Abstract
Severe acute respiratory syndrome (SARS) is a highly contagious and life threatening disease, with a fatality rate of almost 10%. The etiologic agent is a novel coronavirus, severe acute respiratory syndrome coronavirus (SARS-CoV), with animal reservoirs found in bats and other wild animals and thus the possibility of reemergence. In this study, we first investigated at 6 years postinfection whether SARS-specific memory T cells persist in SARS-recovered individuals, demonstrating that these subjects still possess polyfunctional SARS-specific memory CD4+ and CD8+ T cells. A dominant memory CD8+ T cell response against SARS-CoV nucleocaspid protein (NP; amino acids 216 to 225) was then defined in SARS-recovered individuals carrying HLA-B*40:01, a HLA-B molecule present in approximately one-quarter of subjects of Asian ethnicities. To reconstitute such a CD8+ T cell response, we isolated the alpha and beta T cell receptors of the HLA-B*40:01-restricted SARS-specific CD8+ T cells. Using T cell receptor gene transfer, we generated SARS-specific redirected T cells from the lymphocytes of normal individuals. These engineered CD8+ T cells displayed avidity and functionality similar to that of natural SARS-specific memory CD8+ T cells. They were able to degranulate and produce gamma interferon, tumor necrosis factor alpha, and macrophage inflammatory proteins 1α and 1β after antigenic stimulation. Since there is no effective treatment against SARS, these transduced T cells specific for an immunodominant SARS epitope may provide a new avenue for treatment during a SARS outbreak.
INTRODUCTION
No severe human disease associated with coronaviruses was reported until the outbreak of severe acute respiratory syndrome (SARS) in late 2002 in Guangdong, China (22, 29). It affected more than 8,000 patients and caused nearly 800 deaths in more than 30 countries. The etiological agent of the syndrome was identified as a novel coronavirus termed SARS coronavirus (SARS-CoV) (5, 10, 17). The SARS-CoV genome encodes the replicase genes (open reading frame 1a [ORF1a] and ORF1b) and four structural proteins (spike [S], nucleocapsid [NP], membrane [M], and envelope [E]), together with eight other accessory proteins, namely, 3a, 3b, ORF6, 7a, 7b, 8a, 8b, and 9b.
Recent studies have indicated the importance of T cells in viral clearance during a primary infection of SARS-CoV (3, 32). SARS-specific memory T cell responses against the structural proteins, S, M, E, and NP are present in SARS recovered patients (6, 11, 13), but it is unclear whether SARS elicits a lasting memory T cell response.
Most studies focus on the surface glycoprotein S protein, and to date several cytotoxic-T-lymphocyte (CTL) epitopes have been identified in the S protein (15, 25, 26, 27, 28, 33). However, a detailed analysis and epitope definition of T cell responses against SARS 3a (the largest accessory protein and unique to the SARS-CoV) (14, 16) and NP proteins is lacking. Here, we analyzed the presence and function of SARS memory T cell response at 6 years postinfection. We defined a dominant SARS-specific T cell response commonly detectable in Asian individuals and isolated its SARS-specific T cell receptors. We then demonstrated the possibility to generate SARS-specific T cells from lymphocytes of healthy uninfected individuals through T-cell-receptor (TCR) gene transfer. TCR gene transfer has emerged, in several studies, as a way to develop cell-based therapy for chronic viral diseases such as hepatitis C and B (8, 31) and malignancies such as melanoma (21). In addition, adoptive transfer of virus-specific T cells can also protect subjects that undergo immunosuppressive treatment from human cytomegalovirus (HCMV) or Epstein-Barr virus (EBV) reactivation (4, 7). Thus, the identification of SARS epitopes and production of SARS-specific TCR-redirected T cells can provide prophylactic or therapeutic opportunities against this infection.
MATERIALS AND METHODS
Subjects.
Sixteen recovered SARS individuals (6 years postinfection) were enrolled in the present study from the Singapore General Hospital (Singapore). All of the participants had been diagnosed as having SARS based on clinical examination during the period from March to May 2003 according to the World Health Organization's definition of SARS (30). The diagnosis was further confirmed by serological detection of SARS-CoV-specific antibodies detected by enzyme-linked immunosorbent assay and/or reverse transcription-PCR for SAR-CoV mRNA. Five normal subjects without any contact history with SARS patients were used as negative controls. This study was approved by the Centralized Institutional Review Board of the Singapore Health Services Pte, Ltd. (Singapore).
Isolation of PBMC and in vitro expansion of SARS-specific T cells.
Peripheral blood mononuclear cells (PBMC) were isolated from fresh heparinized blood by Ficoll-Hypaque density gradient centrifugation and resuspended in AIM-V medium (Invitrogen, Carlsbad, CA) with 2% pooled human AB serum. T cells were used either directly ex vivo or after a 10-day antigen-specific in vitro stimulation as previously described (24). Briefly, 20% of the PBMC were first stimulated with 10 μg of all the overlapping peptides/ml for 1 h at 37°C and then washed before coculture with the remaining PBMC in AIM-V medium with 2% pooled human AB serum supplemented with interleukin-2 (IL-2; R&D Systems, Abingdon, United Kingdom) at 20 U/ml and seeded at 1 ml/well in 24-well plates.
Synthetic peptides.
Two panels of 15mer peptides overlapping by 10 residues covering NP (total, 83 peptides) and accessory 3a protein (total, 53 peptides) were used. The peptides were purchased from Chiron Mimotopes (Victoria, Australia) at a purity above 80%, and their composition was confirmed by mass spectrometry analysis. The NP peptides were pooled in a 9-by-9 matrix containing 9 or 10 peptides/pool, whereas the 3a peptides were pooled in a 7-by-8 matrix containing 6 to 8 peptides/pool, respectively. All peptides were diluted at 40 mg/ml in dimethyl sulfoxide and then further diluted in RPMI 1640 medium at a working dilution (between 10 and 1 mg/ml).
IFN-γ ELISPOT assay.
Using the panel of 15mer peptides pooled in 33 mixtures as described above, SARS-specific T cell responses were analyzed as previously described (8) in gamma interferon (IFN-γ) enzyme-linked immunospot (ELISPOT) assays either ex vivo using frozen PBMC or short-term polyclonal T cell lines previously expanded in vitro by 10 days of stimulation with the same peptide mixtures. The number of IFN-γ-producing cells was expressed in spot-forming units (SFU) per 1 × 105 cells (ex vivo) or 5 × 104 cells (in vitro). Positive controls consisted of PBMC stimulated with staphylococcal enterotoxin B (SEB) or phytohemagglutinin (PHA). Wells were considered positive if they had values at least two times the background value and the number of spots was greater than 30. A positive peptide was identified as the common peptide belonging to both the “number” and “alphabet” pools that gave positive ELISPOT responses.
Intracellular cytokine staining and degranulation assays.
In vitro-expanded T cells were incubated in medium alone (negative control) or with viral peptides (5 μg/ml) for 5 h in the presence of brefeldin A (10 μg/ml). The cells were stained with anti-CD8 (phycoerythrin [PE]-Cy7) or anti-CD4 PE-Cy7 and anti-CD3 (peridinin chlorophyll protein [PerCP]-Cy5.5) (BD Pharmingen, San Jose, CA) for 30 min at 4°C and washed once and then fixed and permeabilized using Cytofix/Cytoperm fixation/permeabilization (BD Biosciences, San Jose, CA), according to the manufacturer's instructions. Intracellular staining was performed with anti-IFN-γ-PE (BD Biosciences) for 30 min at 4°C, followed by washing and analysis by flow cytometry. To assess the degranulation activity, anti-CD107a (fluorescein isothiocyanate [FITC]) antibody (BD Pharmingen, San Diego, CA) was added to all wells at the beginning of the 5 h of incubation with T cells. After incubation, the cells were washed and stained as described above. Polyfunctional T cell assays were performed using the antibodies against the following: (i) Th1/Th2-associated cytokines (IFN-γ, tumor necrosis factor alpha [TNF-α], IL-2, IL-4, IL-10, and IL-21) or (ii) inflammatory cytokines (IFN-γ, IL-8, macrophage inflammatory protein 1α [Mip1α], Mip1β, granulocyte-macrophage colony-stimulating factor [GM-CSF], and IL-17a).
Major histocompatibility complex restriction for CD8+ T cell responses.
The HLA phenotype for each of 16 individuals who had recovered from SARS (SARS-recovered individuals) was determined by PCR amplification and sequencing based typing method as described previously (23). EBV-transformed lymphoblastoid B cell lines (EBV-LCL) of defined HLA phenotype (kindly provided by Chan Soh Ha, National University of Singapore) were used as antigen-presenting cells.
Isolation of a NP44-specific CD8+ T cell clone and TCR α and β chain DNA.
A NP44-specific CD8+ T cell clone was generated from the HLA-B*4001+ SARS patient 1. Short-term lines were made by stimulating PBMC with 5 μg of NP44 peptide/ml for 10 days. NP44-specific CD8 T cells were labeled with CD107a-PE for 2 h in 37°C, followed by staining with CD8-APC for 30 min at 4°C and purification by fluorescence-activated cell sorting (FACS). CD8 T cells were then cloned by limiting dilution assay, and clonal populations were expanded in AIM-V and 20 U of IL-2, 10 ng of IL-7, and 10 ng of IL-15 (R&D Systems)/ml with 1 μg of PHA (Sigma-Aldrich, Dorset, United Kingdom)/ml, using allogeneic irradiated PBMC as feeder cells.
Total RNA was isolated from the T cell clone using TRIzol (Invitrogen). The SARS NP44 TCR α and β chains were cloned as described previously (8). Briefly, the TCR α and β chains of the NP44 TCR were cloned by RACE (rapid amplification of cDNA ends)-PCR using a GeneRacer kit (Invitrogen). Sequence analysis of the cDNA clones revealed that the T cell clone expressed a single TCR α chain of AV4.0/AJ28.0/AC and a β chain of BV4.3/BJ2.1/BD1.0/BC according to IMGT/V-Quest (http://www.imgt.org) (2). After functional confirmation of the TCR, a codon-optimized, 2A-linked construct was synthesized and cloned into the MP-71 vector for expression in primary human T cells.
Retroviral transduction.
A total of 2 × 106 Phoenix amphotropic packaging cells were seeded into 10-cm tissue culture dishes 24 h prior to transfection. Phoenix cells were transiently cotransfected using calcium phosphate with 18 μg of TCR construct, together with 6 μg of amphotropic envelope for 24 h. Iscove modified Dulbecco medium (IMDM) was replaced with AIM-V supplemented with 2% human AB serum, and phoenix cells were incubated for an additional 24 h before retroviral supernatants were collected for transduction.
PBMC from normal healthy donors were stimulated with 600 U of IL-2 (R&D Systems)/ml and 50 ng of anti-CD3 (OKT-3; eBioscience, San Diego, CA)/ml for 48 h. Untreated 24-well tissue culture plates were coated with 30 μg of retronectin (Takara Bio, Otsu Shiga, Japan)/ml overnight at 4°C 1 day prior to transduction. The wells were washed with Hanks balanced salt solution and then blocked with phosphate-buffered saline plus 2% bovine serum albumin. The lymphocytes were harvested, washed, and counted, and then 5 × 105 cells were plated into retronectin-coated wells and mixed with retroviral supernatants collected as described above. Mock-transduced cells, included as a negative control, were cultured with the supernatant from untransfected phoenix cells. Lymphocytes were incubated for 24 h in the retroviral supernatant, the medium was replaced, and the cells were maintained in AIM-V 2% human AB serum plus 100 U of IL-2/ml. After 3 days, the T cells were stained with anti-CD8 PE-Cy7 (BD Biosciences) and anti-Vβ4.3 PE (Beckman Coulter, Fullerton, CA).
Production of target cells expressing SARS antigens.
For T cell recognition of endogenously expressing SARS antigen, target cells were infected with recombinant vaccinia virus encoding either the SARS NP or S protein at multiplicity of infection (MOI) of 5 for 1 h at 37°C. After infection, the virus was removed, and the cells were washed and resuspended in AIM-V 2% human AB serum at 105 cells/well. Each well was then treated with 10 U of IFN-α and IFN-γ each/ml. An equal number of NP44-specific CD8 T cells or NP44 TCR-transduced T cells (105) was added to the target cells for 5 h in the presence of brefeldin A (10 μg/ml) and stained as described above for degranulation activity with anti-CD107a-FITC antibody.
RESULTS AND DISCUSSION
Memory T cell responses against SARS in recovered individuals.
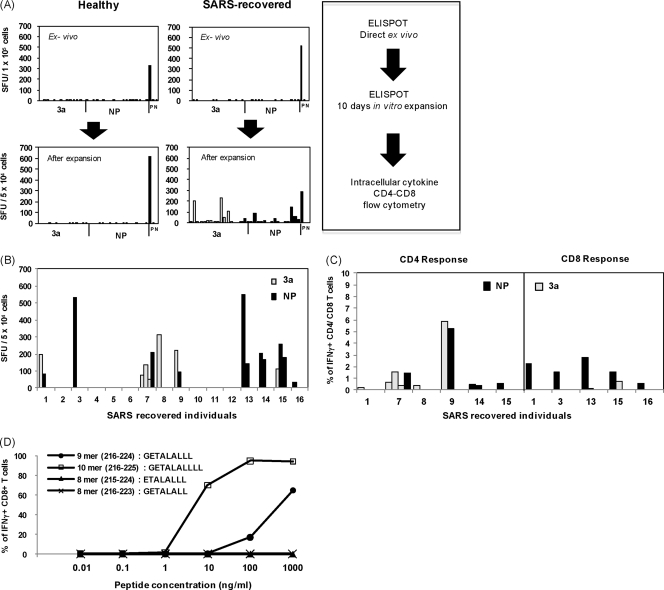
To investigate the presence and functional profile of SARS-specific memory T cell responses against NP and 3a SARS-CoV proteins, peripheral blood of 16 Asian subjects who recovered from SARS-CoV infection was collected 6 years postinfection. As negative controls, PBMC of five healthy individuals with no prior contact history with SARS were included. PBMC were stimulated directly ex vivo or after in vitro expansion with mixtures of synthetic peptides (15mer peptides overlapping by 10 residues) spanning the entire amino acid sequence of the NP and 3a proteins and analyzed using IFN-γ ELISPOT assay. The frequency of IFN-γ-producing SFU did not exceed 10 in the direct ex vivo ELISPOT assay (Fig. 1A), and no significant difference was observed between healthy and SARS-exposed individuals. In contrast, high frequencies of IFN-γ-producing SFU were detected in wells containing in vitro-expanded PBMC from SARS-recovered individuals (Fig. 1A), while no response was found in PBMC from healthy individuals. Figure 1B summarizes the ELISPOT data of the 16 SARS individuals after in vitro expansion of the PBMC. The data showed that 9 of the 16 SARS individuals displayed high frequencies of SFU against the NP protein, the 3a protein, or both proteins. Three individuals had a single response, and others showed multiple responses. These results showed that SARS-specific memory T cells are present in SARS-recovered individuals 6 years after infection, even though the frequency of these cells is low. Interestingly, in contrast to our study, T cell responses can be observed directly in the ex vivo ELISPOT assay conducted in a study by Li et al. (11) in which PBMC were obtained from patients 1 year postinfection. Such comparison suggests that the frequency of SARS-specific memory T cells progressively declines with time. This hypothesis is also supported by another study that monitored immune response in SARS individuals over a 2-year period and showed that CTL responses decline over time at 3, 6, and 18 months postinfection (12).
Fig. 1.
Memory T cell responses in SARS-recovered individuals. (A) ELISPOT analysis was performed on PBMC directly ex vivo (top panels) and after 10 days of in vitro expansion (bottom panels) using the same pools of peptides (3a and NP). The data for one healthy and one SARS-recovered individual are displayed. Each bar represents the response to an individual peptide mixture in SFU per 1 × 105 or 5 × 104 cells. The threshold of positivity is defined as two times the mean SFU of the unstimulated control (N) and >30 SFU. Positive control (P) was included with cells stimulated by SEB. (B) Summary of the positive ELISPOT results for all 16 SARS-recovered individuals after 10 days of in vitro expansion using the same pools of peptides. (C) IFN-γ production by SARS-specific CD8+ and CD4+ T cells determined by intracellular cytokine staining. Distribution of CD4+ (left panel) and CD8+ (right panel) responses between the two proteins tested. The data are expressed as the percentage of the total number of CD4+ or CD8+ T cells. (D) Functional assay measuring IFN-γ production by NP44-specific CD8+ T cells after stimulation with different concentrations of the 8mer (GETALALL) and (ETALALLL), 9mer (GETALALLL), and 10 mer (GETALALLLL) NP44 peptides. Target cells (HLA-B*40:01-positive EBV-LCLs) were pulsed with the indicated peptide concentrations. The data are expressed as the percentage of the total number of CD8+ T cells producing IFN-γ.
To confirm the ELISPOT assay results and to define whether the IFN-γ-producing cells were CD4+ or CD8+ T cells, in vitro-expanded T cell lines of every positive individual were tested with anti-CD4 and anti-CD8 specific antibodies and intracellular staining for IFN-γ. Figure 1C and Table 1 show the complete profile of T cell responses detected in the recruited individuals. More than 50% of the convalescent SARS individuals had positive T cell responses against at least one of the two proteins, and 25% of them had responses against both proteins. CD4+ T cell responses were observed in six of nine responsive individuals, while CD8+ T cell responses were detectable in five subjects. Helper CD4+ T cells recognized both 3a (six different epitopes recognized) and NP (four different epitopes) proteins. Strong responses against the NP or 3a protein were observed equally in individuals with multiple CD4 responses. Although NP or 3a proteins were equally immunogenic for helper T cell responses, CD8+ T cells recognized preferentially the NP protein (six of seven CD8+ T cell responses were specific for epitopes present on NP protein) with a single exception of peptide 3a2 (amino acids [aa] 6 to 20; RFFTLGSITAQPVKI) (Fig. 1C). This peptide was able to elicit both CD8+ and CD4+ responses, as already observed in another study (11). Importantly, three of the five individuals who showed a SARS-specific CD8+ T cell response recognize the aa 216 to 230 region (NP44 peptide) of the SARS-CoV NP protein (Table 1). All of the subjects carried the HLA-B*40:01 allele. The dominant CD8+ T cell response was further characterized using truncated 8-, 9-, and 10mer peptides and HLA-B*40:01-positive and -negative target cells. We verified that the 10mer peptide (GETALALLLL; aa 216 to 225) was the more efficient peptide for activation of NP44-specific CD8+ T cells only when presented by HLA-B*40:01-positive target cells (Fig. 1D).
Table 1.
Summary of CD4+ and CD8+ memory T cell responses in SARS-recovered individuals
| Patient | HLA profile | Peptide(s) | aa positiona | Peptide sequence | Type of response | % IFN-γ+, CD4+, or CD8+ cells |
|---|---|---|---|---|---|---|
| 1 | A*02:07 A*02:01 | 3a11 | 51–65 | AFLAVFQSATKIIAL | CD4 | 0.23 |
| B*46:01 B*40:01 | NP44 | 216–230 | GETALALLLLDRLNQ | CD8 | 2.26 | |
| Cw*01:03 Cw*15:02 | ||||||
| 3 | A*24:02 A*11:02 | NP44 | 216–230 | GETALALLLLDRLNQ | CD8 | 1.54 |
| B*40:01 B*48:01 | ||||||
| Cw*08:03 Cw*07:02 | ||||||
| 7 | A*26:01 A*02:01 | NP66 | 326–340 | TPSGTWLTYHGAIKL | CD4 | 1.48 |
| B*08:01 B*57:01 | 3a2 | 6–20 | RFFTLGSITAQPVKI | CD4 | 0.65 | |
| Cw*07:02 Cw*06:02 | 3a40 | 169–210 | GVKDYVVVHGYFTEV | CD4 | 1.54 | |
| 3a42 | 206–220 | YFTEVYYQLESTQIT | CD4 | 0.43 | ||
| 8 | A*24:07 A*24:07 | 3a2 | 6–20 | RFFTLGSITAQPVKI | CD4 | 0.38 |
| B*15:02 B*58:01 | ||||||
| Cw*03:02 Cw*08:01 | ||||||
| 9 | A*11:01 A*24:07 | NP66 | 326–340 | TPSGTWLTYHGAIKL | CD4 | 5.22 |
| B*15:02 B*40:01 | 3a14 | 66–80 | NKRWQLALYKGFQFI | CD4 | 5.89 | |
| Cw*08:01 Cw*03:04 | ||||||
| 13 | A*31:01 A*02:01 | NP44 | 216–230 | GETALALLLLDRLNQ | CD8 | 2.76 |
| B*13:01 B*40:01 | NP65 | 321–335 | IGMEVTPSGTWLTYH | CD8 | 0.15 | |
| Cw*03:04 Cw*03:04 | ||||||
| 14 | A*31:01 A*02:06 | NP26 | 126–140 | ANKEGIVWVATEGAL | CD4 | 0.49 |
| B*40:01 B*40:01 | NP62 | 306–320 | AQFAPSASAFFGMSR | CD4 | 0.42 | |
| Cw*03:04 Cw*04:01 | ||||||
| 15 | A*33:03 A*11:01 | NP9 | 41–55 | RRPQGLPNNIASWFT | CD8 | 1.54 |
| B*58:01 B*55:02 | NP21 | 101–115 | KMKELSPRWYFYYLG | CD4 | 0.58 | |
| Cw*03:02 Cw*03:03 | 3a2 | 6–20 | RFFTLGSITAQPVKI | CD8 | 0.73 | |
| 16 | A*24:20 A*02:06 | NP53 | 261–275 | QKRTATKQYNVTQAF | CD8 | 0.53 |
| B*15:02 B*15:25 | ||||||
| Cw*08:01 Cw*04:03 |
aa, amino acids.
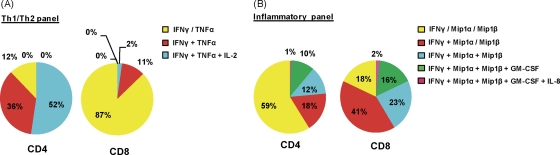
The functionality of SARS-specific CD4+ and CD8+ memory T cells was also analyzed. We used polychromatic flow cytometry to detect the cytokines IFN-γ, TNF-α, IL-2, IL-4, IL-10, and IL-21 (defined as the Th1/Th2 panel) and the cytokines IFN-γ, IL-8, Mip1α, Mip1β, GM-CSF, and IL-17a (defined as the antiviral inflammatory panel). The low frequency of SARS-specific CD4+ and CD8+ T cells in the peripheral blood did not allow us to test their functional profile directly ex vivo, and therefore the cytokine production profile of CD8+ T cells specific for the aa 216 to 230 NP peptide and CD4+ T cells specific for the aa 6 to 20 3a peptide were expanded and tested after 10 days of in vitro expansion. The majority (52%) of SARS-specific memory CD4+ T cells were polyfunctional, producing simultaneously the Th1 cytokines IFN-γ, TNF-α, and IL-2 (Fig. 2A). Instead, a minority of SARS-specific CD4+ cells produce inflammatory chemokines (Mip1α, Mip1β, and GM-CSF) (Fig. 2B), while no CD4+ T cells producing IL-10, IL-4, IL-17a, or IL-21 were detected. SARS-specific CD8+ T cells functional profile was different from CD4+ T cells. Polyfunctional CD8+ T cells producing IFN-γ, TNF-α, Mip1α, and Mip1β dominated the memory SARS-specific CD8+ T cell response, although few CD8+ T cells (<20%) produce IL-2 and TNF-α simultaneously (Fig. 2). Interestingly, the functional profile of SARS-specific memory CD8+ T cells in SARS-recovered individuals is comparable to the one exhibited by virus-specific CD8+ T cells in HIV nonprogressors and in vaccinia virus-vaccinated individuals (1, 19).
Fig. 2.
Cytokine production profile of representative CD4+ and CD8+ T cell responses. Target cells were incubated with 3a2 (aa 6 to 20)-specific CD4+ T cells or NP44 (aa 216 to 225)-specific CD8+ T cells in the presence of brefeldin A and stained with antibodies against Th1/Th2-associated cytokines (A) and inflammatory cytokines (B). Responses are grouped and color-coded according to the number of cytokines produced. The data are summarized in pie charts, with every slice representing the fraction of the total response that consists of CD8+ T cells positive for the cytokines indicated on the right.
Production of SARS-specific TCR-redirected T cells.
The identification of an HLA-B*40:01-restricted SARS NP-specific response has practical interest since HLA-B*40:01 is an HLA-class I allele expressed in 3.0 to 60.0% of the individuals of Asian ethnicities (e.g., Chinese [28.0%], Indians [3.3 to 10.0%], Indonesians [7.2%], Japanese [6.3%], Taiwanese [6.0 to 60.4%], and Filipinos [30.0%]) (9; see also http://allelefrequencies.net/). Importantly, the NP44 (aa 216 to 225) T cell response was not only identified in three of the six HLA-B*40:01-positive individuals analyzed here but was also described in other T cell studies with SARS-CoV-infected individuals (11, 18). For example, in a study by Li et al. (11), 11% of the SARS subjects analyzed gave positive T cell responses against the peptide region containing NP44 (aa 211 to 225). Even though in that study the HLA restriction and fine specificity of the responses were not determined, the data confirmed the association of the NP44 (aa 216 to 225) SARS T cell response with recovery from infection.
Taking into consideration the potential importance of this SARS-specific CD8+ T cells in disease recovery and on the fact that the SARS-CoV NP protein is highly conserved (99%) between different SARS-CoV isolates (22), we sought to clone their α and β T cell receptor chains of NP44 (aa 216 to 225) CD8+ T cells and use them to redirect the specificity of lymphocytes of subjects lacking SARS-specific memory T cells.
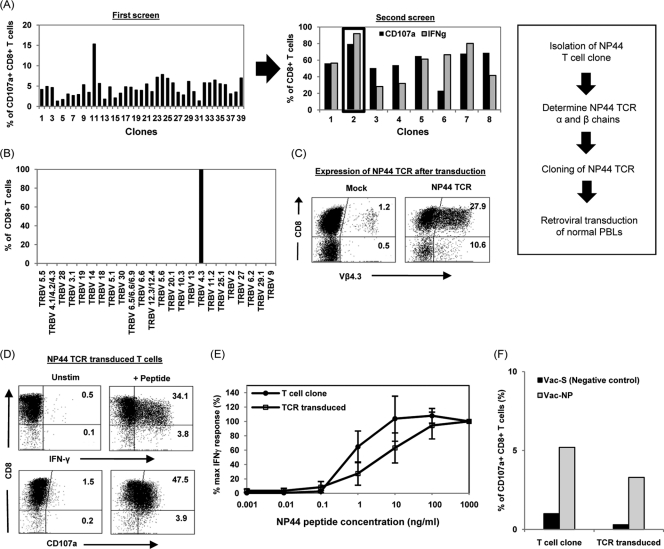
Different CD8+ T cell clones specific for NP44 (aa 216 to 225) were selected through limiting dilution of NP44-specific CD8+ T cells sorted using FACS and expanded from the PBMC of one of the HLA-B*40:01-positive individuals (SARS patient 1) (Fig. 3A). A SARS NP44-specific CD8+ T cell clone (clone 2) was selected based on its high level of responsiveness to NP44 peptide-pulsed target cells (Fig. 3A). The TCR β chain of this T cell clone was then determined using a commercial kit, IO Test Beta Mark (Beckman Coulter) and found to express TCR receptor variable region 4.3 (TRB Vβ4.3) (Fig. 3B). Subsequently, the T cell receptor α and β chains were cloned and further confirmed by sequence analysis of the cDNA clones. The T cell clone expressed a single TCR α chain of Vα4.0/Jα28.0 and a β chain of Vβ4.3/Jβ2.1/Dβ1.0, as determined using IMGT/V-Quest (2; http://www.imgt.org). A recombinant retroviral construct containing the α and β chains of the SARS NP44 TCR was then generated as previously described (8), and the expression and function of the introduced TCR into naive lymphocytes were tested.
Fig. 3.
Production of SARS-specific TCR-redirected T cells. (A) Isolation of NP44-specific T cell clone. NP44-specific CD8+ T cells were purified by FACS and cloned by limiting dilution assay. Clonal populations were expanded and subjected to two rounds of screening for CD107a+ and/or IFN-γ+ CD8+ T cells. (B) Determination of the TCR β chain of the NP44-specific T cell clone by staining for the various TCR β variable regions (TRBV). The percentages of the CD8+ T cells positive for the various TRBV are indicated. Further cloning of the TCR α and β chains of the T cell clone was carried out, and peripheral blood lymphocytes from a healthy donor were transduced with the SARS NP44 TCR retroviral vector. (C) Dot plot of Vβ4.3 expression in mock- or NP44-TCR transduced T cells from a representative healthy donor. The percentages of Vβ4.3+ T cells (CD8+ and CD8−) after gating on the CD3+ fraction of the PBMC are indicated in the upper and lower right quadrants. (D) Dot plot from representative SARS NP44-transduced T cells incubated with target cells loaded with NP44 peptide. The percentages of IFN-γ+ or CD107a+ T cells (CD8+ and CD8−) after gating on the CD3+ fraction of the PBMC are indicated in the upper and lower right quadrants. (E) For functional avidity, NP44-specific T cell clone or SARS NP44 TCR-transduced T cells were incubated with target cells pulsed with the indicated concentrations of NP44 peptide. The percent maximum IFN-γ+ response was plotted against the concentrations of NP44 peptide, whereby the maximum IFN-γ+ response at 1 μg/ml was set as 100%. The data shown are the mean values of three experiments with standard error bars as indicated. (F) For recognition of the endogenous SARS antigen, the SARS NP44-specific T cell clone and NP44 TCR-transduced T cells were incubated with target cells infected with a vaccinia virus construct expressing the SARS spike (S) (negative control) or nucleocapsid (NP) protein at an MOI of 5 and stained for CD107a. The percentages of CD107a+ cells in the total CD8+ population are shown.
Peripheral blood lymphocytes of healthy individuals were transduced and expanded in vitro and stained with anti-Vβ4.3 and anti-CD3. Mock-transduced T cells represent the frequency of endogenous, naturally occurring Vβ4.3+ T cells. After transduction with the SARS NP44 TCR, the T cell population showed a frequency of 38.5% Vβ4.3+ T cells (both CD8+ and CD8− populations) (Fig. 3C). Proper pairing of α and β TCRs was then directly analyzed with functional assays to test the ability of TCR-transduced T cells in recognizing NP44 peptide-pulsed HLA-B*40:01-positive target cells. Figure 3D shows that the introduced TCR endowed SARS specificity to a large proportion of transduced CD8 T cells (34.1% IFN-γ+ and 47.5% CD107a+, respectively).
The functional avidity of SARS-specific TCR-redirected T cells was compared to the one of NP44-specific CD8+ T cell clones isolated from a SARS-recovered patient. Target cells were pulsed with decreasing concentrations of the NP44 aa 216 to 225 peptide, and Fig. 3E shows that the TCR-redirected T cells were activated by HLA-B*40:01 EBV-LCL loaded with 0.1 ng of NP44 peptide/ml, a sensitivity of recognition similar to that seen with the NP44-specific CD8+ T cell clone. These data showed that TCR gene transfer is able to reconstitute SARS-specific T cells with a functional avidity similar to the SARS-memory CD8+ T cells present in subjects recovered from natural infection. Importantly, both TCR-redirected and “natural” SARS-specific CD8+ T cells were able to recognize target cells infected with recombinant vaccinia virus expressing the NP protein (Fig. 3F), indicating that both the CD8+ T cell populations can recognize not only peptide-pulsed cells but also the processed product of endogenously synthesized NP44 SARS antigen.
Functional profiles of natural and TCR-redirected SARS-specific T cells.
T cells producing multiple cytokines (i.e., IFN-γ, TNF-α, Mip1β, and IL-2) are associated with better control of HIV infection and with vaccination efficacy (1, 19), suggesting that T cell polyfunctionality is an important parameter that defines protective antiviral immunity. Furthermore, the CD8+ T cell release of chemokines such as Mip1α and Mip1β has been associated with the recruitment of effector cells and could be important at the peripheral sites of viral infection (20).
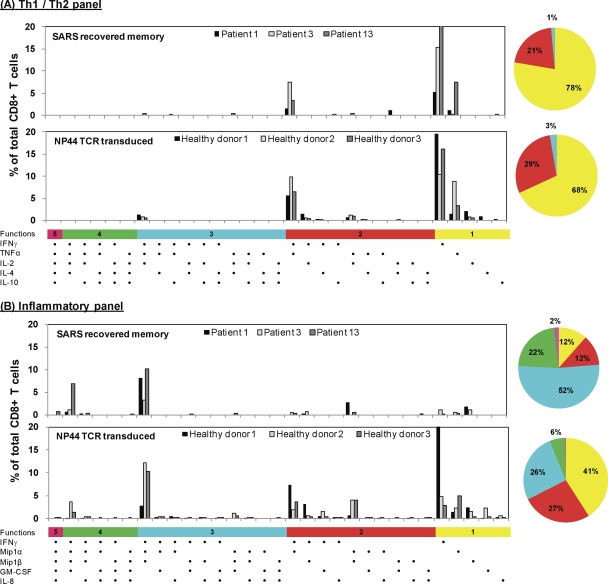
We therefore analyzed whether TCR-redirected T cells display a similar polyfunctional profile compared to SARS-specific memory CD8 T cells. TCR-redirected T cells specific for NP44 (aa 216 to 225) were engineered from PBMC of three different SARS-negative healthy individuals. Polychromatic flow cytometry with the same antibody panels and gating strategy that were used for the analysis of SARS-specific natural CD8+ and CD4+ T cells was utilized. Figure 4 shows (in the lower portion of each panel) that TCR-redirected T cells have a cytokine production profile similar to that of SARS-specific memory CD8+ T cells analyzed in three different patients (upper portion of each panel). IFN-γ, TNF-α, MiP1α, and Mip1β were preferentially produced by TCR redirected T cells, while only very few TCR-redirected T cells produced IL-2, GM-CSF, and IL-8. The polyfunctionality pattern of TCR-redirected T cells showed a decreased frequency of CD8+ T cells able to produce four cytokines in comparison to the natural SARS memory CD8+ T cells, whereas a comparable frequency of CD8+ T cells producing three cytokines (IFN-γ, MiP-1α, and Mip-1β) or two cytokines (IFN-γ and TNF-α) was observed (Fig. 4). Taken together, these data indicate that the NP44 TCR-transduced T cells can achieve functions similar to those obtained directly from a SARS-recovered individual.
Fig. 4.
Cytokine production profile of NP44-specific CD8+ T cells. HLA-B*40:01-positive target cells were pulsed with 1 μg of NP44 peptide/ml or left unpulsed. These cells were then incubated with NP44-specific T cells from a SARS-recovered individual (upper panels) or the SARS NP44 TCR-transduced T cells (lower panels) in the presence of brefeldin A and stained with antibodies against Th1/Th2-associated cytokines (A) or inflammatory cytokines (B). Each graph shows the data for NP44-specific CD8+ T cells from three SARS-recovered individuals or the SARS NP44 TCR-transduced T cells from three healthy donors. The data are expressed as the percentage of the total number of CD8+ T cells expressing the different possible combinations of cytokines. Responses are grouped and color-coded according to the number of cytokines produced. The data are summarized in pie charts, with every slice representing the fraction of the total response that consists of CD8+ T cells positive for the given number of cytokines produced.
In conclusion, we have identified in subjects that recovered from SARS-CoV infection a new immunodominant HLA-B*40:01-restricted SARS-specific CD8+ T cell response. We also demonstrated the feasibility of producing in vitro, from the lymphocytes of healthy subjects, SARS-specific CD8+ T cells that have an avidity and cytokine production profile similar to that of the SARS-specific memory CD8+ T cells present in SARS-recovered individuals. One recent study showed that adoptive transfer of in vitro-cultured SARS-specific CD8+ T cells can significantly enhance survival and virus clearance in SCID mice that were infected with mouse-adapted SARS-CoV (MA15) (32). Moreover, adoptive transfer of virus-specific T cells is becoming an accepted treatment to prevent EBV and HCMV reactivation (4, 7). SARS-specific TCR-redirected T cells thus represent an additional potential prophylactic or therapeutic treatment for this life-threatening infection.
ACKNOWLEDGMENTS
We thank Chan Soh Ha (National University of Singapore, Singapore) for providing the EBV-transformed lymphoblastoid B cell lines. We also thank Falko G. Falkner (Baxter Bioscience, Austria) for sharing Baxter Bioscience's proprietary vaccinia virus expression system and Hans Stauss (Royal Free Hospital and University College Medical School, London, United Kingdom) for the use of the retroviral expression vector.
This study was supported by grants from the Agency for Science Technology and Research (A*STAR), Singapore, and the National Research Foundation (NRF).
Footnotes
Published ahead of print on 3 August 2011.
REFERENCES
- 1. Betts M. R., et al. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 107:4781–4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brochet X., Lefranc M. P., Giudicelli V. 2008. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 36:W503–W508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen J., et al. 2010. Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c mice: CD4+ T cells are important in control of SARS-CoV infection. J. Virol. 84:1289–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Comoli P., et al. 2002. Infusion of autologous Epstein-Barr virus (EBV)-specific cytotoxic T cells for prevention of EBV-related lymphoproliferative disorder in solid organ transplant recipients with evidence of active virus replication. Blood 99:2592–2598 [DOI] [PubMed] [Google Scholar]
- 5. Drosten C., et al. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967–1976 [DOI] [PubMed] [Google Scholar]
- 6. Fan Y. Y., et al. 2009. Characterization of SARS-CoV-specific memory T cells from recovered individuals 4 years after infection. Arch. Virol. 154:1093–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Feuchtinger T., et al. 2010. Adoptive transfer of pp65-specific T cells for the treatment of chemorefractory cytomegalovirus disease or reactivation after haploidentical and matched unrelated stem cell transplantation. Blood 116:4360–4367 [DOI] [PubMed] [Google Scholar]
- 8. Gehring A. J., et al. 2011. Engineering virus-specific T cells that target HBV-infected hepatocytes and hepatocellular carcinoma cell lines. J. Hepatol. 55:103–110 [DOI] [PubMed] [Google Scholar]
- 9. Gonzalez-Galarza F. F., Christmas S., Middleton D., Jones A. R. 2011. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res. 39:D913–D919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ksiazek T. G., et al. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953–1966 [DOI] [PubMed] [Google Scholar]
- 11. Li C. K., et al. 2008. T cell responses to whole SARS coronavirus in humans. J. Immunol. 181:5490–5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li T., et al. 2006. Long-term persistence of robust antibody and cytotoxic T cell responses in recovered patients infected with SARS coronavirus. PLoS One 1:e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu J., et al. 2010. The membrane protein of severe acute respiratory syndrome coronavirus acts as a dominant immunogen revealed by a clustering region of novel functionally and structurally defined cytotoxic T-lymphocyte epitopes. J. Infect. Dis. 202:1171–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu S. J., et al. 2006. Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine 24:3100–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lv Y., Ruan Z., Wang L., Ni B., Wu Y. 2009. Identification of a novel conserved HLA-A*0201-restricted epitope from the spike protein of SARS-CoV. BMC Immunol. 10:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marra M. A., et al. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399–1404 [DOI] [PubMed] [Google Scholar]
- 17. Peiris J. S., Yuen K. Y., Osterhaus A. D., Stohr K. 2003. The severe acute respiratory syndrome. N. Engl. J. Med. 349:2431–2441 [DOI] [PubMed] [Google Scholar]
- 18. Peng H., et al. 2006. Long-lived memory T lymphocyte responses against SARS coronavirus nucleocapsid protein in SARS-recovered patients. Virology 351:466–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Precopio M. L., et al. 2007. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8+ T cell responses. J. Exp. Med. 204:1405–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Price D. A., Klenerman P., Booth B. L., Phillips R. E., Sewell A. K. 1999. Cytotoxic T lymphocytes, chemokines, and antiviral immunity. Immunol. Today 20:212–216 [DOI] [PubMed] [Google Scholar]
- 21. Ray S., et al. 2010. MHC-I-restricted melanoma antigen specific TCR-engineered human CD4+ T cells exhibit multifunctional effector and helper responses, in vitro. Clin. Immunol. 136:338–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rota P. A., et al. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394–1399 [DOI] [PubMed] [Google Scholar]
- 23. Sayer D., et al. 2001. HLA-DRB1 DNA sequencing based typing: an approach suitable for high-throughput typing including unrelated bone marrow registry donors. Tissue Antigens 57:46–54 [DOI] [PubMed] [Google Scholar]
- 24. Tan A. T., et al. 2008. Host ethnicity and virus genotype shape the hepatitis B virus-specific T-cell repertoire. J. Virol. 82:10986–10997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tsao Y. P., et al. 2006. HLA-A*0201 T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus nucleocapsid and spike proteins. Biochem. Biophys. Res. Commun. 344:63–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vita R., et al. 2010. The immune epitope database 2.0. Nucleic Acids Res. 38:D854–D862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang B., et al. 2004. Identification of an HLA-A*0201-restricted CD8+ T-cell epitope SSp-1 of SARS-CoV spike protein. Blood 104:200–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang Y. D., et al. 2004. T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus spike protein elicit a specific T-cell immune response in patients who recover from SARS. J. Virol. 78:5612–5618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weiss S. R., Navas-Martin S. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 69:635–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. World Health Organization 2003. Case definitions for surveillance of severe acute respiratory syndrome (SARS). World Health Organization, Geneva, Switzerland: http://www.who.int/csr/sars/casedefinition/en/ [Google Scholar]
- 31. Zhang Y., et al. 2010. Transduction of human T cells with a novel T-cell receptor confers anti-HCV reactivity. PLoS Pathog. 6:e1001018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhao J., Perlman S. 2010. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J. Virol. 84:9318–9325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou M., et al. 2006. Screening and identification of severe acute respiratory syndrome-associated coronavirus-specific CTL epitopes. J. Immunol. 177:2138–2145 [DOI] [PubMed] [Google Scholar]