Abstract
HIV-1 and HIV-2 are derived from two distinct primate viruses and share only limited sequence identity. Despite this, HIV-1 and HIV-2 Gag polyproteins can coassemble into the same particle and their genomes can undergo recombination, albeit at an extremely low frequency, implying that HIV-1 and HIV-2 RNA can be copackaged into the same particle. To determine the frequency of HIV-1 and HIV-2 RNA copackaging and to dissect the mechanisms that allow the heterologous RNA copackaging, we directly visualized the RNA content of each particle by using RNA-binding proteins tagged with fluorescent proteins to label the viral genomes. We found that when HIV-1 and HIV-2 RNA are present in viral particles at similar ratios, ∼10% of the viral particles encapsidate both HIV-1 and HIV-2 RNAs. Furthermore, heterologous RNA copackaging can be promoted by mutating the 6-nucleotide (6-nt) dimer initiation signal (DIS) to discourage RNA homodimerization or to encourage RNA heterodimerization, indicating that HIV-1 and HIV-2 RNA can heterodimerize prior to packaging using the DIS sequences. We also observed that the coassembly of HIV-1 and HIV-2 Gag proteins is not required for the heterologous RNA copackaging; HIV-1 Gag proteins are capable of mediating HIV-1 and HIV-2 RNA copackaging. These results define the cis- and trans-acting elements required for and affecting the heterologous RNA copackaging, a prerequisite for the generation of chimeric viruses by recombination, and also shed light on the mechanisms of RNA-Gag recognition essential for RNA encapsidation.
INTRODUCTION
The two etiological agents of AIDS, human immunodeficiency virus type 1 (HIV-1) and HIV-2, were introduced into the human population by zoonotic transmission of simian immunodeficiency viruses (SIVs) that naturally infect chimpanzees or gorillas (SIVcpz or SIVgor) and sooty mangabeys (SIVsm), respectively (12, 13, 17, 19, 26, 30, 32). Consequently, these lentiviruses, which are the only two known to infect humans, share only limited sequence identity (∼48% nucleotide [nt] identity at the genome level and ∼60% amino acid identity between their Gag polyproteins). Although both viruses can cause AIDS, HIV-1 is more pathogenic in humans and is distributed worldwide, whereas HIV-2 infection is limited geographically and mostly located in West Africa (20). However, infection with one of the HIVs does not provide protection from infection with the other HIV. Indeed, there is a significant population of people who are dually infected with both HIV-1 and HIV-2 (11, 14). HIV-1 and HIV-2 target the same cell types, using both the same receptor and the same coreceptors for virus entry (1, 31, 34). The possible interplay between these two viruses has been an intriguing topic of research aimed at understanding the extent and the mechanisms of such molecular interactions.
One unique feature of retrovirus replication is that two copies of the full-length RNA are packaged into a viral particle. The specific packaging of the viral RNA, which makes up less than 1% of the total cellular RNA, is mediated by at least two determinants. One determinant is the viral structural protein Gag and especially the nucleocapsid domain in the polyprotein. The second determinant is the packaging signal in the viral RNA, most of which resides at the 5′ end of the viral genome, including part of the 5′ untranslated region, and often extends into part of gag. The interactions between Gag and viral RNA ensure the selection and encapsidation of the proper RNAs into the particles. Retroviral RNA packaging is generally specific and limited to the RNAs of the same virus or closely related viruses. However, cross-packaging does occur, including two known examples of nonreciprocal RNA packaging of distantly related viruses; one set is HIV-1 and HIV-2 (18), and the other set is murine leukemia virus (MLV) and spleen necrosis virus (SNV) (3). SNV Gag polyprotein can efficiently package both SNV and MLV RNA; however, MLV Gag polyprotein does not package SNV RNA. Similarly, it has been shown that HIV-1 Gag can package HIV-2 RNA, whereas HIV-2 Gag does not package HIV-1 RNA (18). However, certain aspects of RNA packaging mechanisms used by HIV-1 and HIV-2 are similar. HIV-1 RNA has been shown to dimerize or select its copackaged RNA partner prior to encapsidation (4, 23, 24). A palindromic sequence located at the top of stem-loop 1 (SL1), termed the dimerization initiation signal (DIS), plays an important role during the RNA partner selection process (6, 8). HIV-1 RNAs that have complementary DIS sequences can copackage together more efficiently than those that do not (7, 23, 28). It is thought that the DIS sequences of the two RNAs form intermolecular base pairings to initiate the dimerization or the RNA partner selection process. Our recent studies showed that the RNA packaging mechanisms of HIV-2 have many features similar to those of HIV-1, including RNA partner selection prior to encapsidation and the involvement of a palindromic sequence in this process (27).
It has been shown that HIV-1 and HIV-2 can interact molecularly in several ways. For example, despite sharing a low level of amino acid sequence identity, the Gag polyproteins from HIV-1 and HIV-2 can coassemble into the same particle and complement each other's functions to carry out infection (2). It has also been shown in a cell culture system that HIV-1 and HIV-2 can recombine and generate hybrid genomes, albeit at a rate far lower than that occurring between two HIV-1 or between two HIV-2 genomes (25). Retroviral recombination is the result of the reverse transcriptase (RT) switching between the two copackaged RNA templates during DNA synthesis. Therefore, the observation that HIV-1 and HIV-2 can recombine indicates that the RNA genomes of these two different viruses must be able to be packaged into the same particle (heterologous RNA copackaging). However, it was unclear how frequently these heterologous RNAs can be copackaged and whether elements in the viral RNAs exist that constrain such copackaging. Furthermore, it is unclear whether the coassembly of the HIV-1 and HIV-2 Gag polyproteins is required to promote copackaging of the heterologous RNAs by each interacting with its own RNA.
In this study, we used the single-virion analysis for direct measurement of the frequency of HIV-1 and HIV-2 RNA copackaging. This system, based on labeling HIV-1 and HIV-2 genomes with fluorescent protein-tagged RNA-binding proteins, offers a direct measurement of viral RNA content at single-virion resolution. Unlike standard biochemical assays that measure the efficiency with which a given RNA is packaged, this microscopy-based method allows us to determine whether two RNAs are copackaged into the same virion. We found that when HIV-1 and HIV-2 RNAs are packaged at similar levels, ∼10% of the particles contained both HIV-1 and HIV-2 RNA. The heterologous RNA copackaging frequency can be influenced by the identities of the DIS sequences, indicating that HIV-1 and HIV-2 RNAs can heterodimerize using the DIS sequences prior to packaging. We also determined that HIV-1 Gag alone is sufficient for heterologous RNA copackaging.
MATERIALS AND METHODS
Viral vectors and plasmid construction.
The HIV-1 constructs GagCeFP-MS2SL and Gag-MS2SL have been previously described (4); for clarity, in this report these plasmids are referred to as HIV-1–GagCeFP-MS2SL and HIV-1–Gag-MS2SL, respectively. Briefly, these two constructs were derived from the molecular clone NL4-3; portions of the pol, vif, vpr, vpu, and env genes were deleted, rendering the encoded proteins nonfunctional. Additionally, 24 copies of the stem-loop recognized by the bacteriophage MS2 coat protein were inserted in the pol gene. All cis-acting elements required for HIV-1 replication, including the packaging signal, are present in these genomes. The two plasmids have similar structures, except the HIV-1–GagCeFP-MS2SL expresses Gag tagged with cerulean fluorescent protein (CeFP), whereas HIV-1–Gag-MS2SL expresses untagged Gag. In all experiments, tagged Gag-CeFP and untagged Gag are coexpressed to avoid possible distortions of viral particle morphology. For brevity, only GagCeFP-expressing constructs are mentioned.
The mutant with altered DIS sequences, HIV-1–DIS6C–GagCeFP-MS2SL, was derived from HIV-1–GagCeFP-MS2SL by changing the GCGCGC sequence in the DIS into CCCCCC. This was done by replacing the SphI-XhoI DNA fragment in 6CDIS–GagCeFP-BglSL with the corresponding fragment from HIV-1–GagCeFP-MSL; both the Gag-CeFP and the untagged Gag versions were generated (4). A Gag mutant, HIV-1–noGag-MSL, which contains a stop codon after amino acid 109 of CA, was generated by introducing a 4-bp frameshift mutation in the gag gene of HIV-1–Gag-MS2SL. This was achieved by digesting HIV-1–Gag-MS2SL with SpeI, followed by a fill-in reaction using the Klenow fragment of Escherichia coli polymerase and DNA ligation.
The HIV-2 constructs 2-GagCeFP-BglSL, 2-Gag-BglSL, and 2-6G-GagCeFP-BSL have been previously described (27); for clarity, these constructs are referred to as HIV-2–GagCeFP-BglSL, HIV-2–Gag-BglSL, and HIV-2–DIS6G-GagCeFP-BglSL in this report. These two constructs were derived from the ROD12 molecular clone and contained all cis-acting elements important for viral replication and inactivating mutations in pol, vpr, and env. Additionally, both constructs contained 18 copies of stem-loops (BglSL) in pol recognized by the E. coli BglG protein. Construct HIV-2–GagCeFP-BglSL expresses Gag tagged with CeFP, whereas HIV-2–Gag-BglSL expresses untagged Gag (27). As with HIV-1, only the GagCeFP construct is mentioned, although both CeFP-tagged HIV-2 Gag and wild-type HIV-2 Gag were coexpressed in all experiments.
Construct HIV-2–GagCeFP-BglSL-noTatRev was generated by digesting HIV-2–GagCeFP-BglSL plasmid DNA with BsmBI, followed by a fill-in reaction using the Klenow fragment from E. coli DNA polymerase and DNA ligation. This procedure generated an inactivating frameshift mutation in both tat and rev. The HIV-2–DIS6G-GagCeFP-BglSL-noTatRev construct was generated using the same procedure by starting with HIV-2–DIS6G-GagCeFP-BglSL plasmid. The HIV-2–noGag-BglSL was derived from a previously described 2*T-17 mutant that contained two stop codons in gag, one at codon 17 of Gag in MA and one at codon 171 of CA (27). DIS mutants in the HIV-2–noGag-BglSL context were constructed using 2-step PCR. Primers harboring the DIS mutations were used to amplify the HIV-2–noGag-BglSL template, yielding a 2.4-kb DNA fragment containing DIS mutations and both stop codons in Gag that was cloned into the HIV-2–GagCeFP-BglSL backbone by the use of the NgoMIV and SwaI restriction sites. The general structures of the newly generated plasmids were determined by restriction enzyme mapping, and regions generated by PCR were characterized by DNA sequencing to avoid inadvertent mutations.
The MS2-yellow fluorescent protein (MS2-YFP) construct was a generous gift from Robert Singer (Albert Einstein Medical College); the Bgl-mCherry plasmid has been previously described (4).
Cell culture, transfection, and virus production.
The 293T human embryonic kidney cell line was grown in a humidified 37°C incubator with 5% CO2 and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin (50 U/ml), and streptomycin (50 μg/ml). In all experiments, plasmids expressing Gag and GagCeFP were cotransfected at a 1:1 molar ratio; DNA transfections were performed using FuGeneHD (Roche) according to manufacturer's recommendations. Supernatants were harvested from 293T cells 19 to 20 h posttransfection, clarified through a 0.45-μm-pore-size filter, and either stored at −80°C or used immediately for image acquisition.
Single-virion analyses.
To prepare particles for microscopy, clarified culture supernatant was mixed with Polybrene and the mixture was plated on a 35-mm-diameter glass-bottom plate (MatTek Corp.) and incubated at 37°C for 2 h. An inverted Nikon Eclipse Ti microscope with a 100× oil objective and a numerical aperture of 1.40 and an X-Cite 120 system (EXFO Photonic Solutions Inc.) were used for image capture as previously described (27). The filter settings were 427/10 nm and 480/40 nm (CeFP), 504/12 nm and 542/27 nm (YFP), and 577/25 nm and 632/60 nm (mCherry) for excitation and emission, respectively. Digital images were acquired by the use of an AndOr technology iXon camera and NIS Element AR software (Nikon). Custom software developed with Matlab was used to identify particles and their colocalization with RNA.
RESULTS
Determining HIV-1 and HIV-2 RNA genome copackaging frequency.
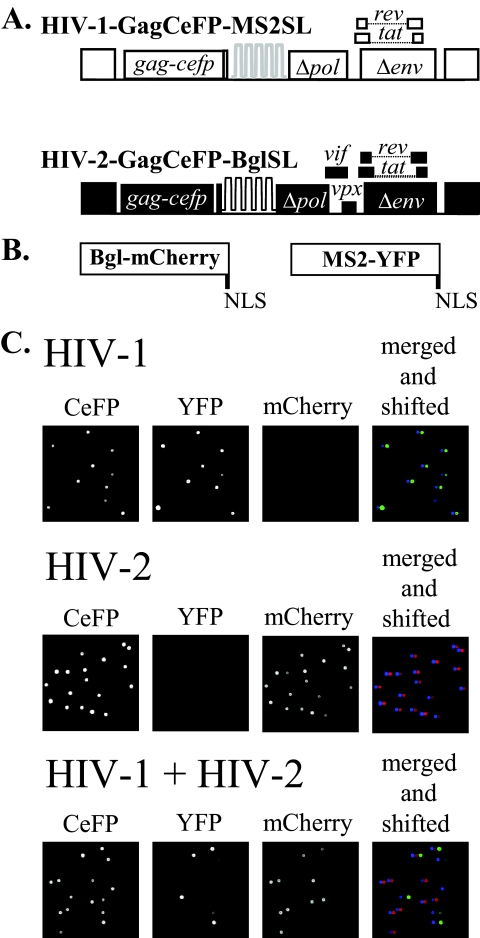
To measure the frequency of HIV-1 and HIV-2 RNA copackaging, we used two previously described constructs that express near-full-length HIV-1 and HIV-2 genomes containing stem-loop sequences recognized by RNA-binding proteins (Fig. 1A) (4, 27). These viral genomes allowed us to fluorescently tag and directly visualize encapsidated RNA in individual viral particles. The modified HIV-1 genome HIV-1–GagCeFP-MS2SL expresses HIV-1 Gag tagged with CeFP, Tat, and Rev; additionally, this construct also contains 24 copies of MS2SL, the stem-loop sequence specifically recognized by the bacteriophage MS2 coat proteins. The modified HIV-2 genome, HIV-2–GagCeFP-BglSL, expresses viral gene products, including HIV-2 Gag tagged with CeFP, Tat, and Rev; additionally, it contains 18 copies of BglSL, the stem-loop sequence specifically recognized by the E. coli BglG protein. Two versions of each viral construct were generated; one expresses wild-type Gag and the other expresses Gag tagged with CeFP. In all experiments, GagCeFP and Gag were coexpressed to preserve normal particle morphology; for simplicity, only the names of the GagCeFP constructs are mentioned.
Fig. 1.
System used to determine HIV-1 and HIV-2 RNA copackaging efficiency. (A) General structures of the HIV constructs. HIV-1 construct HIV-1–GagCeFP-MS2SL contains in its RNA stem-loops recognized by the bacteriophage MS2 coat protein. HIV-2 construct HIV-2–GagCeFP-BglSL contains in its RNA stem-loops recognized by the E. coli BglG protein. Both constructs express Gag tagged with CeFP. In all experiments, these constructs were coexpressed with sister constructs that were identical except that they expressed wild-type Gag proteins without CeFP. For simplicity, the constructs expressing wild-type Gag are not shown. White boxes represent HIV-1 sequences, whereas black boxes represent HIV-2 sequences. (B) General structures of RNA-binding proteins tagged with fluorescent proteins containing nuclear localization signals (NLS) at the C terminus. (C) Representative images of single-virion analyses. MS2-YFP and Bgl-mCherry were cotransfected along with viral constructs in all samples. All three channels were merged and shifted (the YFP and mCherry channels were shifted 5 and 10 pixels to the right, respectively) as shown in the fourth column to illustrate the RNA signals associated with each particle.
To examine the RNA content in the HIV particles, viral constructs were transfected into 293T cells along with two plasmids that express fluorescently tagged RNA-binding proteins, MS2-YFP and Bgl-mCherry (Fig. 1B). The supernatant was harvested 19 to 20 h posttransfection and clarified, and images of viral particles were obtained using fluorescence microscopy. Examples of images of viral particles obtained from 293T cells transfected with HIV-1–GagCeFP-MS2SL, MS2-YFP, and Bgl-mCherry are shown in the upper panels of Fig. 1C. As a portion of the HIV-1 Gag polyproteins were tagged with CeFP, viral particles can be identified by their signals in the CeFP channel. The full-length RNA expressed by HIV-1–GagCeFP-MS2SL contained sequences recognized by the MS2 coat protein; as a result, the majority (>90%) of the CeFP+ particles also contained YFP signals but not mCherry signals (summarized in Table 1). In contrast, HIV-2 construct HIV-2–GagCeFP-BglSL contains sequences recognized by Bgl proteins; hence, most of the CeFP+ particles generated from cotransfection of HIV-2–GagCeFP-BglSL, MS2-YFP, and Bgl-mCherry also contained mCherry signals but not YFP signals (Fig. 1C, middle panels; Table 1). These results showed that most of the HIV particles contained viral RNA genomes; furthermore, the signals observed from MS2-YFP or Bgl-mCherry were specific to the stem-loops in the viral genomes and had little background and nonspecific labeling.
Table 1.
Single-virion analyses of HIV-1 and HIV-2 RNA copackaging
| Coexpressed constructsa | CeFP+ particles |
RNA labeling efficiencyc | |||
|---|---|---|---|---|---|
| No. analyzed | YFP+ (%) | mCherry+ (%) | YFP+ + mCherry+ (%)b | ||
| HIV-1–GagCeFP-MS2SL | |||||
| Expt 1 | 1,005 | 93.2 | 0.0 | 0.0 | 93.2 |
| Expt 2 | 1,772 | 93.7 | 0.1 | 0.3 | 94.1 |
| Expt 3 | 1,786 | 92.9 | 0.0 | 0.1 | 93.0 |
| Expt 4 | 2,369 | 92.0 | 0.0 | 0.0 | 92.0 |
| Expt 5 | 5,032 | 92.8 | 0.0 | 0.0 | 92.8 |
| Expt 6 | 5,060 | 91.8 | 0.0 | 0.0 | 91.8 |
| Expt 7 | 4,568 | 91.6 | 0.0 | 0.0 | 91.6 |
| Expt 8 | 5,578 | 91.5 | 0.0 | 0.2 | 91.7 |
| Expt 9 | 7,505 | 91.3 | 0.0 | 0.3 | 91.6 |
| Expt 10 | 3,655 | 92.3 | 0.0 | 0.1 | 92.4 |
| Expt 11 | 8,138 | 93.0 | 0.0. | 0.0 | 93.0 |
| Expt 12 | 20,159 | 91.4 | 0.0 | 0.4 | 91.8 |
| Expt 13 | 16,632 | 92.0 | 0.0 | 0.0 | 92.0 |
| HIV-2–GagCeFP-BglSL | |||||
| Expt 1 | 1,644 | 0.1 | 92.7 | 0.4 | 93.2 |
| Expt 2 | 2,902 | 0.0 | 93.7 | 0.2 | 93.9 |
| Expt 3 | 5,708 | 0.0 | 93.8 | 0.1 | 93.9 |
| Expt 4 | 4,738 | 0.0 | 93.3 | 0.3 | 93.6 |
| Expt 5 | 6,467 | 0.0 | 92.7 | 0.5 | 93.2 |
| Expt 6 | 1,077 | 0.0 | 91.8 | 1.0 | 92.8 |
| Expt 7 | 8,240 | 0.0 | 92.8 | 0.4 | 93.2 |
| Expt 8 | 10,027 | 0.0 | 95.6 | 0.3 | 95.9 |
| Expt 9 | 15,232 | 0.0 | 91.4 | 0.2 | 91.6 |
| Expt 10 | 15,107 | 0.0 | 94.1 | 0.0 | 94.1 |
| Expt 11 | 2,384 | 0.0 | 92.1 | 0.5 | 92.6 |
| HIV-1–GagCeFP-MS2SL + HIV-2- GagCeFP-BglSL | |||||
| Expt 1 | 1,025 | 43.8 | 44.4 | 6.1 | 94.3 |
| Expt 2 | 1,251 | 45.2 | 42.0 | 5.7 | 92.9 |
| Expt 3 | 1,651 | 53.0 | 34.0 | 5.6 | 92.6 |
| Expt 4 | 3,072 | 32.8 | 50.7 | 9.5 | 93.0 |
| Expt 5 | 5,983 | 50.4 | 33.6 | 8.0 | 92.0 |
| Expt 6 | 12,462 | 36.6 | 46.0 | 10.2 | 92.8 |
| Expt 7 | 15,366 | 44.2 | 37.6 | 10.9 | 92.7 |
| Expt 8 | 13,505 | 49.9 | 32.1 | 10.9 | 92.9 |
| Expt 9 | 7,259 | 52.1 | 33.0 | 8.0 | 93.1 |
| Expt 10 | 12,956 | 37.4 | 41.1 | 13.1 | 91.6 |
| Expt 11 | 12,245 | 36.9 | 42.6 | 12.9 | 92.4 |
| Expt 12 | 10,749 | 35.4 | 44.3 | 12.1 | 91.8 |
| Expt 13 | 10,431 | 36.0 | 42.4 | 12.7 | 91.1 |
Although not indicated, both Bgl-mCherry and MS2-YFP were coexpressed in all experiments.
Average plus standard deviation (SD) for HIV-1–GagCeFP-MS2SL + HIV-2–GagCeFP-BglSL, 9.7 ± 2.8.
Calculated by adding the values from the three columns under the CeFP+ heading.
We then generated particles by coexpressing HIV-1–GagCeFP-MS2SL, HIV-2–GagCeFP-BglSL, MS2-YFP, and Bgl-mCherry (Fig. 1C, lower panels). Most of the CeFP+ particles also had either YFP or mCherry signals, indicating that they contained either HIV-1 or HIV-2 RNA, respectively; some of the CeFP+ particles were positive for YFP and mCherry signals, indicating that they contained both HIV-1 and HIV-2 RNA. As the relative levels of abundance of these two viral RNAs affect the copackaging frequency, in our experiments, we used only results from experiments that showed similar levels of HIV-1 and HIV-2 RNA signals. In 13 independent experiments, between 6% and 13% of the particles in the viral population exhibited CeFP, YFP, and mCherry signals, with an average of 9.7% and standard deviation of 2.8% (Table 1).
Examining the influence of DIS on copackaging of heterologous viral RNAs.
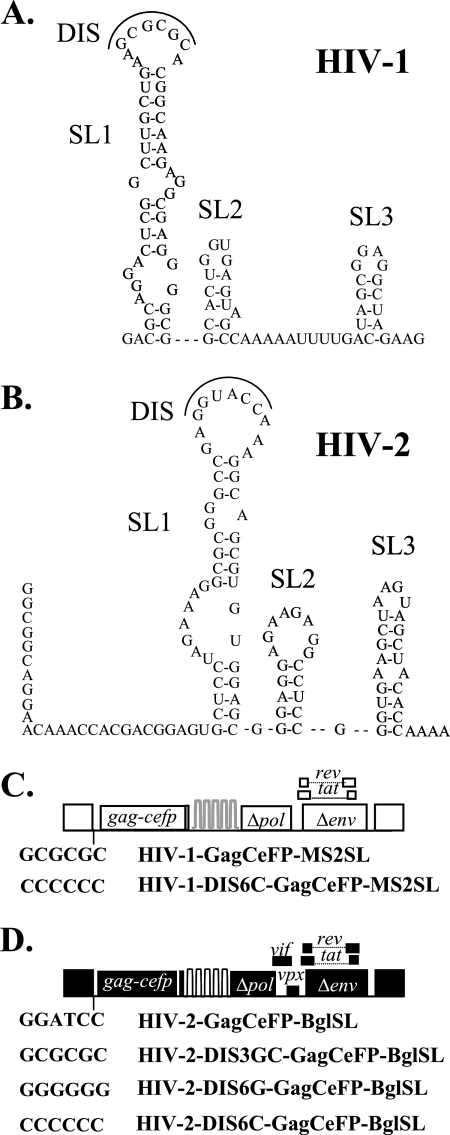
In both HIV-1 and HIV-2, RNA partner selection occurs prior to the encapsidation of the viral genome; additionally, the DIS sequences play a major role in the selection process (4, 6, 8, 23, 24, 27). However, previous experiments examined copackaging of RNAs derived from the same viral species; it is unclear whether DIS can affect copackaging of heterologous viral RNAs. We sought to determine whether RNAs from genetically distinct viruses can interact through DIS sequences and be copackaged. The sequences of the 5′ untranslated regions of HIV-1 and HIV-2 have little homology; the two molecular clones we used had 42% nucleotide sequence identity, but the two sequences had similar predicted RNA structures (Fig. 2A and B). The DIS sequences for HIV-1 and HIV-2 are discordant; the subtype B HIV-1 used in this study has a GCGCGC sequence whereas HIV-2 has a GGTACC sequence (Fig. 2). Using an in vitro assay, it was previously shown that RNA dimerization occurs only when HIV-1 and HIV-2 RNA contain the same DIS (9). To examine whether the frequency of heterologous RNA copackaging can be altered, we generated viral constructs containing various DIS sequences and tested the abilities of their RNAs to be copackaged with RNA from another virus. We first mutated the HIV-2 DIS sequence to GCGCGC so that this mutant (HIV-2–DIS3GC-GagCeFP-BglSL; Fig. 2D) had the same DIS sequence as the HIV-1 construct HIV-1–GagCeFP-MS2SL. We found that the presence of matching and complementary DIS sequences did not have a significant effect on the frequency of heterologous RNA copackaging; approximately 12% of the particles contained both HIV-1 RNA and HIV-2 RNA with GCGCGC at the DIS (range in 7 experiments, 10% to 14%) (Table 2).
Fig. 2.
Examining the role of the DIS in copackaging of HIV-1 and HIV-2 RNA. (A) Predicted secondary structure of a portion of the 5′ leader sequences of HIV-1, containing stem-loop 1, stem-loop 2, and stem-loop 3 (labeled SL1, SL2, and SL3) (33). (B) Predicted secondary structure of a portion of the 5′ leader of HIV-2 containing stem-loop 1, stem-loop 2, and stem-loop 3 (labeled SL1, SL2, and SL3) (9). DIS, dimer initiation signal. Nucleotides that comprise the DIS are outlined. (C) General structures of HIV-1 constructs and their DIS sequences. The wild-type DIS sequence (GCGCGC) of HIV-1–gagCeFP-MS2SL is shown for comparison. (D) General structures of HIV-2 constructs and their DIS sequences. The wild-type HIV-2 DIS sequence (GGUACC) is shown as a reference.
Table 2.
The effects of DIS sequences on HIV-1 and HIV-2 RNA copackaging
| Coexpressed constructsa | CeFP+ particles |
RNA labeling efficiencyc | |||
|---|---|---|---|---|---|
| Total no. analyzed | YFP+ (%) | mCherry+ (%) | YFP+ + mCherry+ (%)b | ||
| HIV-1–GagCeFP-MS2SL + HIV-2–DIS3GC-GagCeFP-BglSL | |||||
| Expt 1 | 10,420 | 35.0 | 45.9 | 12.4 | 93.3 |
| Expt 2 | 7,558 | 42.6 | 38.6 | 12.2 | 93.4 |
| Expt 3 | 5,866 | 47.9 | 32.5 | 11.3 | 91.7 |
| Expt 4 | 8,644 | 47.6 | 33.1 | 12.5 | 93.2 |
| Expt 5 | 9,474 | 49.7 | 33.1 | 9.6 | 92.4 |
| Expt 6 | 11,034 | 39.2 | 37.5 | 13.6 | 90.3 |
| Expt 7 | 11,664 | 33.4 | 45.6 | 12.6 | 91.6 |
| HIV-1–DIS6C-GagCeFP-MS2SL + HIV-2–DIS6G-GagCeFP-BglSL | |||||
| Expt 1 | 2,762 | 26.2 | 34.2 | 31.9 | 92.3 |
| Expt 2 | 2,853 | 24.1 | 40.7 | 28.6 | 93.4 |
| Expt 3 | 2,896 | 28.7 | 33.4 | 31.9 | 94.0 |
| Expt 4 | 6,605 | 36.6 | 26.4 | 31.0 | 94.0 |
| Expt 5 | 6,647 | 36.0 | 25.0 | 33.0 | 94.0 |
| Expt 6 | 6,387 | 32.4 | 29.2 | 32.3 | 93.9 |
| Expt 7 | 5,095 | 37.7 | 26.6 | 28.5 | 92.8 |
| Expt 8 | 4,806 | 35.2 | 29.3 | 28.5 | 93.0 |
| Expt 9 | 5,702 | 36.3 | 26.2 | 31.1 | 93.6 |
| HIV-1–DIS6C-GagCeFP-MS2SL + HIV-2–DIS6C-GagCeFP-BglSL | |||||
| Expt 1 | 3,085 | 35.4 | 38.7 | 16.9 | 91.0 |
| Expt 2 | 3,374 | 36.7 | 37.3 | 17.7 | 91.7 |
| Expt 3 | 3,259 | 35.0 | 40.7 | 15.1 | 90.8 |
| Expt 4 | 6,100 | 35.7 | 40.6 | 14.4 | 90.7 |
| Expt 5 | 4,209 | 39.8 | 35.2 | 16.3 | 91.3 |
| Expt 6 | 4,609 | 41.0 | 33.7 | 15.3 | 90.0 |
| Expt 7 | 8,688 | 34.1 | 42.2 | 14.8 | 91.1 |
| Expt 8 | 9,626 | 33.2 | 42.5 | 16.6 | 92.3 |
Although not indicated, both Bgl-mCherry and MS2-YFP were coexpressed in all experiments.
Average ± SD for HIV-1–GagCeFP-MS2SL + HIV-2–DIS3GC-GagCeFP-BglSL, 12.0 ± 1.3; average ± SD for HIV-1–DIS6C-GagCeFP-MS2SL + HIV-2–DIS6G-GagCeFP-BglSL, 30.8 ± 1.8; average ± SD for HIV-1–DIS6C-GagCeFP-MS2SL + HIV-2–DIS6C-GagCeFP-BglSL, 15.9 ± 1.2.
Calculated by adding the values from the three columns under the CeFP+ heading.
Although mutating the HIV-2 DIS sequence to GCGCGC may have allowed base pairing of the DIS sequences in the HIV-1 and HIV-2 RNA, this mutation did not interfere with the ability of two RNAs from the same virus to base pair at the DIS sequences. To simultaneously promote heterodimerization and discourage homodimerization of the viral RNAs, we mutated the HIV-1 DIS sequence to CCCCCC (HIV-1–DIS6C–GagCeFP-MS2SL; Fig. 2C) and the HIV-2 DIS sequence to GGGGGG (HIV-2–DIS6G-GagCeFP-BglSL; Fig. 2D), coexpressed these constructs with RNA-binding proteins, and examined the resulting viruses. Indeed, the copackaging frequency of these two RNAs increased to ∼31% (range in 9 independent experiments, 28% to 33%) (Table 2). These results suggested that the increased copackaging of HIV-1 and HIV-2 RNAs is the result of heterodimerization that occurs before RNA encapsidation and is not the result of the packaging of one HIV-1 and one HIV-2 RNA each as a monomer.
The increased copackaging of HIV-1–DIS6C–GagCeFP-MS2SL and HIV-2–DIS6G-GagCeFP-BglSL can occur through two distinct mechanisms. The presence of CCCCCC in HIV-1 RNA and GGGGGG in HIV-2 RNA can encourage RNA heterodimerization or discourage RNA homodimerization or both. To distinguish among those possible mechanisms, we measured the copackaging frequency of HIV-1 and HIV-2 RNAs when both genomes contained CCCCCC in their DIS sequences, that is, HIV-1–DIS6C-GagCeFP-MS2SL and HIV-2–DIS6C–GagCeFP-BglSL (Fig. 2D). Both RNAs derived from these viruses were discouraged from homodimerization; however, the lack of complementarity of the two DIS sequences also do not encourage heterodimerization of these two RNAs. In 8 experiments using the particles generated by coexpressing the two constructs containing CCCCCC in DIS sequences, we observed a modest increase in the heterologous RNA copackaging frequency to ∼16% (range, 14% to 18%) (Table 2).
Taken together, these data indicate that the copackaging of HIV-1 and HIV-2 RNAs can be influenced by the sequence at the DIS. Simply allowing intermolecular interaction by changing the HIV-2 DIS to the same sequence as the HIV-1 DIS had little effect on the copackaging frequency, indicating that other elements are present that favor RNA homodimerization. The level of heterologous RNA copackaging was increased slightly when the DIS was mutated to discourage RNA homodimerization. However, it was when the two DIS sequences were mutated to simultaneously discourage RNA homodimerization and encourage RNA heterodimerization that we observed the largest increase of the copackaging frequency of HIV-1 and HIV-2 RNAs.
Determining the roles of HIV-1 and HIV-2 Gag polyproteins in heterologous RNA copackaging.
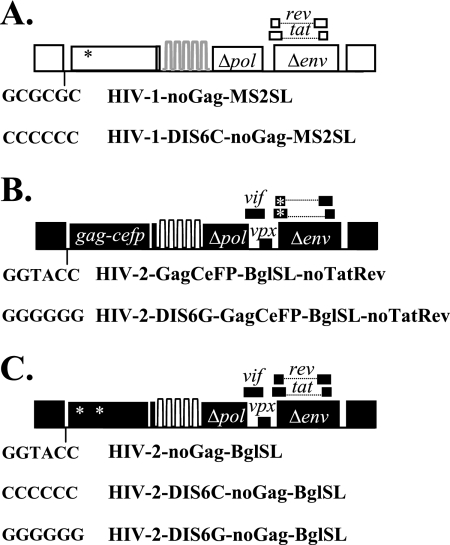
We previously showed that HIV-1 and HIV-2 Gag can coassemble into the same particle (2). It is possible that the coassembly of these two Gag proteins, by each interacting with its corresponding RNA, can promote the copackaging of HIV-1 and HIV-2 RNAs. To investigate the individual and collective contributions of HIV-1 and HIV-2 Gag proteins in heterologous RNA copackaging, we examined the effects of eliminating one of the Gag proteins in the system. We first examined the RNA encapsidation that resulted when only HIV-2 Gag was present in the system. It was previously shown that HIV-2 Gag does not package HIV-1 RNA (18); however, it is unclear whether HIV-1 RNA packaging can be promoted by interactions with HIV-2 RNA. For this purpose, we introduced an inactivating frameshift mutation into the gag gene of the HIV-1 construct to generate HIV-1–noGag-MS2SL (Fig. 3A), which can produce near-full-length viral RNA with the packaging signal but does not express functional Gag proteins. As expected, HIV-1–noGag-MS2SL did not generate CeFP+ particles but its RNA was able to be packaged into particles when coexpressed with another HIV-1 that expresses Gag (data not shown). We then tested whether heterologous RNA copackaging can be observed when only HIV-2 Gag was present. HIV-1–noGag-MS2SL, HIV-2–GagCeFP-BglSL, MS2-YFP, and Bgl-mCherry were cotransfected into 293T cells, and the resulting viruses were examined by single-virion analysis. We found that over 90% of the CeFP+ particles had mCherry signals, indicating that HIV-2 RNA was packaged efficiently in these viruses; however, we did not observe a significant level of YFP signals, indicating that HIV-1 RNA was not packaged efficiently, and copackaging was not observed (Table 3).
Fig. 3.
General structures of constructs used to examine the roles of HIV-1 and HIV-2 Gag proteins in heterologous RNA copackaging. (A) HIV-1 constructs that do not express functional Gag. These constructs harbor a frameshift mutation in gag. (B) HIV-2 constructs that do not express functional Tat and Rev. These constructs harbor a frameshift mutation that simultaneously prevents expression of functional HIV-2 Tat and that of functional Rev. (C) HIV-2 constructs that do not express functional Gag and their DIS sequences. Each construct contains two mutations in gag to prevent expression of functional Gag. DIS sequences of the constructs are indicated. Frameshift mutations are indicated by asterisks.
Table 3.
Single-virion analyses of RNAs packaged by HIV-2 Gag
| Coexpressed constructsa | CeFP+ particles |
RNA labeling efficiencyc | |||
|---|---|---|---|---|---|
| Total no. analyzed | YFP+ (%) | mCherry+ (%) | YFP+ + mCherry+ (%)b | ||
| HIV-1–noGagCeFP-MS2SL + HIV-2–GagCeFP-BglSL | |||||
| Expt 1 | 2,405 | 0.0 | 92.6 | 0.9 | 93.5 |
| Expt 2 | 835 | 0.0 | 96.3 | 0.2 | 96.5 |
| Expt 3 | 1,227 | 0.0 | 92.3 | 2.1 | 94.4 |
| Expt 4 | 1,089 | 0.0 | 93.8 | 1.0 | 94.8 |
| Expt 5 | 909 | 0.0 | 97.4 | 0.9 | 98.3 |
| Expt 6 | 614 | 0.0 | 98.4 | 0.0 | 98.4 |
| Expt 7 | 1,548 | 0.0 | 91.5 | 1.2 | 92.7 |
| HIV-1–noGagCeFP-MS2SL + HIV-2G–agCeFP-BglSL-noTatRev | |||||
| Expt 1 | 495 | 0.0 | 97.0 | 0.6 | 97.6 |
| Expt 2 | 428 | 0.0 | 93.9 | 1.4 | 95.3 |
| Expt 3 | 391 | 0.0 | 98.0 | 0.3 | 98.3 |
| Expt 4 | 1,176 | 0.0 | 90.3 | 0.7 | 91.0 |
| HIV-1–DIS6C-noGagCeFP-MS2SL + HIV-2–DIS6G-GagCeFP-BglSL-noTatRev | |||||
| Expt 1 | 687 | 0.0 | 91.7 | 0.9 | 92.6 |
| Expt 2 | 527 | 0.0 | 91.7 | 0.8 | 92.5 |
| Expt 3 | 1,146 | 0.0 | 94.9 | 1.6 | 96.5 |
| Expt 4 | 810 | 0.0 | 90.6 | 1.5 | 92.1 |
Although not indicated, both Bgl-mCherry and MS2-YFP were coexpressed in all experiments.
Average ± SD for HIV-1–noGagCeFP-MS2SL + HIV-2–GagCeFP-BglSL, 0.9 ± 0.7; average ± SD for HIV-1–noGagCeFP-MS2SL + HIV-2–GagCeFP-BglSL-noTatRev, 0.8 ± 0.5; average ± SD for HIV-1–DIS6C-noGagCeFP-MS2SL + HIV-2-DIS6G-GagCeFP-BglSL-noTatRev, 1.2 ± 0.4.
Calculated by adding the values from the three columns under the CeFP+ heading.
To address the possibility that HIV-1 RNA may not have been expressed in the cells transfected with HIV-2 and therefore not packaged, we generated HIV-2–GagCeFP-BglSL-noTatRev, a construct that contains a frameshift mutation in the overlapping region of the HIV-2 tat and rev genes, to abolish the expression of functional Tat and Rev (Fig. 3B). Lacking functional Tat and Rev, HIV-2–GagCeFP-BglSL-noTatRev does not produce Gag and therefore does not generate viral particles (data not shown). However, when cotransfected with HIV-1–noGag-MS2SL that expressed functional Tat and Rev, HIV-2–GagCeFP-BglSL-noTatRev was able to produce viral particles. This strategy ensures the expression of HIV-1 RNA in the virus-producing cells. Analyses of these particles revealed abundant mCherry signals, indicating efficient packaging of HIV-2 RNA but very little YFP signals, indicating the lack of encapsidation of HIV-1 RNA (Table 3). To investigate whether HIV-1 RNA packaging can be rescued by base pairing in the DIS sequences with HIV-2 RNA, we generated HIV-1–DIS6C-noGag-MS2SL and HIV-2–DIS6G-GagCeFP-BSL-noTatRev, two variants containing DIS sequences that, when both HIV-1 and HIV-2 Gag proteins were expressed, promoted heterologous RNA packaging. Single-virion analyses revealed that the complementary DIS mutations were not sufficient to promote HIV-1 RNA packaging to detectable levels (Table 3).
Next, we examined the effects of eliminating HIV-2 Gag in heterologous RNA copackaging experiments. We generated HIV-2–noGag-MS2SL, which contains two inactivating mutations in the gag gene: a substitution mutation in the MA-encoding region that generated a stop codon and a frameshift in the CA-encoding region that disrupts the Gag reading frame. We then analyzed particles generated by coexpressing HIV-1–GagCeFP-MS2SL, HIV-2–noGag-MS2SL, MS2-YFP, and Bgl-mCherry and observed a heterologous RNA copackaging frequency of ∼10% (range, 9% to 11%) (Table 4), which is similar to the frequency observed when both HIV-1 and HIV-2 Gag were present (Table 1). We then generated HIV-2 constructs that contained mutations in DIS sequences (Fig. 3C) and examined whether these mutations had the same effects on RNA copackaging when only HIV-1 Gag was present in the system. Along with MS2-YFP and Bgl-mCherry, coexpression of HIV-1–DIS6C–GagCeFP-MS2SL and HIV-2–DIS6C-noGag-BglSL resulted in ∼16% heterologous RNA copackaging (Table 4), whereas coexpression of HIV-1–DIS6C–GagCeFP-MS2SL and HIV-2–DIS6G-noGag-BglSL resulted in ∼39% RNA copackaging (Table 4). The introduction of DIS mutations in the absence of HIV-2 Gag influenced copackaging in a manner parallel to that observed when both HIV-1 and HIV-2 Gag were present. Therefore, HIV-1 Gag alone is most likely responsible for HIV-1/HIV-2 RNA copackaging; although coassembly of HIV-1 and HIV-2 Gag occurs frequently, this event is not driving heterologous RNA copackaging.
Table 4.
Single-virion analyses of RNAs packaged by HIV-1 Gag
| Coexpressed constructsa | CeFP+ particles |
RNA labeling efficiencyc | |||
|---|---|---|---|---|---|
| Total no. analyzed | YFP+ (%) | mCherry+ (%) | YFP+ + mCherry+ (%)b | ||
| HIV-1–GagCeFP-MS2SL + HIV-2–noGagCeFP-BglSL | |||||
| Expt 1 | 1,243 | 46.3 | 33.8 | 10.1 | 90.2 |
| Expt 2 | 349 | 22.6 | 57.9 | 9.5 | 90.0 |
| Expt 3 | 1,399 | 45.6 | 36.7 | 10.1 | 92.4 |
| Expt 4 | 1,405 | 47.0 | 35.9 | 9.1 | 92.0 |
| Expt 5 | 1,240 | 42.5 | 40.9 | 9.0 | 92.4 |
| Expt 6 | 2,839 | 27.2 | 53.9 | 8.9 | 90.0 |
| Expt 7 | 2,376 | 45.5 | 37.7 | 9.9 | 93.1 |
| Expt 8 | 4,052 | 27.8 | 54.8 | 9.7 | 92.3 |
| Expt 9 | 2,016 | 47.2 | 37.1 | 9.0 | 93.3 |
| Expt 10 | 4,701 | 53.9 | 26.9 | 10.3 | 91.1 |
| Expt 11 | 5,218 | 58.0 | 23.7 | 10.5 | 92.2 |
| Expt 12 | 6,564 | 48.9 | 31.7 | 10.5 | 91.1 |
| Expt 13 | 6,789 | 42.7 | 38.7 | 9.3 | 90.7 |
| HIV-1–DIS6C-GagCeFP-MS2SL + HIV-2–DIS6C-noGagCeFP-BglSL | |||||
| Expt 1 | 3,152 | 45.4 | 34.1 | 12.7 | 92.2 |
| Expt 2 | 4,006 | 47.8 | 30.3 | 15.4 | 93.5 |
| Expt 3 | 5,282 | 48.0 | 29.2 | 17.5 | 94.7 |
| Expt 4 | 5,703 | 45.6 | 33.1 | 15.8 | 94.5 |
| Expt 5 | 6,045 | 44.7 | 27.5 | 18.5 | 90.7 |
| HIV-1–DIS6C-GagCeFP-MS2SLL + HIV-2–DIS6G-noGagCeFP-BglSL | |||||
| Expt 1 | 3,881 | 29.4 | 28.7 | 36.7 | 94.8 |
| Expt 2 | 4,589 | 33.7 | 21.5 | 39.8 | 95.0 |
| Expt 3 | 5,334 | 34.4 | 22.7 | 38.3 | 95.4 |
| Expt 4 | 5,020 | 29.6 | 26.1 | 38.8 | 94.5 |
| Expt 5 | 4,862 | 24.8 | 28.5 | 41.1 | 94.4 |
Although not indicated, both Bgl-mCherry and MS2-YFP were coexpressed in all experiments.
Average ± SD for HIV-1–GagCeFP-MS2SL + HIV-2–noGagCeFP-BglSL, 9.7 ± 0.6; average ± SD for HIV-1–DIS6C-GagCeFP-MS2SL + HIV-2–DIS6C-noGagCeFP-BglSL, 16.0 ± 2.2; average ± SD for HIV-1–DIS6C-GagCeFP-MS2SLL + HIV-2–DIS6G-noGagCeFP-BglSL, 38.9 ± 1.6.
Calculated by adding the values from the three columns under the CeFP+ heading.
DISCUSSION
During the assembly process of all known retroviruses, Gag proteins select from a large pool of total cellular RNAs and package two copies of full-length, single-stranded viral RNA into viral particles. In some viruses, such as HIV-1 and HIV-2, RNA selects its copackaged partner (or dimerizes) prior to encapsidation (4, 23, 27). Considering the specificity of this process, the RNA-RNA and RNA-protein interactions involved are likely to be highly regulated and coordinated events. The packaging of two distinct retroviral RNAs into the same virus particle requires at least a partial overlap, spatially as well as temporally, of the two RNAs in the producer cell, as well as similar packaging determinants and mechanisms of the two viruses. The work described in this report explored the factors that influence the copackaging of two divergent human retroviruses, HIV-1 and HIV-2. The copackaging of the heterologous RNAs is a determinant for potential genetic interactions; furthermore, this system allowed us to study the mechanisms of retroviral RNA dimerization and packaging.
In this study we found that, within a population of particles containing similar proportions of HIV-1 and HIV-2 RNAs, about 10% of the particles contained both HIV-1 and HIV-2 RNAs. Equal expression and random assortment of RNAs from two proviruses predict that 50% of the virions would be heterozygous particles, containing one RNA derived from each provirus. The 10% copackaging frequency implies that HIV-1 and HIV-2 RNA packaging mechanisms have sufficient overlap to yield a modest but reproducible level of copackaging but are divergent enough that HIV-1 and HIV-2 RNAs still prefer to homodimerize. We have previously measured recombination betwen HIV-1 and HIV-2 using two modified genomes, each carrying a mutated green fluorescent protein (gfp) gene; recombination between the two mutated genes could reconstitute a functional gfp gene, and its expression could be measured. We found that a very small percentage (∼0.5%) of infected cells had a GFP+ phenotype (25), which is much lower than that generated between two HIV-1 or two HIV-2 viruses (∼7%) (5, 29). In all three studies, the gfp gene was used as a target sequence for reporting recombination; thus, regardless of the sequence diversity elsewhere in the viral genomes, the target regions between the mutations in the gfp genes contain identical sequences. Results from this study indicate that ∼10% of the particles have copackaged HIV-1 and HIV-2 RNAs and that the low copackaging frequency was a contributing factor but not the only one that caused the lower recombination rate. Together, these studies revealed that there is a block or barrier in the reverse transcription step that limits the completion of DNA synthesis or recombination in the target gfp gene between these heterologous genomes. The details of the mechanism for this block or barrier are currently unknown. It is possible that the two copackaged RNAs have different structures, thereby inhibiting DNA synthesis or preventing RT from switching templates; alternatively, it is also possible that the low sequence homology flanking the gfp genes reduces RT template switching within the gfp gene. Further experimentation is needed to distinguish between these possibilities.
In this report, we studied factors that may restrict the heterodimerization of HIV-1 and HIV-2 RNA and thus restrict copackaging. We found that changing the DIS to the same palindrome (GCGCGC) to allow the DIS sequences of HIV-1 and HIV-2 RNA to form base pairs did not increase the copackaging, most likely because the two RNAs still homodimerize efficiently. The largest improvement of heterologous RNA copackaging was observed when we simultaneously encouraged RNA heterodimerization and discouraged homodimerization by changing the HIV-1 DIS to CCCCCC and the HIV-2 DIS to GGGGGG. These results not only revealed that DIS sequences can mediate dimerization of the heterologous RNAs but also shed light on the mechanisms of Gag-RNA interactions.
Although RNA partner selection occurs prior to the encapsidation of HIV-1 or HIV-2 RNA, it is unclear how HIV Gag proteins distinguish between monomeric and dimeric RNAs. Studies of MLV packaging showed that the monomeric and dimeric RNAs have very different secondary structures (10, 15, 21). Furthermore, dimerization of MLV RNAs exposes high-affinity binding sites for nucleocapsids, which is critical for RNA packaging (16, 22). It is thought that exposure of these sites allows Gag binding, leading to the incorporation of RNA into virus particles. Despite its attractiveness, this model has been difficult to extend to the RNA packaging mechanisms of HIV-1, mainly because structural analyses of HIV-1 RNA have not revealed a drastic difference between monomeric RNA and dimeric RNA (35); hence, the relationship between switching the RNA states and RNA-Gag molecular recognition is not well defined. Our study revealed surprising insights into this process. Despite the low homology of HIV-1 and HIV-2 leader sequences, when DIS sequences were manipulated to form base pairing between these two heterologous RNAs but not between homologous RNAs, we observed increased copackaging. If the HIV-1 and MLV Gag-RNA recognition mechanisms are similar, then our results suggest that base pairing of the DIS between these two heterologous RNAs can expose the hidden high-affinity binding sites recognized by HIV-1 Gag. The limited homology between the 5′ leader sequences of HIV-1 and HIV-2 makes it less likely that there are many other conserved interactions between these two RNAs; hence, this hypothesis implies that base pairing of DIS provides a molecular switch that directly or indirectly exposes Gag-binding sites.
A possible model for the mechanism of heterologous RNA copackaging is that such events are driven by the coassembly of Gag polyproteins from different viruses. However, our results indicated that HIV-1 Gag, in the absence of HIV-2 Gag, can mediate heterologous RNA copackaging at levels similar to those observed with both HIV-1 and HIV-2 Gag proteins. This observation defines the trans-acting element required for the heterologous RNA copackaging, that is, the Gag protein that recognizes its own RNA and the heterologous RNA.
Replication strategies used by retroviruses ensure frequent generation of recombinants, as one viral particle has two full-length viral genomes and RT switches templates during DNA synthesis. Although copackaged RNAs are derived from the same types of viruses most of the time, there are infrequent occurrences of heterologous RNA copackaging, which provides the backdrop for the generation of chimeric viruses from two distantly related viruses. Delineating the requirements and factors that influence heterologous RNA copackaging allows us to understand the potential for the genetic interactions between distantly related viruses and also provide insights into the general mechanisms of RNA-RNA and RNA-Gag interactions that lead to encapsidation of viral genomes.
ACKNOWLEDGMENTS
We thank Vinay K. Pathak for discussions throughout the project and critical reading of the manuscript.
This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research; IATAP funding, NIH.
Footnotes
Published ahead of print on 17 August 2011.
REFERENCES
- 1. Ariën K. K., et al. 2005. The replicative fitness of primary human immunodeficiency virus type 1 (HIV-1) group M, HIV-1 group O, and HIV-2 isolates. J. Virol. 79:8979–8990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boyko V., et al. 2006. Coassembly and complementation of Gag proteins from HIV-1 and HIV-2, two distinct human pathogens. Mol. Cell 23:281–287 [DOI] [PubMed] [Google Scholar]
- 3. Certo J. L., Shook B. F., Yin P. D., Snider J. T., Hu W. S. 1998. Nonreciprocal pseudotyping: murine leukemia virus proteins cannot efficiently package spleen necrosis virus-based vector RNA. J. Virol. 72:5408–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen J., et al. 2009. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc. Natl. Acad. Sci. U. S. A. 106:13535–13540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen J., Powell D., Hu W. S. 2006. High frequency of genetic recombination is a common feature of primate lentivirus replication. J. Virol. 80:9651–9658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chin M. P., Chen J., Nikolaitchik O. A., Hu W. S. 2007. Molecular determinants of HIV-1 intersubtype recombination potential. Virology 363:437–446 [DOI] [PubMed] [Google Scholar]
- 7. Chin M. P., et al. 2008. Long-range recombination gradient between HIV-1 subtypes B and C variants caused by sequence differences in the dimerization initiation signal region. J. Mol. Biol. 377:1324–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chin M. P., Rhodes T. D., Chen J., Fu W., Hu W. S. 2005. Identification of a major restriction in HIV-1 intersubtype recombination. Proc. Natl. Acad. Sci. U. S. A. 102:9002–9007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dirac A. M., Huthoff H., Kjems J., Berkhout B. 2002. Requirements for RNA heterodimerization of the human immunodeficiency virus type 1 (HIV-1) and HIV-2 genomes. J. Gen. Virol. 83:2533–2542 [DOI] [PubMed] [Google Scholar]
- 10. D'Souza V., Summers M. F. 2004. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature 431:586–590 [DOI] [PubMed] [Google Scholar]
- 11. Evans L. A., et al. 1988. Simultaneous isolation of HIV-1 and HIV-2 from an AIDS patient. Lancet ii:1389–1391 [DOI] [PubMed] [Google Scholar]
- 12. Gao F., et al. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436–441 [DOI] [PubMed] [Google Scholar]
- 13. Gao F., et al. 1992. Human infection by genetically diverse SIVSM-related HIV-2 in West Africa. Nature 358:495–499 [DOI] [PubMed] [Google Scholar]
- 14. George J. R., et al. 1992. Prevalence of HIV-1 and HIV-2 mixed infections in Cote d'Ivoire. Lancet 340:337–339 [DOI] [PubMed] [Google Scholar]
- 15. Gherghe C., Leonard C. W., Gorelick R. J., Weeks K. M. 2010. Secondary structure of the mature ex virio Moloney murine leukemia virus genomic RNA dimerization domain. J. Virol. 84:898–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gherghe C., et al. 2010. Definition of a high-affinity Gag recognition structure mediating packaging of a retroviral RNA genome. Proc. Natl. Acad. Sci. U. S. A. 107:19248–19253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hirsch V. M., Olmsted R. A., Murphey-Corb M., Purcell R. H., Johnson P. R. 1989. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 339:389–392 [DOI] [PubMed] [Google Scholar]
- 18. Kaye J. F., Lever A. M. 1998. Nonreciprocal packaging of human immunodeficiency virus type 1 and type 2 RNA: a possible role for the p2 domain of Gag in RNA encapsidation. J. Virol. 72:5877–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lemey P., et al. 2003. Tracing the origin and history of the HIV-2 epidemic. Proc. Natl. Acad. Sci. U. S. A. 100:6588–6592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McCutchan F. E. 2006. Global epidemiology of HIV. J. Med. Virol. 78(Suppl. 1):S7–S12 [DOI] [PubMed] [Google Scholar]
- 21. Miyazaki Y., et al. 2010. An RNA structural switch regulates diploid genome packaging by Moloney murine leukemia virus. J. Mol. Biol. 396:141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miyazaki Y., et al. 2010. Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography. J. Mol. Biol. 404:751–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moore M. D., et al. 2007. Dimer initiation signal of human immunodeficiency virus type 1: its role in partner selection during RNA copackaging and its effects on recombination. J. Virol. 81:4002–4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moore M. D., et al. 2009. Probing the HIV-1 genomic RNA trafficking pathway and dimerization by genetic recombination and single virion analyses. PLoS Pathog. 5:e1000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Motomura K., Chen J., Hu W. S. 2008. Genetic recombination between human immunodeficiency virus type 1 (HIV-1) and HIV-2, two distinct human lentiviruses. J. Virol. 82:1923–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murphey-Corb M., et al. 1986. Isolation of an HTLV-III-related retrovirus from macaques with simian AIDS and its possible origin in asymptomatic mangabeys. Nature 321:435–437 [DOI] [PubMed] [Google Scholar]
- 27. Ni N., et al. 2011. Mechanisms of human immunodeficiency virus type 2 RNA packaging: efficient trans packaging and selection of RNA copackaging partners. J. Virol. 85:7603–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nikolaitchik O. A., Galli A., Moore M. D., Pathak V. K., Hu W. S. 2011. Multiple barriers to recombination between divergent HIV-1 variants revealed by a dual-marker recombination assay. J. Mol. Biol. 407:521–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rhodes T., Wargo H., Hu W. S. 2003. High rates of human immunodeficiency virus type 1 recombination: near-random segregation of markers one kilobase apart in one round of viral replication. J. Virol. 77:11193–11200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Santiago M. L., et al. 2005. Simian immunodeficiency virus infection in free-ranging sooty mangabeys (Cercocebus atys atys) from the Tai Forest, Cote d'Ivoire: implications for the origin of epidemic human immunodeficiency virus type 2. J. Virol. 79:12515–12527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sattentau Q. J. 1988. The role of the CD4 antigen in HIV infection and immune pathogenesis. AIDS 2(Suppl. 1):S11–S16 [DOI] [PubMed] [Google Scholar]
- 32. Van Heuverswyn F., et al. 2006. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature 444:164. [DOI] [PubMed] [Google Scholar]
- 33. Watts J. M., et al. 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460:711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Weiss R. A., et al. 1988. Human immunodeficiency viruses: neutralization and receptors. J. Acquir. Immune Defic. Syndr. 1:536–541 [PubMed] [Google Scholar]
- 35. Wilkinson K. A., et al. 2008. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 6:e96. [DOI] [PMC free article] [PubMed] [Google Scholar]