceed under the exposure of 50 nM AIC246 (∼10× EC50) until a CPE developed in one or more of the compound-treated wells (indicative of resistant virus breakthrough). Noninfected and nontreated cells served as controls on each plate. Mutant virus amplification was accomplished after cultures achieved maximum CPE by the passage of cell-free supernatant virus in the presence of 50 nM AIC246. The resultant AIC246-resistant progeny virus mutants were plaque purified three times by limiting dilutions in the presence of AIC246. The stability of resistance was tested by serially passaging plaque-purified viruses without selective pressure (8 to 10 times). The resistant phenotype of the mutants was confirmed by CPE reduction assays. The sequencing of the open reading frames (ORFs) UL56, UL89, and UL104 revealed the respective UL56 mutations indicated in Table 3.
Table 3.
Genotyping of AIC246-resistant viruses
| HCMV strain | Clone | UL56 |
UL89 |
UL104 |
|||
|---|---|---|---|---|---|---|---|
| DNAa | AAb | DNA | AA | DNA | AA | ||
| AD169 | —c | — | — | — | — | — | |
| rAIC246-1 | 5C9ppG9 | t723c | L241P | — | — | — | — |
| rAIC246-2 | Mt7E8 | g1107c | R369S | — | — | — | — |
Nucleotide exchange.
Amino acid exchange.
—, not applicable.
Real-time PCR.
For real-time PCR, DNA was extracted from virus-infected cells (MOI, 0.1 to 1 PFU/cell) that were grown in the presence of placebo, AIC246 (10 nM), or GCV (20 μM) using the DNeasy tissue 96 kit (Qiagen, Hilden, Germany). Real-time PCR was performed as described elsewhere (34) with the following exceptions: 200 nM concentrations of each primer and 200 nM probe directed against the HCMV IE region were used. The respective sequences of probe and primers were published previously (34). To calculate the genomic equivalents/cell ratio, primers 5′ TCA CTG TGT GTA AAC ATG ACT TCC A and 3′ TTC ACA CAG AGC TGC AGA AAT CA, and the 6-carboxyfluorescein/6-carboxytetramethylrhodamine (FAM/TAMRA)-labeled probe CTG GCC GTG GCT CTC TTG GCA G directed against the cellular interleukin-8 (IL-8) gene, were used to quantify cellular DNA. The CMV DNA standard for quantification was prepared by the PCR amplification of HCMV DNA extracted from the supernatant of AD169-infected cells (primers 5′cmv-TCT CAG CCA CAA TTA CTG AGG ACA GAG GGA and 3′cmv-GGT CAC TAG TGA CGC TTG TAT GAT GAC CA), and the IL-8 DNA standard was prepared from total DNA extracted from peripheral blood mononuclear cells (PBMCs). The supernatant of the infected cells was stored at −80°C and used to titrate progeny virus yield by means of IE1p72 fluorescence as described above.
Antibodies, indirect immunofluorescence, and Western blot analysis.
The monoclonal antibody (MAb) 63-27 (IE1, UL123), MAb 65-33 (pp65, UL83), MAb 28-4 (MCP, UL86), and MAb 41-18 (pp28, UL99) and the polyclonal antisera against IE2 and UL69 were described elsewhere and were kindly provided by W. Britt (University of Birmingham, AL), M. Mach (University of Erlangen, Germany), and T. Stamminger (University of Erlangen, Germany) (29, 31, 45). The monoclonal antibody MAb84 (UL84) was purchased from Novus Biologicals (Cambridge, United Kingdom), and the antibody sc-69744 (UL44) was obtained from Santa Cruz (Santa Cruz, CA). Endogenous β-actin was detected by using the MAb AC-15 purchased from Sigma-Aldrich (Deisenhofen, Germany). Anti-mouse, anti-rabbit horseradish peroxidase-conjugated secondary antibodies were obtained from Dianova (Hamburg, Germany), while Alexa 488- and Alexa 555-conjugated secondary antibodies were purchased from Invitrogen (Karlsruhe, Germany).
For indirect immunofluorescence analysis, HCMV AD169 (MOI, 0.1 PFU/cell)-infected NHDF cells were grown on coverslips either in the absence of drug or in the presence of ∼10 times the EC50 for AIC246 (50 nM) or GCV (20 μM). At 8, 48, or 72 h postinfection (p.i.), cells were fixed with ice-cold methanol, and the indicated proteins were detected by indirect immunofluorescence analysis according to a standard protocol (30). Cell nuclei were counterstained using 4′,6′-diamidino-2-phenylindole (DAPI), and dishes were analyzed with a Leica DM-RB microscope.
For Western blotting, whole-cell extracts of mock-infected or HCMV AD169 (MOI, 0.1 PFU/cell)-infected NHDF cells, grown in the presence or absence of 50 nM AIC246, were prepared at the indicated time points p.i. using radioimmunoprecipitation assay (RIPA) lysis buffer (Sigma-Aldrich, Deisenhofen, Germany). Protein lysates were separated on 10% Bis-Tris polyacrylamide gels (Novex NuPAGE; Invitrogen, Karlsruhe, Germany) and transferred to nitrocellulose membranes (Schleicher & Schuell, Dassel, Germany). Antibody incubation and chemiluminescence detection were done according to the manufacturer's protocol (Amersham ECL Western blotting detection kit; GE Healthcare, Munich, Germany).
En passant BAC mutagenesis and reconstitution of virus mutants.
The HCMV AD169-derived bacterial artificial chromosome (BAC) pHG served as the backbone for the construction of the two mutant viruses reported in this study (9). The markerless introduction of point mutations into pHG was done using a two-step recombination protocol according to the en passant method previously published by Tischer et al. (46). Briefly, a recombination fragment containing the I-SceI-AphAI cassette from plasmid pEPKan-S was obtained by PCR using primers containing ∼60 bp of homology to the intended integration site in ORF UL56 of BAC pHG and ∼20 bp specific for pEPKan-S (Table 1). This fragment was electroporated into Escherichia coli GS1783 (generous gift of Greg Smith, Northwestern University, Chicago, IL) (3) that harbored the pHG BAC. The first Red recombination resulted in a selectable BAC with an ISceI restriction site and a kanamycin cassette flanked by a duplication of the UL56 target sequence. After successful kanamycin selection, all non-HCMV sequences were removed by an intrabacterial ISceI digest and a subsequent second Red recombination, resulting in the scarless repair of the mutated UL56 gene in the background of BAC pHG. The integrity of the generated BACs designated pHG-UL56-L241P and pHG-UL56-R369S was confirmed by restriction enzyme digestion and sequencing. To reconstitute virus mutants, the recombinant BACs were transfected into permissive MRC5 cells basically as described elsewhere (45). Reconstituted mutant viruses were termed RV-HG-UL56-L241P and RV-HG-UL56-R369S. At least two independent virus clones per mutation were generated and evaluated.
Table 1.
Oligonucleotides used for the construction of recombinant BACs
| Primer | Sequencea |
|---|---|
| 5′UL56 L241P | 5′-CAGTCCGATGTGAATATCCAGACGGTGGAGCAGGACCTGCCGGACCTGACAACGCGCATCAGGATGACGACGATAAGTAGGG-3′ |
| 3′UL56 L241P | 5′-AGGACTCCAGCCAAGTGGGGGATGCGCGTTGTCAGGTCCGGCAGGTCCTGCTCCACCGTCCAACCAATTAACCAATTCTGATTAG-3′ |
| 5′UL56 R369S | 5′-GAGCGCATGTTCGTGGGGTCGGTCTTTGCCGCCCCCAACAGCATCATCGACCTCATCACATCAGGATGACGACGATAAGTAGGG-3′ |
| 3′UL56 R369S | 5′-CGAAAGCTTGAATGCTGAGGGATGTGATGAGGTCGATGATGCTGTTGGGGGCGGCAAAGACAACCAATTAACCAATTCTGATTAG-3′ |
The underlined sequence anneals to plasmid pEPkan-S.
Viral DNA cleavage assay.
Functional viral DNA cleavage assays were performed basically as previously described (10, 40). Briefly, HELF cells (5 × 107) were infected either with AD169 or AD169rAIC246-1 at an MOI of 1 PFU/cell. After virus adsorption, cells were treated for 96 h either with ∼50-fold the EC50 of BAY 38-4766 (25 μM), BDCRB (20 μM), or with the indicated multiples of the AIC246 EC50 (4 nM). One sample was kept untreated and served as a positive control. After the 96-h incubation period, DNA was extracted from infected cells using the blood and cell culture DNA kit (Qiagen, Hilden, Germany). Viral DNA was quantified by real-time PCR or slot-blot analysis, and comparable amounts of HCMV DNA were digested with 20 U of KpnI for 2 h, size fractionated on a 0.7% agarose gel by electrophoresis, and blotted to nylon membranes (Millipore, Schwalbach, Germany) via Southern transfer. After UV cross-linking, Southern blot analysis was performed according to standard protocols. Terminase-cleaved and uncleaved DNA fragments were visualized using a randomly primed, digoxigenin-labeled PCR fragment (HCMV genome positions 703 to 1524). Detection was carried out by luminescence as instructed by the manufacturer (New England BioLabs, Ipswich, MA).
Electron microscopy.
Transmission electron microscopy was performed as described in reference 21. In brief, placebo- or AIC246 (50 nM)-treated, late-stage AD169-infected HFF cells were fixed primarily in buffered 2.5% glutaraldehyde solution for 15 min at 37°C, followed by 45 min of fixation at 4°C. After extensive washing, cells were postfixed in buffered 1% osmium tetroxide for 1 h at room temperature. The third fixation was done by incubation with 1% aqueous uranyl acetate. After a final washing step, samples were dehydrated and embedded in epoxy resin according to standard protocols. Ultrathin sections were stained with uranyl acetate and lead citrate and subjected to electron microscopic analysis.
RESULTS
Effect of AIC246 on HCMV-DNA synthesis and gene expression.
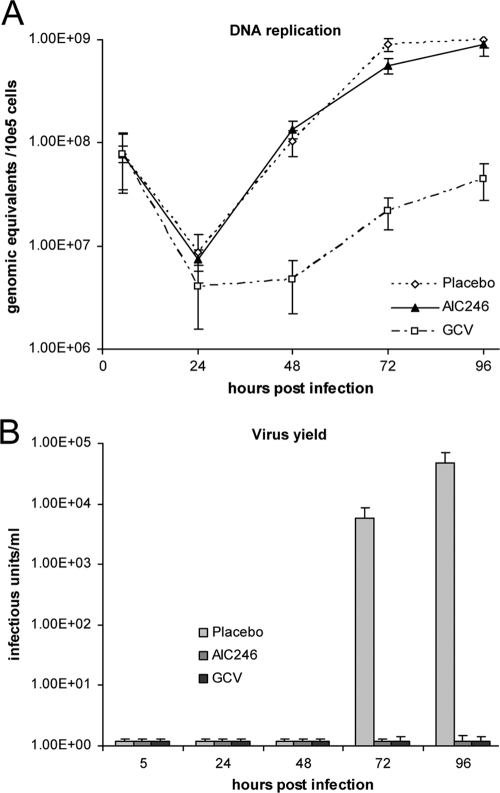
Prior studies in our laboratories raised the hypothesis that the novel HCMV inhibitor AIC246 targets a process in the HCMV replication cycle that occurs later than DNA synthesis (28). To verify this, the kinetics of viral DNA accumulation in infected cells was monitored in the presence of AIC246 or the specific polymerase inhibitor GCV. NHDF cells were infected with HCMV AD169 and overlaid with medium containing either DMSO (placebo) or inhibitory concentrations (∼10× the EC50) of AIC246 or GCV. Total DNA was extracted from infected cells at the indicated time points postinfection, and viral DNA was quantified using real-time PCR (Fig. 1A). As expected, the polymerase inhibitor GCV exerted a marked inhibitory effect on the de novo synthesis of progeny DNA. In contrast, comparable HCMV DNA synthesis kinetics were observed for cells treated with AIC246 or placebo. Virus yield measurements confirmed the efficacy of the used antivirals, since no infectious virus particles were detected in the supernatant of cells treated either with AIC246 or GCV (Fig. 1B). Taken together, these data support our initial hypothesis and point to a late (L) antiviral target for AIC246 inhibition.
Fig. 1.
Synthesis of HCMV DNA in the presence of AIC246 or ganciclovir. (A) HCMV AD169-infected cells were treated with placebo, AIC246, or ganciclovir (GCV). Total intracellular DNA was harvested during a period of 96 h postinfection as indicated, and viral progeny DNA was measured by quantitative real-time PCR. All experiments were performed in triplicate, and standard deviations are depicted by error bars. (B) Production of progeny virus at the indicated time points monitored via virus yield measurements.
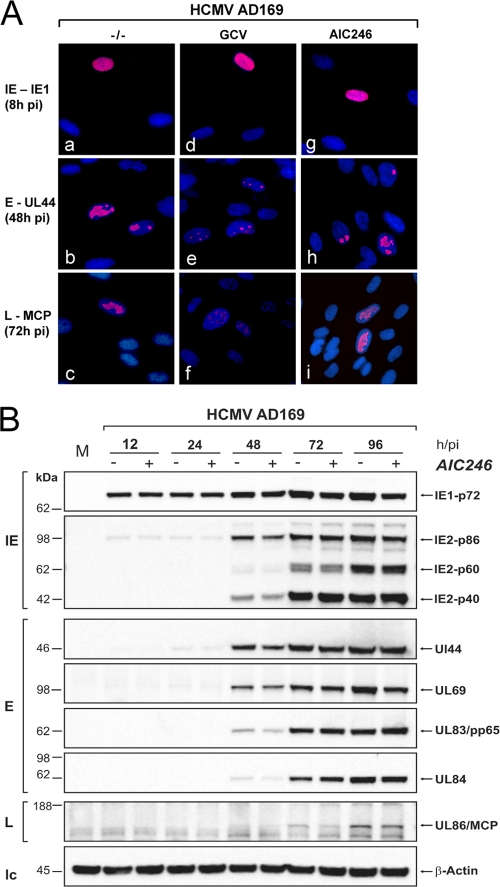
To further explore the mode of action of AIC246, we determined whether the antiviral activity of the compound is accompanied by a delayed or a reduced expression of viral proteins. To this end, we first analyzed the HCMV antigen expression kinetics in nontreated and compound-treated cells by immunofluorescence (Fig. 2A). Coverslip cultures were infected with AD169 in the presence or absence of inhibitory concentrations of GCV, which served as a control compound, or AIC246. Cells were fixed at the indicated time points and then stained for representative immediate-early (IE), early (E), and L antigens. Under these conditions, no difference was found regarding the time points of the appearance of IE (IE1), E (UL44), and L (MCP) antigens in untreated (Fig. 2A, images a to c) and AIC246-treated (Fig. 2A, images g to i) cell cultures. IE antigen appeared at 8 h, early antigen appeared at 48 h, and late antigen appeared at 72 h after infection. In contrast, a significant delay of E and L antigen expression was observed in cells treated with the control compound GCV (Fig. 2A, images e and f). These findings indicate that in AIC246-treated cells, HCMV gene expression proceeds normally toward the late phase of replication. To more closely explore the effect of AIC246 on HCMV protein expression, we also asked whether the drug exerts effects on the expression levels of viral proteins. For this, Western blotting at various times p.i. was performed for a panel of representative virus-encoded proteins of the IE, E, and L classes of genes. As depicted in Fig. 2B, all IE (IE1 and IE2), E (UL44, UL69, UL83, and UL84), and L (UL86) proteins analyzed after 96 h showed expression kinetics that accumulated to comparable levels either in the absence or presence of the inhibitor, thus confirming the results of our immunofluorescence experiments. We concluded from these collective data that differences in neither HCMV DNA synthesis nor protein expression can account for the antiviral activity of AIC246.
Fig. 2.
Effect of AIC246 on expression of representative immediate-early (IE), early (E), and late (L) proteins of HCMV. (A) Immunofluorescence analyses. HCMV AD169-infected fibroblast cells were treated with either GCV (d to f) or AIC246 (g to i) or were kept untreated (a to c). Cells were fixed at the indicated time postinfection (pi) and stained for IE1 (a, d, and g), UL44 (b, e, and h), or MCP (c, f, and i) expression (red signals). Cell nuclei were counterstained using DAPI (blue signals). (B) Western blot analyses. NHDF cell monolayers were infected with HCMV strain AD169 at an MOI of 0.1 PFU/cell. The infection was allowed to proceed in the absence (−) or presence (+) of AIC246. At different time points pi (h/pi), cells were harvested and processed for Western blotting. Representative HCMV IE, E, or L proteins indicated at the right of each panel were visualized using the respective antibodies. Cellular β-actin served as a loading control (lc). Size markers (in kDa) are indicated on the left. M, mock-infected cells.
Isolation and characterization of AIC246-resistant viruses.
To gain deeper insight into the molecular mechanism of AIC246 antiviral activity and to facilitate the identification of the putative viral target of the drug, we next sought to isolate mutant viruses that escape compound inhibition. By applying a one-step selection procedure, we independently isolated two resistant HCMV strains that were capable of growing in the presence of 10 times the EC50 of AIC246. The resulting AD169 mutant strains, designated rAIC246-1 and rAIC246-2, were compared to the parent strain AD169 for their ability to replicate in the presence of AIC246 in a CPE reduction assay. GCV was included in these experiments as a reference compound. The results are summarized in Table 2 and demonstrate that both virus mutants were highly resistant to AIC246, with EC50s ∼220- or ∼65-fold higher than those obtained for the parental virus. Moreover, both viruses retained sensitivity to GCV, which further strengthens the assumption that the target site of AIC246 is different from that of GCV.
Table 2.
Isolation of mutant viruses that escape AIC246 inhibition
| HCMV strain | Clone | EC50a (μM) for: |
RIb | |
|---|---|---|---|---|
| AIC246 | GCV | |||
| AD169 | 0.0056 ± 0.0016 | 3.5 ± 1.5 | — | |
| rAIC246-1 | 5C9ppG9 | 1.24 ± 0.38 | 1.2 ± 0.2 | 221 |
| rAIC246-2 | Mt7E8 | 0.37 ± 0.07 | 4.0 ± 0.9 | 66 |
EC50s were determined by a CPE reduction assay. Data are means from at least three independent experiments and are expressed with standard deviations.
The resistance index (RI) is the AIC246 EC50 for mutant virus divided by the AIC246 EC50 for wild-type virus. —, not applicable.
Identification of mutations putatively conferring resistance to AIC246.
We next aimed to identify the mutated viral genes responsible for the AIC246-resistant phenotype via sequence analysis. Genes for sequencing were selected based upon our previous finding that the time point of action of AIC246 exactly coincides with that of the chemically unrelated HCMV inhibitor BAY 38-47766 in time-of-drug-addition studies, which raises the possibility of a common viral target (10, 28). Mutations conferring resistance to BAY 38-4766 have been mapped to the HCMV genes UL56 and UL89, which encode the two subunits of the viral terminase. Additional mutations were detected in ORF UL104 which, however, were not related to BAY 38-4766 resistance (10). Nevertheless, it is known that the interaction of the terminase subunit pUL56 with the putative portal protein pUL104 is a further prerequisite for the proper cleavage and packaging of HCMV DNA into procapsids. Therefore, ORF UL104 was included in our genotyping analyses (14, 23). As depicted in Table 3, a comparison of the UL56, UL89, and UL104 nucleotide sequences from the resistant viruses to that of the parental AD169 strain revealed that both resistant viruses harbored only one distinct point mutation in ORF UL56, implicating this protein as being involved in the AIC246 mechanism of action.
Recombinant phenotyping by marker transfer.
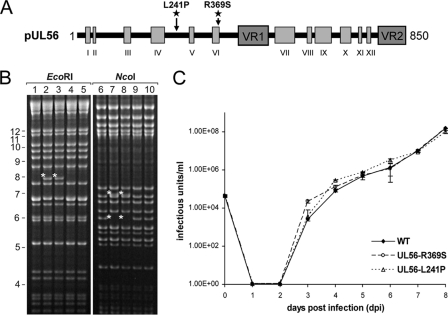
To obtain direct evidence that the identified UL56 mutations were necessary and sufficient to cause resistance to AIC246, the marker transfer of the respective mutations to HCMV strain AD169 was performed. Applying markerless BAC mutagenesis, we independently introduced the UL56 nucleotide mutations t723c (L241P) and g1107c (R369S) into the GFP-expressing, AD169-derived BAC pHG (9, 28). The structural integrity of the resulting BACs pHG-UL56-L241P and pHG-UL56-R369S was examined by restriction enzyme cleavage and sequencing (Fig. 3A and B). Infectious virus was reconstituted by transfecting BAC DNA into permissive MRC5 cells. We then characterized the growth properties of the obtained recombinant viruses in one-step growth curves. As shown in Fig. 3C, the parental virus RV-HG and the two virus mutants RV-HG-UL56-L241P and RV-HG-UL56-R369S exhibited similar growth kinetics in this experiment. Hence, the putative resistance mutations had little or no effect on the growth properties of the recombinants in vitro.
Fig. 3.
Markerless introduction of point mutations into ORF UL56 via BAC mutagenesis (marker transfer). (A) Schematic representation of the UL56 domain organization according to Champier et al. (11). Conserved regions are indicated as light gray boxes (I to XII) and variable regions (VR1 and VR2) as dark gray boxes. The localizations of the independently inserted UL56 mutation t723c (L241P) and g1107c (R369S) are highlighted by asterisks. (B) EcoRI or NcoI restriction analysis of WT BAC pHG (lanes 1 and 6) and recombinant BACs pHG-UL56-L241P (lanes 2, 4, 7, and 9) and pHG-UL56-R369S (lanes 3, 5, 8, and 10), respectively. DNA was size fractionated by a 0.9% agarose gel. Asterisks indicate new EcoRI or NcoI restriction fragments resulting as intermediates from recombination. (C) Single-step growth curve of reconstituted viruses RV-HG (WT), RV-HG-UL56-R369S (UL56-R369S), and RV-HG-UL56-L241P (UL56-L241P). NHDF cultures were infected at an MOI of 0.1 PFU/cell. At the indicated time points postinfection, supernatants were harvested and titrated via IE1 staining. Standard deviations are derived from three independent experiments.
To precisely elucidate the role of the UL56 L241P and R369S substitution in drug resistance, we next determined the drug susceptibility profile for the two newly constructed virus mutants by fluorescence reduction assays. The data summarized in Table 4 show that the two RV-HG-UL56 mutants were resistant to AIC246 at levels similar to those of the initial AIC246-resistant isolates (compare Tables 2 and 4). Mutation L241P conferred an ∼160-fold increase and mutation R369S an ∼38-fold increase in the respective EC50s. As expected, the sensitivity of the two mutants to approved control compounds that act by a different mechanism (GCV, CDV, and FOS) did not differ from the sensitivity of the parental wild-type virus RV-HG (Table 4). The finding that mutations within one terminase subunit confer resistance to AIC246 prompted us to examine whether the AIC246-resistant viruses demonstrate cross-resistance against sulfonamides (e.g., BAY 38-4766) or benzimidazoles (e.g., BDCRB), the only two chemical classes of HCMV inhibitors that were reported to target the viral terminase (10, 26, 40, 47, 48). Interestingly, we noticed that both compounds were comparably active against the parental wild-type HCMV (RV-HG) as well as against the two AIC246-resistant strains (Table 4). Taken together, these findings (i) confirm that a single conservative L241P or R369S amino acid substitution is necessary and sufficient to produce AIC246 resistance in vitro, (ii) suggest that AIC246 inhibits HCMV replication through a specific antiviral mechanism that involves the viral gene product UL56, and (iii) imply that AIC246 exerts its effects at the molecular level via a mechanism that is distinct from that of other compound classes known to target the HCMV terminase.
Table 4.
Susceptibility of reconstituted WT virus and constructed UL56 mutant viruses to AIC246, approved polymerase inhibitors, and drugs targeting the viral terminase
| Drug | EC50a (μM) for: |
RIb | ||
|---|---|---|---|---|
| RV-HG | RV-HG UL56-L241P | RV-HG UL56-R369S | ||
| AIC246 | 0.0029 ± 0.001 | 0.47 ± 0.06 | 0.11 ± 0.04 | 160, 38 |
| GCVc | 1.79 ± 0.76 | 1.72 ± 0.36 | 2.02 ± 1.39 | — |
| CDVc | 0.22 ± 0.15 | 0.16 ± 0.02 | 0.15 ± 0.06 | — |
| FOSc | 113 ± 57 | 76 ± 34 | 56 ± 20 | — |
| BDCRBd | 0.32 ± 0.12 | 0.29 ± 0.02 | 0.20 ± 0.03 | — |
| Bay38-4766d | 0.36 ± 0.07 | 0.27 ± 0.08 | 0.52 ± 0.13 | — |
EC50s were determined by the fluorescence reduction assay. Data are means from at least three independent experiments and are expressed with standard deviations.
The resistance index (RI) is the EC50 for the indicated RV-HG mutant divided by the EC50 of the RV-HG WT. RI values given are for L241P and R369S mutations, respectively. —, not applicable.
Approved polymerase inhibitor.
Discontinued terminase inhibitor.
Effect of AIC246 on maturation of viral progeny DNA.
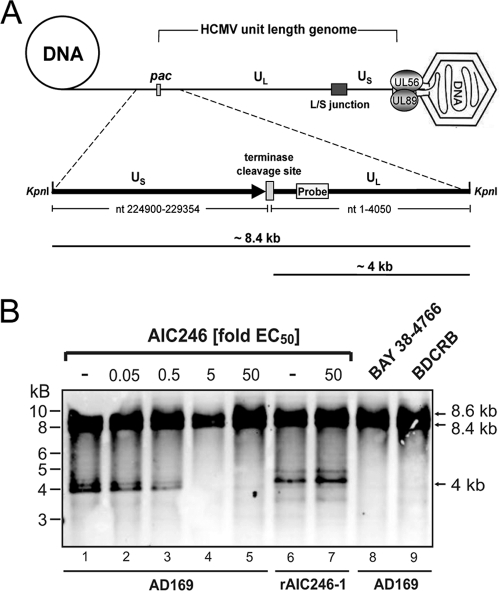
The presented experiments raised the possibility that AIC246 inhibition exerts some effects on the activity of the viral terminase complex that is involved in cleavage packaging of HCMV DNA. Consequently, we analyzed the impact of AIC246 on the activity of this enzyme by employing a functional viral DNA cleavage assay (10, 40). For this, total DNA was isolated from untreated or inhibitor-treated late-stage AD169-infected cells and was subjected to KpnI restriction followed by Southern blotting using an HCMV-specific probe that binds to the 5′-terminal end of the virus genome. As illustrated schematically in Fig. 4A and consistently with previous studies by Reefschlaeger et al. (40), the KpnI restriction digest of isolated HCMV DNA gives two possible patterns using this probe: (i) uncleaved concatemeric HCMV DNA formed in infected cells gives rise to an ∼8.4-kb restriction fragment spanning the terminase cleavage site (7, 41), and (ii) in contrast, concatemeric DNA that has been cleaved to mature unit-length genomes by the viral terminase enzyme exhibits a free genomic terminus of ∼4 kb upon KpnI cleavage that is derived from the left end of the viral genome. Thus, the presence of a 4-kb KpnI fragment in cells at the late stage of infection is indicative of the proper processing of viral DNA (Fig. 4A and B, lane 1). Notably, because terminal genome sequences also are found at the internal region where the long and short arms of the HCMV genomes join (L/S junction) (Fig. 4A), an additional KpnI fragment with a molecular size of 8.6 kb hybridized to the terminal probe used in this study (Fig. 4B). As shown in Fig. 4B, lanes 1 to 5, AIC246 inhibited the formation of the ∼4-kb terminal fragment in a concentration-dependent manner, indicating that the drug allowed the accumulation of viral concatemeric DNA while further maturation to mature unit-length genomes was prevented. Consistently with this interpretation, a high concentration of AIC246 (50× the EC50) did not change the DNA restriction pattern of the AIC246-resistant virus rAIC246-1 that was used as a specificity control (Fig. 4B, lanes 6 and 7). For the purpose of comparison, we also assayed the susceptibility of WT AD169 to high concentrations (50× the EC50) of BAY 38-4766 and BDCRB (Fig. 4B, lanes 8 and 9). Consistently with previous reports, cells treated with these drugs contained concatemeric DNA but virtually no 4-kb DNA fragment derived from mature unit-length genomes (36, 40, 48). These findings therefore demonstrate that although not cross-resistant to other terminase inhibitors, AIC246 activity is comparably related to the processing (cleavage/packaging) of concatemeric DNA, a process catalyzed by the viral terminase.
Fig. 4.
Functional viral DNA cleavage assay. (A) Schematic model of HCMV DNA synthesis and packaging. The HCMV terminase complex (UL56/UL89) and the HCMV genome structure are illustrated. Long and short arms are comprised of unique long (UL) and unique short (US) regions separated by a region designated the L/S junction. pac indicates the terminase cleavage site at the genome terminus. Below is an expansion of the location of the KpnI restriction sites at genomic transitions and the terminase cleavage site. A gray rectangle indicates the position of the terminal DNA probe. Following restriction digestion, concatemeric viral DNA yields an ∼8.4-kb KpnI fragment, whereas terminase-cleaved concatemeric DNA results in a shortened ∼4-kb fragment. (B) Southern blot analysis of viral DNA. HELF cells were infected with HCMV AD169 or AD169-rAIC246-1 and maintained at the AIC246 concentrations indicated. BAY 38-4766- and BDCRB-treated cells served as positive controls. Seventy-two h after infection, viral DNA was isolated, digested with KpnI, and size fractionated by gel electrophoresis. After blotting, hybridization was carried out using the terminal DNA probe depicted in panel A. Terminase-cleaved and uncleaved genomic HCMV DNA can be distinguished by the size of the respective restriction fragment.
Effect of AIC246 on capsid formation.
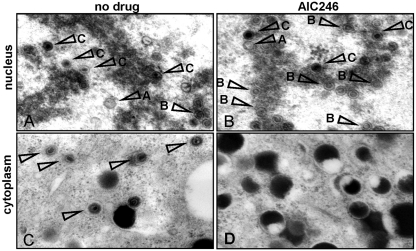
Since the cleavage and packaging of HCMV genomic DNA are tightly linked processes (14, 38), we next investigated whether AIC246 affects virus particle maturation. To examine this, we analyzed late-stage AD169-infected fibroblast cells grown in the absence or the presence of AC246 by electron microscopy (Fig. 5). As expected, all types of HCMV capsids were readily detected in the nuclei of placebo-treated cells: DNA-filled C capsids, B capsids that contain proteolytically cleaved scaffold proteins, and A capsids that appear to be empty (Fig. 5A and Table 5) (for a review, see references 19, 37 and 38). Importantly, C capsids (and consequently C capsid-derived A capsids) were markedly reduced after AIC246 treatment, while B capsids became significantly more abundant (Fig. 5B and Table 5). Even more striking was the effect of AIC246 on virus maturation when we examined the cytoplasm of the infected cells (Fig. 5C and D). Although HCMV virions were easily detectable in the cytoplasm of placebo-treated cells (Fig. 5C), not a single virus particle was found in the cytoplasm of cells that were treated with AIC246 (Fig. 5D). This suggests that nuclear egress, even of C-capsid-like particles that were observed occasionally in the nucleus of AIC246-treated cells, was efficiently blocked. However, nuclear egress and secondary envelopment were not fundamentally affected by AIC246, since a large number of noninfectious particles, mainly enveloped dense bodies, was released into the cytoplasm of drug-treated cells (Fig. 5D). Thus, in combination with the results of the concatemer cleavage assay, these data support the conclusion that AIC246 inhibits HCMV replication in cell culture by interfering with the proper cleavage/packaging of HCMV progeny DNA.
Fig. 5.
Transmission electron micrographs of thin-section preparations of virus-infected placebo- or AIC246-treated cells. Placebo (A and C)- or AIC246 (B and D)-treated, AD169-infected HFF cells were fixed 5 days postinfection, and thin sections were prepared for electron microscopic analysis. Panels A and B show a representative image of a cell nucleus, and panels C and D show an image of the respective cytoplasm. Arrows in the nucleus indicate A, B, and C capsids, respectively; arrows in the cytoplasm indicate virions.
Table 5.
Effect of AIC246 on HCMV capsid formation
| Virus and treatment | No. of nuclei counted | Capsid types per nucleus (%) |
||
|---|---|---|---|---|
| A | B | C | ||
| AD169 alone | 5 | 21 | 41 | 38 |
| AD169 with AIC246 | 5 | 4 | 75 | 21 |
DISCUSSION
Given the tolerability issues associated with current HCMV antivirals and the emergence of drug resistance, there is an unmet need for a novel drug acting through a different mechanism. AIC246 is a representative of a novel class of anti-HCMV drugs (dihydroquinazolinyl-acetic acids) that is currently being evaluated clinically in a phase IIb trial (clinicaltrials.gov). Several lines of evidence from our earlier in vitro studies indicated that AIC246 has a mode of action that is different from that of the drugs currently approved for clinical use (28). The results presented here extend these findings and show that AIC246 interferes with DNA concatemer maturation and exerts its effects through the HCMV terminase enzyme complex.
Herpesvirus DNA replication results in the formation of long, branched, head-to-tail DNA concatemers. Maturation into unit-length molecules involves site-specific cleavage at distinct DNA packaging signals termed pac motifs (6, 38). The HCMV terminase complex is a two-subunit enzyme that is responsible for the postsynthesis cleavage of concatemeric progeny DNA to unit-length genomes and the packaging of those genomes into preformed procapsids. Although this multifunctional process is not completely understood, it is assumed that the small terminase subunit, pUL89, is required for DNA duplex nicking, whereas pUL56, the large subunit, mediates sequence-specific binding to concatemeric viral DNA and provides energy for DNA translocation via its ATPase activity (6, 7). In addition, pUL56 is involved in the association of the terminase complex with the viral procapsid due to a direct protein-protein interaction of pUL56 with the putative HCMV portal protein pUL104 (14). Portal proteins are macromolecules that assemble into ring-like structures that form a channel for the entry of viral DNA into the nucleocapsid (13). Given their important activities, it is obvious that the proteins mentioned above, together with a number of additional viral proteins also implicated in the cleavage/packaging of progeny DNA (UL51, UL52, and UL77), are essential for virus replication and therefore represent attractive targets for antiviral intervention (1, 9, 49).
Indeed, potent and selective antiviral compounds that are believed to interfere with the functions performed by the HCMV terminase have been described previously. These include the benzimidazole ribonucleosides (e.g., BDCRB and TCRB) as well as the structurally distinct sulfonamides (e.g., BAY 38-4766) (26, 40, 47, 48). Resistance to these chemically unrelated compounds has been mapped to the HCMV terminase genes UL56 and UL89. Although it remains to be elucidated exactly how these compounds exert their inhibitory effects, it is believed that both drugs effectively block HCMV genome maturation by a mechanism preventing the cleavage of high-molecular-weight viral DNA concatemers before the packaging of monomeric genomes into procapsids (26, 36, 40, 48). Unfortunately, despite their excellent in vitro activity against HCMV, these compounds proved unsatisfactory for clinical development. Several lines of evidence from the present study strongly suggest that AIC246 acts in a manner similar to that of the cleavage/packaging inhibitors of the benzimidazole-ribonucleoside or sulfonamide class.
First, experiments to test the effects of AIC246 on HCMV protein expression or DNA replication showed that none of these processes was affected by the drug, excluding the possibility that AIC246 interferes with a mechanism directly or indirectly involved in viral genome replication.
Second, the genetic mapping of AIC246 resistance to HCMV ORF UL56 strongly implies that the viral terminase complex is involved in the AIC246 mechanism of action. Marker transfer experiments confirmed that the revealed UL56 mutations were sufficient to confer AIC246 resistance and therefore do not represent polymorphisms or reporting errors. In cell culture, none of the mutations substantially impaired virus replication, which, however, may not necessarily predict the in vivo situation. Interestingly, mutations which confer resistance reside in different regions of ORF UL56 for AIC246 (L241P and R369S), BDCRB (Q204R) (17, 26) or BAY 38-4766 (A662V) (10). This finding is remarkable given the strong differences within the chemical structures of these three compounds and the complexity of the drug target. It is particularly noteworthy that the AIC246-resistant viruses demonstrated a lack of cross-resistance to both BDCRB and BAY 38-4766. Likewise, no cross-resistance between BDCRB and BAY 38-4766 has been observed (10, 17). Taken together, these findings imply that the three drug classes do not share the same putative binding domain on the pUL56 subunit and thus have distinct physical interactions with the viral terminase complex. In line with this, we could not identify AIC246 resistance mutations in ORF UL89 as reported for BDCRB and BAY 38-4766 (10, 26, 40, 48). However, due to the limited number of AIC246-resistant viruses analyzed, we cannot exclude the possibility that viral gene products other than UL56 also serve as targets for the drug.
The third piece of evidence supporting the conclusion that the viral terminase is involved in AIC246 activity is the potent inhibition of the formation of properly processed unit-length genomes in a functional terminase cleavage assay.
Nevertheless, we were surprised by the effects of AIC246 on particle formation. As expected, the treatment of AD169-infected cells with AIC246 resulted in a complete block of nuclear egress. However, although the formation of dense core capsids in the nucleus (indicative of viral DNA packaging) was markedly reduced, it was not completely abolished. This observation is not consistent with the assumption that AIC246 treatment results in concatemers remaining uncut and not being encapsidated. Recent findings of McVoy and Nixon may offer an explanation for this phenomenon (36, 39). While analyzing the effect of BDCRB on HCMV genome maturation, the authors confirmed that BDCRB predominantly acts via blocking the processing of viral concatemer DNA to monomeric genome lengths (36, 48). Despite this, they reported that BDCRB treatment also permits the formation of a limited amount of incorrectly processed HCMV unit-length genomes that are heterogeneously truncated at the long (left) arm end. Moreover, evidence was provided that these abnormal, premature genomes are packaged and give rise to immature, C-capsid-like particles in the nucleus. These particles, however, are unable to egress from, and thus remain within, the nucleus.
At this stage, we cannot resolve whether AIC246 completely blocks the cleavage of DNA concatemers at viral intergenomic transitions, induces the inexact premature cleavage of DNA concatemers, or exerts its effects via a combination of both processes. However, given that prematurely cleaved, left-end-truncated genomes are not visible in our terminase cleavage assay (due to a lack of the probe binding site) and regarding the evidence provided by our electron micrographs, an AIC246 activity similar to that described for BDCRB is conceivable. Further experiments are under way to clarify this hypothesis.
Finally, during our electron microscopy studies we made a second intriguing observation: AIC246 activity was accompanied by the cytoplasmic accumulation of large amounts of subviral, noninfectious particles termed dense bodies (DB) (Fig. 5B and D). Since similar observations were made previously for BAY 38-4766-treated cells, this further emphasizes similar mechanistic phenotypes of HCMV cleavage/packaging inhibitors (10). Notably, DB currently are being evaluated as HCMV vaccine candidates in preclinical studies (4). Hence, one can speculate that the release of noninfectious immunogenic DB during AIC246 therapy is beneficial for patients, since it facilitates the reconstitution of an antiviral immune response following immunosuppression. However, clinical data are needed to prove this hypothesis.
In summary, we have presented evidence which strongly suggests that the mechanism of action of AIC246 is unique in relation to the currently approved HCMV antivirals, in that AIC246 interferes with the activities of the viral terminase complex rather than with that of the viral DNA polymerase. This mechanism of action is expected to offer an advantage over current therapies given its antiviral efficacy without the target-related toxicity seen for the marketed nucleosidic anti-CMV drugs, since mechanism-based side effects are unlikely to be due to the lack of a mammalian counterpart of the viral terminase enzyme. In addition, the different modes of action of AIC246 should provide new treatment options for patients infected with resistant virus strains for which no effective therapy is currently available.
ACKNOWLEDGMENTS
We thank Thomas Stamminger, Michael Mach, William Britt, and Greg Smith for providing reagents, antibodies, and plasmids, Eva Borst and Martin Messerle for HCMV BAC pHG, and Christian Sinzger for help with electron microscopy. The excellent technical assistance of Marion Heidtmann, Kerstin Pixberg, Wiebke Schulze, Christine Hempel, and Bernd Schulz is gratefully acknowledged.
T.G., H.R.-S., H.Z., and P.L. are employees of AiCuris GmbH & Co. KG; G.H. has served as a consultant to AiCuris GmbH & Co. KG.
Footnotes
Published ahead of print on 13 July 2011.
REFERENCES
- 1. Andrei G., De C. E., Snoeck R. 2009. Drug targets in cytomegalovirus infection. Infect. Disord. Drug Targets 9: 201–222 [DOI] [PubMed] [Google Scholar]
- 2. Andreoni M., Faircloth M., Vugler L., Britt W. J. 1989. A rapid microneutralization assay for the measurement of neutralizing antibody reactive with human cytomegalovirus. J. Virol. Methods 23: 157–167 [DOI] [PubMed] [Google Scholar]
- 3. Antinone S. E., Smith G. A. 2010. Retrograde axon transport of herpes simplex virus and pseudorabies virus: a live-cell comparative analysis. J. Virol. 84: 1504–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Becke S., et al. 2010. Optimized recombinant dense bodies of human cytomegalovirus efficiently prime virus specific lymphocytes and neutralizing antibodies without the addition of adjuvant. Vaccine 28: 6191–6198 [DOI] [PubMed] [Google Scholar]
- 5. Biswas S., Swift M., Field H. J. 2007. High frequency of spontaneous helicase-primase inhibitor (BAY 57-1293) drug-resistant variants in certain laboratory isolates of HSV-1. Antivir. Chem. Chemother. 18: 13–23 [DOI] [PubMed] [Google Scholar]
- 6. Bogner E. 2002. Human cytomegalovirus terminase as a target for antiviral chemotherapy. Rev. Med. Virol. 12: 115–127 [DOI] [PubMed] [Google Scholar]
- 7. Bogner E., Radsak K., Stinski M. F. 1998. The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity. J. Virol. 72: 2259–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Borst E. M., Messerle M. 2005. Analysis of human cytomegalovirus oriLyt sequence requirements in the context of the viral genome. J. Virol. 79: 3615–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Borst E. M., Wagner K., Binz A., Sodeik B., Messerle M. 2008. The essential human cytomegalovirus gene UL52 is required for cleavage-packaging of the viral genome. J. Virol. 82: 2065–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Buerger I., et al. 2001. A novel nonnucleoside inhibitor specifically targets cytomegalovirus DNA maturation via the UL89 and UL56 gene products. J. Virol. 75: 9077–9086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Champier G., et al. 2008. Putative functional domains of human cytomegalovirus pUL56 involved in dimerization and benzimidazole D-ribonucleoside activity. Antivir. Ther. 13: 643–654 [PubMed] [Google Scholar]
- 12. Coen D. M., Furman P. A., Gelep P. T., Schaffer P. A. 1982. Mutations in the herpes simplex virus DNA polymerase gene can confer resistance to 9-beta-D-arabinofuranosyladenine. J. Virol. 41: 909–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dittmer A., Bogner E. 2005. Analysis of the quaternary structure of the putative HCMV portal protein PUL104. Biochemistry 44: 759–765 [DOI] [PubMed] [Google Scholar]
- 14. Dittmer A., Drach J. C., Townsend L. B., Fischer A., Bogner E. 2005. Interaction of the putative human cytomegalovirus portal protein pUL104 with the large terminase subunit pUL56 and its inhibition by benzimidazole-D-ribonucleosides. J. Virol. 79: 14660–14667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Drew W. L. 2010. Cytomegalovirus resistance testing: pitfalls and problems for the clinician. Clin. Infect. Dis. 50: 733–736 [DOI] [PubMed] [Google Scholar]
- 16. Eid A. J., Razonable R. R. 2010. New developments in the management of cytomegalovirus infection after solid organ transplantation. Drugs 70: 965–981 [DOI] [PubMed] [Google Scholar]
- 17. Evers D. L., et al. 2002. Interactions among antiviral drugs acting late in the replication cycle of human cytomegalovirus. Antivir. Res. 56: 61–72 [DOI] [PubMed] [Google Scholar]
- 18. Fowler K. B., Boppana S. B. 2006. Congenital cytomegalovirus (CMV) infection and hearing deficit. J. Clin. Virol. 35: 226–231 [DOI] [PubMed] [Google Scholar]
- 19. Gibson W. 2006. Assembly and maturation of the capsid, p. 231–243 In Reddehase M. J. (ed.), Cytomegaloviruses: molecular biology and immunology. Horizon Scientific Press, Hethersett, Norwich, United Kingdom [Google Scholar]
- 20. Griffiths P. D., Walter S. 2005. Cytomegalovirus. Curr. Opin. Infect. Dis. 18: 241–245 [DOI] [PubMed] [Google Scholar]
- 21. Jiang X. J., et al. 2008. UL74 of human cytomegalovirus contributes to virus release by promoting secondary envelopment of virions. J. Virol. 82: 2802–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaul D. R., et al. First report of successful treatment of multidrug-resistant cytomegalovirus disease with the novel anti-cmv compound aic246. Am. J. Transplant., in press [DOI] [PubMed] [Google Scholar]
- 23. Komazin G., Townsend L. B., Drach J. C. 2004. Role of a mutation in human cytomegalovirus gene UL104 in resistance to benzimidazole ribonucleosides. J. Virol. 78: 710–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kotton C. N., et al. 2010. International consensus guidelines on the management of cytomegalovirus in solid organ transplantation. Transplantation 89: 779–795 [DOI] [PubMed] [Google Scholar]
- 25. Kropeit D., et al. 2010. Phase I safety and PK data of the novel anti-HCMV terminase inhibitor AIC246, abstr. 1994. Abstr. 50th Intersci. Conf. Antimicrob. Agents Chemother. [Google Scholar]
- 26. Krosky P. M., et al. 1998. Resistance of human cytomegalovirus to benzimidazole ribonucleosides maps to two open reading frames: UL89 and UL56. J. Virol. 72: 4721–4728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li C. R., Greenberg P. D., Gilbert M. J., Goodrich J. M., Riddell S. R. 1994. Recovery of HLA-restricted cytomegalovirus (CMV)-specific T-cell responses after allogeneic bone marrow transplant: correlation with CMV disease and effect of ganciclovir prophylaxis. Blood 83: 1971–1979 [PubMed] [Google Scholar]
- 28. Lischka P., et al. 2010. In vitro and in vivo activities of the novel anticytomegalovirus compound AIC246. Antimicrob. Agents Chemother. 54: 1290–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lischka P., Rosorius O., Trommer E., Stamminger T. 2001. A novel transferable nuclear export signal mediates CRM1-independent nucleocytoplasmic shuttling of the human cytomegalovirus transactivator protein pUL69. EMBO J. 20: 7271–7283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lischka P., Sorg G., Kann M., Winkler M., Stamminger T. 2003. A nonconventional nuclear localization signal within the UL84 protein of human cytomegalovirus mediates nuclear import via the importin alpha/beta pathway. J. Virol. 77: 3734–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lischka P., Toth Z., Thomas M., Mueller R., Stamminger T. 2006. The UL69 transactivator protein of human cytomegalovirus interacts with DEXD/H-box RNA helicase UAP56 to promote cytoplasmic accumulation of unspliced RNA. Mol. Cell. Biol. 26: 1631–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lischka P., Zimmermann H. 2008. Antiviral strategies to combat cytomegalovirus infections in transplant recipients. Curr. Opin. Pharmacol. 8: 541–548 [DOI] [PubMed] [Google Scholar]
- 33. Ljungman P., Hakki M., Boeckh M. 2010. Cytomegalovirus in hematopoietic stem cell transplant recipients. Infect. Dis. Clin. N. Am. 24: 319–337 [DOI] [PubMed] [Google Scholar]
- 34. Lorz K., et al. 2006. Deletion of open reading frame UL26 from the human cytomegalovirus genome results in reduced viral growth, which involves impaired stability of viral particles. J. Virol. 80: 5423–5434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lurain N. S., Chou S. 2010. Antiviral drug resistance of human cytomegalovirus. Clin. Microbiol. Rev. 23: 689–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McVoy M. A., Nixon D. E. 2005. Impact of 2-bromo-5,6-dichloro-1-beta-D-ribofuranosyl benzimidazole riboside and inhibitors of DNA, RNA, and protein synthesis on human cytomegalovirus genome maturation. J. Virol. 79: 11115–11127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mettenleiter T. C., Klupp B. G., Granzow H. 2009. Herpesvirus assembly: an update. Virus Res. 143: 222–234 [DOI] [PubMed] [Google Scholar]
- 38. Mocarski E. S., Shenk T., Pass R. 2007. Cytomegalovirus. In Knipe D. M., et al. (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 39. Nixon D. E., McVoy M. A. 2004. Dramatic effects of 2-bromo-5,6-dichloro-1-beta-D-ribofuranosyl benzimidazole riboside on the genome structure, packaging, and egress of guinea pig cytomegalovirus. J. Virol. 78: 1623–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reefschlaeger J., et al. 2001. Novel non-nucleoside inhibitors of cytomegaloviruses (BAY 38-4766): in vitro and in vivo antiviral activity and mechanism of action. J. Antimicrob. Chemother. 48: 757–767 [DOI] [PubMed] [Google Scholar]
- 41. Scheffczik H., Savva C. G., Holzenburg A., Kolesnikova L., Bogner E. 2002. The terminase subunits pUL56 and pUL89 of human cytomegalovirus are DNA-metabolizing proteins with toroidal structure. Nucleic Acids Res. 30: 1695–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schreiber A., et al. 2009. Antiviral treatment of cytomegalovirus infection and resistant strains. Expert. Opin. Pharmacother. 10: 191–209 [DOI] [PubMed] [Google Scholar]
- 43. Scott G. M., Weinberg A., Rawlinson W. D., Chou S. 2007. Multidrug resistance conferred by novel DNA polymerase mutations in human cytomegalovirus isolates. Antimicrob. Agents Chemother. 51: 89–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Snydman D. R. 2010. Leflunomide: a small step forward in meeting the urgent need for treatment of drug-resistant cytomegalovirus infection. Transplantation 90: 362–363 [DOI] [PubMed] [Google Scholar]
- 45. Tavalai N., Kraiger M., Kaiser N., Stamminger T. 2008. Insertion of an EYFP-pp71 (UL82) coding sequence into the human cytomegalovirus genome results in a recombinant virus with enhanced viral growth. J. Virol. 82: 10543–10555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tischer B. K., von Kaufer E. J., Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40: 191–197 [DOI] [PubMed] [Google Scholar]
- 47. Townsend L. B., Devivar R. V., Turk S. R., Nassiri M. R., Drach J. C. 1995. Design, synthesis, and antiviral activity of certain 2,5,6-trihalo-1-(beta-D-ribofuranosyl)benzimidazoles. J. Med. Chem. 38: 4098–4105 [DOI] [PubMed] [Google Scholar]
- 48. Underwood M. R., et al. 1998. Inhibition of human cytomegalovirus DNA maturation by a benzimidazole ribonucleoside is mediated through the UL89 gene product. J. Virol. 72: 717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yu D., Silva M. C., Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci.U. S. A. 100: 12396–12401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zimmermann H., et al. 2009. A novel non-nucleoside compound with activity against human cytomegalovirus-overview of clinical trials and resistance breaking activity, abstr. V1256b. Abstr. 49th Intersci. Conf. Antimicrob. Agents Chemother. [Google Scholar]