Abstract
The molecular epidemiology of HIV-1 is constantly changing, mainly as a result of human migratory flows and the high adaptive ability of the virus. In recent years, Spain has become one of Europe's main destinations for immigrants and one of the western European countries with the highest rates of HIV-positive patients. Using a phylogeographic approach, we have analyzed the relationship between HIV-1 variants detected in immigrant and native populations of the urban area of Madrid. Our project was based on two coincidental facts. First, resistance tests were extended to naïve and newly diagnosed patients, and second, the Spanish government legislated the provision of legal status to many immigrants. This allowed us to obtain a large data set (n = 2,792) from 11 Madrid hospitals of viral pol sequences from the two populations, and with this unique material, we explored the impact of immigration in the epidemiological trends of HIV-1 variants circulating in the largest Spanish city. The prevalence of infections by non-B HIV-1 variants in the studied cohort was 9%, rising to 25% among native Spanish patients. Multiple transmission events involving different lineages and subsubtypes were observed in all the subtypes and recombinant forms studied. Our results also revealed strong social clustering among the most recent immigrant groups, such as Russians and Romanians, but not in those groups who have lived in Madrid for many years. Additionally, we document for the first time the presence of CRF47_BF and CRF38_BF in Europe, and a new BG recombinant form found in Spaniards and Africans is tentatively proposed. These results suggest that the HIV-1 epidemic will evolve toward a more complex epidemiological landscape.
INTRODUCTION
Molecular epidemiology analyses of the nucleotide sequences of viral genomes have led to the classification of HIV-1 into four groups, M, N, O, and P (40, 50). The M group, the main culprit in the global HIV-1 pandemic, is further divided into 9 subtypes (A to D, F to H, J, and K). In those places where the prevalence of infections by multiple HIV-1 variants is high, the frequency of coinfections or superinfections with different HIV-1 subtypes should also be high (51), facilitating the appearance of a myriad of recombinant forms (circulating recombinant forms [CRFs] and unique recombinant forms [URFs]) (http://www.hiv.lanl.gov/content/sequence/HIV/CRFs/CRFs.html). The sub-Saharan African region is particularly interesting for understanding the evolutionary dynamics of HIV because it is where the virus originated and disseminated from and the only area where it has been circulating for more than half a century (25, 55). Moreover, sub-Saharan human populations have the highest rates of HIV infection, which, along with viral characteristics, such as high HIV-1 replication, mutation, and recombination rates (27, 31, 37), have given rise to high levels of viral diversification (53). The last Joint United Nations Program on HIV/AIDS (UNAIDS) report (http://www.unaids.org/documents/20101123_GlobalReport_em.pdf) suggests that a continuous increase in viral diversification should be expected in the future, at least in Africa (42, 56). According to this suggestion, the diversification process should also be observed in the destination countries of African immigrants.
At the beginning of the AIDS pandemic, some African HIV-1 variants were introduced into previously unexposed human populations out of Africa. This led to the local dissemination of a few variants in particular regions (founder effect), such as subtype F in Romania (2), CRF01_AE and subtype B in Thailand (13), or subtype B in many European countries (14), North America, and Japan (20). These ancestral strains have continued evolving in a diversification process leading to the emergence of particular subpopulations, which has allowed the association of particular clusters with specific geographical regions (6) and the tracing of viral migrations using phylogeographic approaches (35). Therefore, the population genetics structure of HIV-1 could change in countries with high rates of HIV-infected immigrants, leading to complex local epidemiological landscapes (39). Such a complex scenario can be expected in Spain, the European country with the highest immigration rates in the last decade. Immigrants currently represent ∼10% of the Spanish population (21), and their main sources are Africa (the continent with the highest diversification rate), South America (with a high rate of BF recombinant forms), and Eastern Europe (with a high prevalence of subtypes A and F) (52), whereas native Spaniards (http://www.ecdc.europa.eu/en/publications/publications/0812_SUR_HIV_AIDS_Surveillance _in_Europe.pdf) were traditionally infected by HIV-1 subtype B (23, 24). Moreover, Spain is also one of the main tourist destinations in the world, receiving around 90 million travelers each year.
Since 2005, two important events have allowed us to gain access to a large number of HIV-1 sequences in native and immigrant populations and to perform phylogeographical analyses. First, genotypic resistance tests for HIV-1 based on sequencing were extended to naïve and newly diagnosed patients as part of standard components of clinical care, following international guidelines (45). Second, the Spanish government passed a law to provide legal status to a large number of immigrants living in Spain. During the 2005 to 2007 period, the number of legal immigrants entitled to use the public health system increased 300% compared to the 2002 to 2003 period, and Madrid was the Spanish city with the highest proportion of immigrants (∼20% [http://extranjeros.mtin.es]). These two events have permitted us to estimate the viral diversity present in the HIV-infected population living in Madrid and to establish the current molecular epidemiological status of HIV-1 infection.
International guidelines recommend carrying out molecular epidemiology studies for the accurate definition of vulnerable groups and the identification of clusters of dissemination in the general population as an important tool for combating HIV/AIDS (http://unaids.org/en/media/unaids/contentassets/documents/unaidspublication/2010/jc2034_unaids_strategy_en.pdf; UNAIDS strategy 2011–2015). Consequently, our aim was to apply molecular phylogenetic methods to analyze HIV-1 sequences obtained in routine genotypic tests of resistance in order to estimate the impact of immigration on the molecular epidemiology landscape of HIV-1 in Madrid, a large urban receptor area for many immigrants and travelers, to detect the entry of new HIV-1 variants into this population and to relate the emergence and spread of these new variants with social or risky behavior groups. Two particular features of Spain, high immigration and tourism rates, suggest that the country must be one of the hot spots in the research into the molecular epidemiology of HIV. To our knowledge, this is the first study on the epidemiology of HIV in Spain using a phylogeographic approach. Moreover, this approach provides additional information on the prevalence and epidemiological trends of HIV-1 variants over time based on the general population, helping to define the dynamics of transmission. These findings can be used as a valuable asset for public health officials and in AIDS prevention programs.
MATERIALS AND METHODS
Study population and work design.
pol gene sequence data from 2,792 HIV-infected patients were collected between 2005 and 2007 from clinical and routine drug resistance analyses done in 10 general hospitals of the Public Health System of Madrid (SERMAS). Each hospital is in charge of the diagnosis and monitoring of patients corresponding to its geographically delimited health care area in Madrid (Table 1). Sequences were also recovered from patients studied in an area-independent hospital (Hospital Carlos III, dependent on the Ministry of Health of Spain) where patients from the whole city of Madrid and people not registered in the health system were attended.
Table 1.
Distribution of HIV sequences from different health care areas of Madrid included in the study
| Health care area | Hospitala | No. of sequencesb | Known HIV+ population | Method of sequencing |
|---|---|---|---|---|
| 1 | HGM | 407 | 1,800 | Trugened |
| 2 | HP | 87 | NDf | Trugene |
| 3 | HPA | 36 | 750 | Trugene |
| 4 | HRyC | 298 | 1,950 | Viroseqe |
| 5 | HLP | 528 | 1,700 | Trugene |
| 6 | HPH | 24 | ND | Trugene |
| 7 | FJD | 329 | 2,700 | Trugene |
| HSC | 317 | 1,500 | Trugene | |
| 10 | HG | 67 | 600 | Trugene |
| 12 | HDO | 434 | 2,050 | Viroseq |
| –c | HCIII | 265 | Trugene/Viroseq |
HGM, Hospital Gregorio Marañón; HP, Hospital Princesa; HPA, Hospital Principe de Asturias; HRyC, Hospital Ramón y Cajal; HLP, Hospital La Paz; HPH, Hospital Puerta de Hierro; FJD, Fundación Jiménez-Díaz; HSC, Hospital Clínico San Carlos; HG, Hospital General Getafe; HDO, Hospital Doce de Octubre; HCIII, Hospital Carlos III.
The numbers of sequences include multiple sequences from the same patient (total, 2,792 sequences).
−, no health care area assigned.
Trugene HIV Genotyping System; Siemens Medical Solutions Diagnostics, Tarrytown, NY.
ViroSeq HIV Genotyping System; Abbott Molecular Inc., Des Plaines, IL.
ND, not determined.
Sequence lengths ranged between 918 and 1,302 nucleotides (nt), depending on the commercial procedure used for routine genotypic drug resistance testing (Trugene HIV Genotyping System [Siemens Medical Solutions Diagnostics, Tarrytown, NY] or ViroSeq HIV Genotyping System [Abbott Molecular Inc., Des Plaines, IL], respectively). All sequences were trimmed to 918 nt for analysis. This region included the complete fragment coding for the protease (PR) and a partial fragment coding for the reverse transcriptase (RT). Demographic data collected from those patients carrying HIV-1 non-B subtypes (and recombinant forms) were requested, including country of birth, antiretroviral treatment experience, and risk group.
Phylogenetic reconstructions.
Nucleotide sequences were virtually translated and aligned using the ClustalW algorithm implemented in MEGA, thus maintaining the correct phase of the reading frame. In the first approach for HIV-1 subtype characterization, phylogenetic trees were constructed using the neighbor-joining (NJ) method (44) and HIV-1 reference pol sequences available in the Los Alamos HIV Sequence Database (http://www.hiv.lanl.gov). A total of 30 reference sequences from HIV group M (15 reference sequences from the A to D subtypes, 11 from the F to H subtypes, and 4 from the J and K subtypes) and at least 1 sequence of the 47 CRFs with known sequences (http://www.hiv.lanl.gov/content/sequence/HIV/CRFs/CRFs.html) were included in the subtype characterization. One group N sequence was used as the outgroup. Evolutionary distances were estimated with the maximum-likelihood (ML) distance, and 1,000 bootstrap replicates were performed to evaluate the support for the inferred clades.
Detection of intersubtype recombination events.
Initially, sequences not clustering with any known subtype or CRF with bootstrap support of >70% were defined as unclassified (U) and were selected to further determine possible recombination events. Unclassified sequences were analyzed using Recombination Detection Program 3beta27 (RDP3beta27) (19), identifying the subtypes involved in eventual recombination events and defining the hypothetical recombination breakpoints within reference sequences. To further confirm the putative recombination events detected in U sequences, new phylogenetic analyses were performed using the fragments assigned to different subtypes according to the proposed breakpoint position(s). The topologies obtained with each fragment were compared with Shimodaira-Hasegawa (SH), expected likelihood weight (ELW), and Kishino-Hasegawa (KH) tests using the TREE-PUZZLE 5.2 program, and differences were considered significant if α was <5%. In the positive cases, the recombinant sequences were redefined as URFs; otherwise, they were denoted U. URFs found in at least three epidemiologically unrelated patients and showing apparently identical parental subtypes and breakpoints were candidates to be considered new CRFs.
Inference of phylogeographic relationships among HIV-1 non-B variants circulating in Madrid.
A phylogeographic approach was used to infer groups of closely related pol sequences sharing specific geographic associations (6, 15) with all sequences previously classified as non-B variants (non-B subtypes, CRFs, and URFs). For this purpose, sequences of a specific subtype or CRF were analyzed separately with a set of reference sequences of the same genotype (subtype or CRF) downloaded from the Los Alamos HIV Sequence Database (http:www.hiv.lanl.gov), which are annotated by country of origin according to the classification used by UNAIDS (20). This reference data set included 1,822 sequences with the following distribution: 97 subtype A (including all subsubtypes), 470 subtype C, 60 subtype D, 312 subtype F, 143 subtype G, 4 subtype H, and, among recombinant forms, 241 CRF01_AE, 57 CRF02_AG, 68 CRFs B and G, 337 CRFs B and F, and 33 other CRFs. Additionally, 54 subtype C sequences from the United Kingdom database, kindly provided by Robert J. Gifford (10), were included in the analyses.
Phylogenetic trees were reconstructed by ML with PhyML 3.0 (17) using the general time reversible plus proportion of invariable sites plus gamma distribution parameter (GTR) plus I plus G evolutionary model selected with jModeltest 0.1 (41) and a BIONJ starting tree. Heuristic tree searches under the ML optimality criterion were performed using the nearest neighbor interchange (NNI) branch-swapping algorithm. The approximate likelihood ratio test (aLRT) based on a Shimodaira-Hasegawa-like procedure was used as a statistical test to calculate branch support (1). Only aLRT support values of >90% were considered statistically significant and displayed at the tree nodes. We considered that if a majority of sequences could be grouped in a single and well-supported cluster (monophyletic clade), they would indicate with confidence a single-point introduction in the studied population. In this case, if most patients with pol sequences in the same cluster were Spanish natives, it would suggest a local transmission chain in Madrid. In contrast, the detection of polyphyletic clades would imply multiple HIV-1 introductions, strongly suggesting separate origins of the infections in other regions or countries.
Nucleotide sequence accession numbers.
The non-B sequences described in this work have been deposited in GenBank with the following accession numbers: EU255306 to EU255310, EU255328, EU255331, EU255342, EU255345, EU255346, EU255349, EU255354, EU255355, EU255358, EU255360, EU255363, EU255373 to EU255391, EU255415, EU255442, EU255444, EU255456, EU255465, EU255467, EU255468, EU255477, EU255489, EU255503, EU255507, EU255509, EU255510, EU255513, EU255514, EU255518, EU255522 to EU255531, EU255540, EU255542, EU255544, EU255548, EU255553, EU362926, EU545188, and JF929015 to JF929179.
RESULTS
Surveillance and geographical distribution of non-B HIV-1 subtypes in the metropolitan area of Madrid.
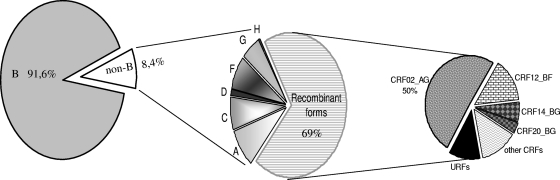
During the studied period, 2005 to 2007, a total of 2,802 HIV-1 pol sequences were available from the participating hospitals. Exploratory phylogenetic analyses identified 22 identical sequences, all but 2 of which (a mother-child transmission) corresponded to duplicates from the same patient. Therefore, 2,792 sequences were finally included in the phylogenetic analyses. The study population represented 10 to 13% of all HIV-1-infected individuals living in Madrid at the end of 2007, according to the Madrid Public Healthcare System (http://www.madrid.org/cs). According to the NJ tree (http://www.hrc.es/images/FigS1.jpg), 2,558 of the 2,792 sequences (91.6%) belonged to subtype B (Fig. 1). Among the 234 sequences (8.4%) classified as non-B variants, 31% were pure non-B subtypes and 69% were recombinant forms, including 14 sequences initially identified as U. Seven of the nine pure subtypes and 17 of the 47 known CRFs were present in the sampled population. The frequency (f) of non-B variants in the non-area hospital (f = 69/265 = 0.26) was higher than in the area-based hospitals as a whole (f = 165/2527 = 0.065). However, genetic diversity (D = 1 − Σf 2, a measure of variability that takes into account the frequencies of the variants) of non-B variants in the non-area hospital (D = 0.816) was not significantly different from that of the area-based hospitals together (D = 0.828). A high prevalence of non-B variants was detected in the northeast region, followed by the southeast and western regions (χ2 = 6.40; P = 0.01) (Fig. 2 a). On the other hand, several variants (such as subtypes A, C, and F, or recombinant forms, such as CRF02_AG, CRF12_BF, and the BG complex) were widely distributed throughout the metropolitan area (Fig. 2b).
Fig. 1.
Distribution of HIV-1 subtypes and recombinants in Madrid. Other CRFs are CRF01_AE, CRF06_cpx, CRF10_CD, CRF11_cpx, CRF19_cpx, CRF22_01A1, CRF24_BG, CRF25_cpx, CRF29_BF, CRF30_cpx, CRF38_BF, CRF44_BF, and CRF47_BF. URFs are recombinants AG, BA, BC, BF, BG, 02C, 02G, 01U, GU, and U.
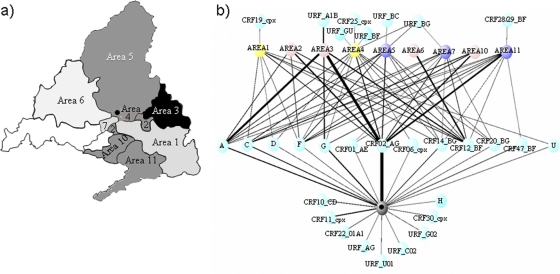
Fig. 2.
(a) Incidence of non-B subtype HIV-1 in Madrid. The different shadings indicate the different incidences of non-B variants (subtypes and recombinant forms): black, >20%; gradient of gray, between 5 and 10%; and white, unknown. The black dot corresponds to the area-independent hospital. (b) Geographical distribution of each non-B subtype and recombinant form, based on the distribution of health care areas in Madrid. The different colors indicate the different numbers of patients: gray, >50; blue, 30 to 50; yellow, 20 to 30; pink, <20. The thickness of the lines is proportional to the number of patients.
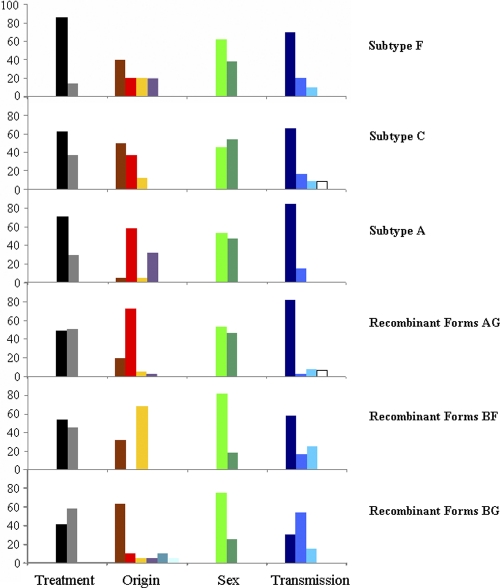
The most frequent epidemiological profile for patients carrying HIV-1 non-B subtypes corresponded to an African origin (47%) and heterosexual behavior (49%). The most relevant features observed in the subtypes analyzed were as follows: (i) there was a high proportion of heterosexual Africans carrying subtype A and CRF02_AG, (ii) BG recombinant forms were more common in Spanish intravenous drug users (IDUs), and (iii) BF recombinant forms were found only in South Americans and Spaniards (Fig. 3). On the other hand, approximately 25% of non-B variants were found in Spaniards. Remarkably, the proportion of naïve versus nonnaïve patients carrying non-B variants was significantly higher in Spaniards than in foreigners (χ2 = 9.58; P = 0.002), suggesting that non-B HIV-1 infections among Spaniards could be more recent than among foreigners.
Fig. 3.
Epidemiological data for the most prevalent non-B variants found. Four large modules are presented: treatment (the black bars correspond to naïve patients), origin (brown, Spaniards; red, Africans; yellow, South Americans; pink, eastern Europeans; orange, western Europeans; magenta, Caribbean natives), sex (light green, men; dark green, women), and transmission route (dark blue, heterosexual; blue, intravenous drugs users; light blue, MSM; white, mother to child).
Inferring the mobility of HIV-1 non-B variants. (i) Pure subtypes.
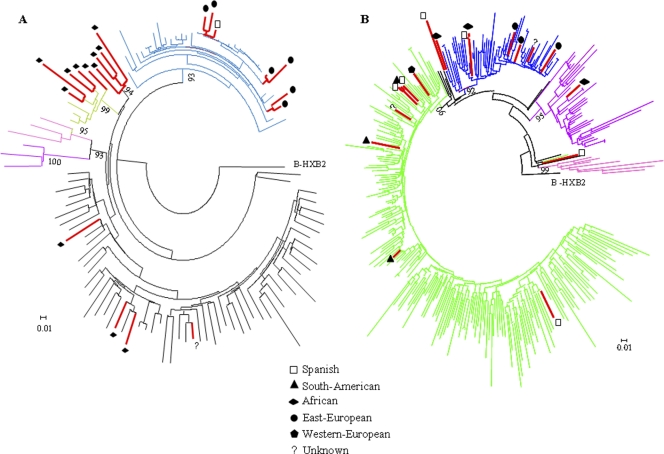
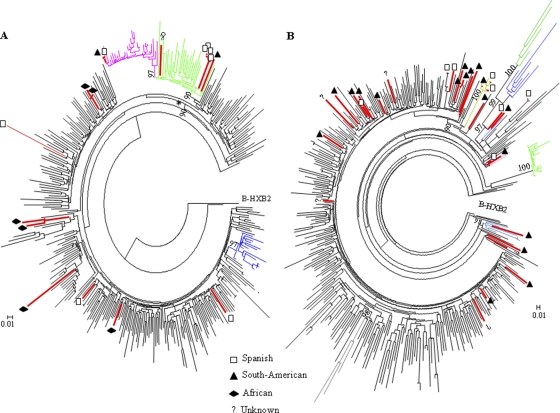
An ML phylogenetic tree was obtained with the 20 subtype A sequences found in this study and 97 reference sequences, which included all subsubtypes described (A1 to A4) and all lineages proposed (African and Former Soviet Republics [FSR]). Eleven sequences grouped with subsubtype A1: seven sequences found in eastern European patients (only one in a Spaniard) clustered with the FSR lineage, whereas the remaining four sequences grouped with the African lineage and were found in Africans (Fig. 4 A). Five sequences found in people from two western African regions (Cameroon, Equatorial Guinea, and Guinea) corresponded to subsubtype A3, which was initially described in Senegalese patients. Four sequences from patients from Mali and Equatorial Guinea clustered in a separate and as yet uncharacterized group. An almost perfect match between the geographical area of origin (eastern Europe and West Africa) and the prevalence of subtype A variants was observed (18/20 cases). Moreover, these variants have low prevalence in Spain, which suggests that those patients were probably infected in their home countries.
Fig. 4.
Maximum-likelihood (GTR plus I plus G; aLR ≥ 90%) trees inferred for subtype A (A) and subtype F (B) using reference sequences from different origins. (A) The black and blue branches correspond to African and FSR lineages, respectively, belonging to subsubtype A1. The violet, light-green, and pink branches correspond to the A2, A3, and A4 subsubtypes, respectively. The red branches correspond to sequences found circulating in Madrid from 2005 to 2007. (B) The green and blue lines correspond to South American and Angolan-Romanian lineages, respectively, belonging to subsubtype F1. The black branches correspond to non-Angolan African. The violet and pink branches correspond to subsubtype F2 and subtype, respectively. The red branches correspond to sequences found circulating in Madrid from 2005 to 2007.
Although subtype F represents <1% of HIV-1 infections worldwide, it was the second most prevalent subtype in our sampling. All but one sequence previously defined as subtype F corresponded to subsubtype F1 (17/18 sequences). Among these, eight clustered with the South American lineage and eight others grouped with the Angolan-Romanian lineage (Fig. 4B). This lineage was defined as a monophyletic clade (16), and all subtype F sequences found in patients from these countries grouped in this clade, suggesting that these patients had been infected in their home countries. Five sequences from Spanish patients grouped in the Angolan-Romanian (2 cases) and South American (3 cases) lineages. Only one sequence, found in a heterosexual patient from Equatorial Guinea, was related to subsubtype F2.
Seventeen sequences were initially identified as subtype C, which was the third most prevalent pure subtype detected in our sampling. Six of 17 subtype C sequences were closely related to sequences of the African lineage (Fig. 5 A), but the majority (11/17) clustered with the East African/South American lineage, and most of them were assigned to the United Kingdom/South American clade (8/11 sequences). Moreover, a strain found in Spanish men who have sex with men (MSM) clustered with sequences assigned to the recently described “UK-MSM” clade (10). No sequence closely related to the Indian clade (26) was found.
Fig. 5.
Maximum-likelihood tree (GTR plus I plus G; aLR ≥ 90%) inferred for subtype C (A) and for recombinant forms BF (B) using known sequences from different countries downloaded from the Los Alamos database. (A) The black branches correspond to African sequences; the blue branches show the Indian clade, and the asterisk indicates the East African/South American lineage. Within this lineage, the green branches correspond to South American sequences, and the fuchsia branches correspond to the UK-MSM clade, redefined as the United Kingdom/South American clade by de Oliveira et al. (10). The red branches correspond to sequences found in Madrid from 2005 to 2007. (B) Recombinant forms BF (CRF12_BF and CRF17_BF, black; CRF28_BF and CRF29_BF, steel blue; CRF38_BF, pink; CRF39_BF, gray; CRF40_BF, green; CRF42_BF, light green; CRF44_BF, purple; CRF46_BF, blue; and CRF47_BF, burgundy). The red branches correspond to sequences found in Madrid from 2005 to 2007.
(ii) Recombinant forms.
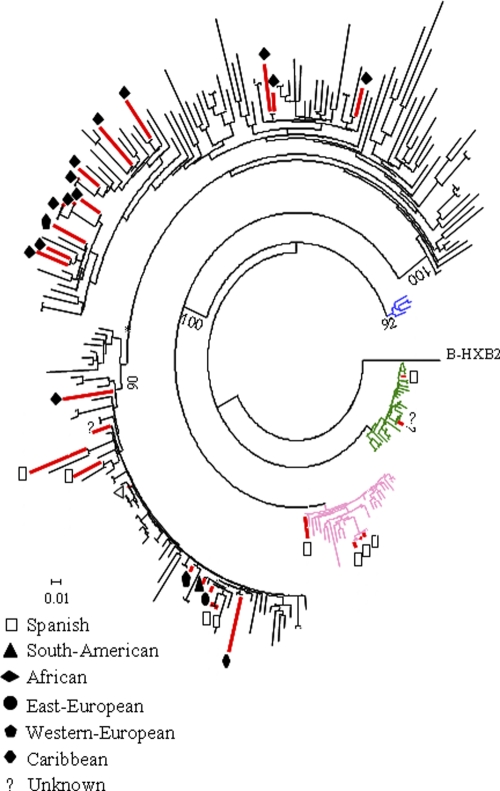
In our epidemiological survey, ∼70% of non-B subtypes were classified as recombinant forms, representing ∼6% of the total sample. The most prevalent recombinant forms found in this study (36% of CRF02_AG and 10% of CRF12_BF) corresponded to the most prevalent recombinant forms in the most common regions of origin of immigrants to Madrid, namely, West Africa and South America (18). The third most prevalent recombinant form was the complex recombinant BG (CRF14_BG, representing ∼6%), probably originating in the Iberian Peninsula (9).
Five of the 11 BF recombinant forms described so far were found circulating in Madrid (CRF12_BF, CRF28_BF/CRF29_BF; CRF38_BF, CRF44_BF, and CRF47_BF), suggesting a complex epidemiological setting of BF forms, although for two types, CRF28_BF/CRF29_BF and CRF44_BF, only one representative was found. The most prevalent were CRF12_BF isolates (77%) (Fig. 5B). The recombinant forms CRF38_BF, CRF44_BF, and CRF47_BF found in this surveillance were described as new CRFs during 2009 and 2010. The patient infected with CRF44_BF was born in Chile, where this recombinant form was recently described (8). CRF38_BF, initially confined to Uruguay (43), was found in two patients from Argentina and Spain. This is the first report describing CRF38_BF among Spaniards. The recombinant form CRF47_BF originated in Spain (12), and in our surveillance program, it was found in a Spaniard and two South American individuals (from Brazil and Peru), although at least one of them was infected in Spain, suggesting that the transmission route could also be from Spain to South America. Although 11 sequences showed the closest phylogenetic relationship with CRF14_BG, they could not be distinguished from G sequences circulating in Spain and Portugal (9) in the analyzed region of the pol gene (Fig. 6), as both belonged to the same monophyletic cluster. Consequently, these sequences have been denoted G/CRF14_BG. The sequences belonging to the G/CRF14_BG cluster were found in all the immigrant groups differentiated in this surveillance study, suggesting a wide spread of the variants, whereas all the sequences of the G cluster (Fig. 6) were found in patients from Africa. Moreover, two types of recombinant forms, CRF20_BG and CRF24_BG, were detected among Spaniards, although they were originally described in Caribbean patients and seldom outside Cuba (46).
Fig. 6.
Maximum-likelihood (GTR plus I plus G; aLR > 90%) tree inferred for subtype G and recombinant forms BG using reference sequences from different countries. The pink branches correspond to CRF20_BG, the blue branches to CRF23_BG, the green branches to CRF24_BG, and the black branches to subtype G. Indiscriminate G/CRF14_BG sequences in the analyzed fragment of the pol gene are identified with asterisks. The red branches correspond to sequences found circulating in Madrid from 2005 to 2007.
(iii) Unique recombinant forms.
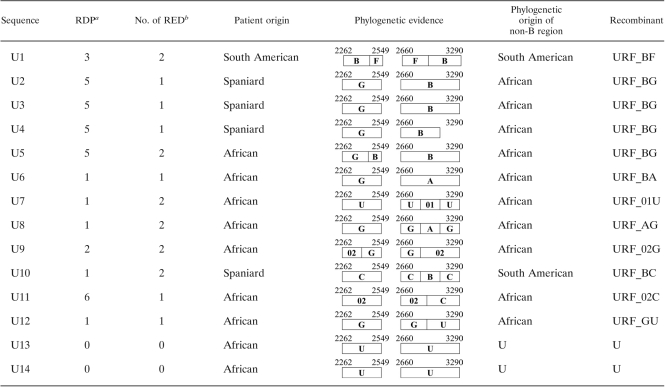
Only 14 sequences (<5%) were identified as URFs in our study, without clear relationship to any previously reported lineage and thus representing novel HIV-1 diversity (Table 2 ). Four sequences showed recombination events not previously described between B and G subtypes (URF_BG). According to the analysis with RDP, three of these sequences had the same breakpoints and thus could tentatively be considered a new CRF. Moreover, four sequences (URF_AB, URF_AG, URF_BC, and URF_BF) were described as recombinant forms between two pure subtypes, and two sequences involved pure subtype G or C and the recombinant form CRF02_AG (URF_02G and URF_02C). Finally, four sequences derived from African immigrants from Equatorial Guinea and Angola had fragments that could not be classified with any HIV-1 type or subtype (URF_01U, URF_GU, and 2 U sequences). In summary, many new recombinant forms from Africa and URF_BF from South America have recently arrived in Madrid. Nevertheless, transmission events from native people to immigrants might also have occurred, as represented by the URF_BG variant described in two Spanish and one African IDUs.
Table 2.
Unique recombinant forms detected circulating in Madrid
Number of recombination analysis methods capable of detecting recombination. RDP, Recombination Detection Program.
RED, recombination events detected.
DISCUSSION
Many of the studies describing the epidemiology of HIV-1 in a local area have been performed using too few sequences or a poorly representative sampling (11, 24, 36, 54). The current availability of large numbers of sequences from any particular region is revealing more epidemiological information. Phylogeographic approaches using a sufficiently large number of sequences have provided a clearer picture of the patterns of viral transmission and have helped to detect vulnerable groups in which new variants could be disseminated (4, 6, 10, 15, 30, 35, 48). These studies are especially interesting in regions with high migration rates because they might reveal how the local epidemiology of HIV is altered by immigration (39). Spain is especially appropriate for the study of these epidemiological changes because it is one of the countries with the highest rates of immigration (52): approximately 40% and 18% of all South Americans and Africans, respectively, now residing in Europe live in Spain, and many immigrants from eastern European countries, especially Romania, have also arrived recently in Spain (21). On the other hand, Spain and Greece attract many central and northern European travelers and tourists who could become sources for the spread of new HIV-1 variants back into their home countries (35). Therefore, Spain constitutes an excellent model for evaluating the impact of the inflow of immigrants from geographical areas with a high prevalence of non-B HIV-1 subtypes into a region previously dominated by subtype B. Additionally, changes in the molecular epidemiology of HIV-1 in Spain could also have effects in other European countries.
Despite the recent increase in immigration rates, the incidence of non-B variants circulating in Madrid (∼9%) during the period 2005 to 2007 has not increased in comparison with previously published data (32). Nevertheless, our phylogeographic analysis has revealed a very complex epidemiological scenario in all the subtypes studied in detail. A second interesting observation was that approximately 25% of non-B subtypes were found in native Spaniards. It is remarkable that the proportion of naïve versus nonnaïve patients was significantly higher in Spanish than in foreign patients infected with non-B variants (χ2 = 9.58; P = 0.002). This result suggests that many infections of Spaniards with non-B HIV-1 have occurred recently. The best example is the spread of subtype C. Among all patients carrying HIV-1 subtype C, 50% were Spaniards, whereas in previous studies, this subtype was found mainly in immigrants (22, 24), suggesting that, similarly to other countries, such as the United Kingdom or South America, the incidence of subtype C in Spain will increase in the future (47, 49).
The detailed analysis of non-B variants of HIV-1 has revealed strong social clustering (57) in the most recent immigrant groups coming to Spain. For instance, sequences from the subsubtype A1 FRS lineage were practically found only in immigrants from Russia and the Ukraine (Fig. 4A). However, some cases of transmission between natives and foreigners belonging to immigrant groups who have been living in Madrid for many years could also be identified. For instance, transmission events in subtype F1 between native people and South Americans (Fig. 4B) or independent introductions of subtype C from South Americans and Africans to Spaniards (Fig. 5A) were detected. South Americans share linguistic and cultural links with the Spanish population, making them very good candidates among immigrant groups for spreading imported variants. The lack of historical and cultural interchanges with other immigrant groups, most notably sub-Saharan Africans and eastern Europeans, although it does not represent an absolute barrier to the exchange of HIV-1 variants with the native population, certainly limits it. This, along with a more recent and rapidly increasing flow of these immigrant groups, can explain the relative paucity of transmission events involving native Spaniards and African and eastern European natives. Curiously, no transmission events were detected between South Americans and Africans, although other possible explanations, such as host restriction, cannot be excluded (33).
The frequencies of non-B subtypes and CRFs found in this study were more related to migratory flows than to their worldwide prevalence. Among the pure subtypes most frequently found in this study, subtype A sequences were closely related to sequences from African and eastern European countries, subtype F sequences to those from Romania, and subtype C to the United Kingdom/South American clade. Among the most prevalent recombinant forms circulating in Madrid, CRF02_AG, the CRF_BF complex, and the CRF_BG complex were more closely related to sequences from Africa, South America, and Spain, respectively. All CRF20_BG and CRF24_BG variants were found in Spanish individuals, supporting the idea that BG recombinant forms are amply distributed in Spain (9), although these variants originated in Caribbean countries (46) and have barely been described outside Cuba, with only one case of CRF24_BG having been described in Europe (7). On the other hand, some of the most recently described forms in South America (2009 and 2010) were already circulating in Madrid in the period 2005 to 2007, such as CRF44_BF (described for the first time in Spain) and CRF47_BF (12), indicating that BF recombinant forms originating in South America could be detected in Spaniards shortly after and suggesting that Spain could be an intermediate stage in their dissemination to other European countries.
Based on the origin of HIV-infected patients with URFs found in this study, an increase in the arrival of new variants in Spain from Africa and South America could be predicted (Table 2). Unclassified sequences (recombinant and nonrecombinant forms, such as the suggested new cluster within subtype A) were always found linked to Africa, where a clear distinction between subtypes is lost (4, 28). This could reflect the huge potential for new HIV variants appearing and spreading in the future (29). For instance, a new possible subsubtype A close to subsubtype A3 could be suggested in individuals from Equatorial Guinea and Mali living in Madrid (Fig. 4A). Moreover, a new tentative recombinant form between subtypes B and G is also proposed, because three different isolates with identical recombination patterns were found in native patients from different health care areas.
This is the first work documenting the genetic diversity of HIV-1 using a phylogeographic approach in a large number of patients living in Madrid, the largest city in Spain, the country with the highest rate of immigration in Europe. This approach has revealed that founder effects, characteristic of the HIV-1 epidemic in developed countries (typically subtype B), are being blurred by immigration and are evolving toward a more complex landscape with multiple clusters in each subtype or recombinant forms leading to polyphyletic scenarios. This kind of analysis also strengthens the importance of continuous surveillance programs (15), because they allow the early detection of new variants spreading in the population before they become more prevalent, since they might be fitter than their ancestral nonrecombinant forms, such as subtypes B and G (34) or F (3), by being more virulent (5, 38) or having a higher transmission rate (58). This high-quality information is necessary to evaluate the public health responses to the better control of HIV-1 transmission in European countries.
ACKNOWLEDGMENTS
J.M.G.-A. is supported by a fellow research contract from the European Commission (PAR-241476). A.H. is supported by Agencia Laín Entralgo. F.G.-C.'s work was partly funded by project BFU2008-03000 from the Ministerio de Ciencia e Innovación.
Footnotes
Published ahead of print on 27 July 2011.
REFERENCES
- 1. Anisimova M., Gascuel O. 2006. Approximate likelihood ratio test for branches: a fast, accurate and powerful alternative. Syst. Biol. 55:539–552 [DOI] [PubMed] [Google Scholar]
- 2. Apetrei C., et al. 1997. HIV type 1 subtype F sequences in Romanian children and adults. AIDS Res. Hum. Retroviruses 13:363–365 [DOI] [PubMed] [Google Scholar]
- 3. Aulicino P. C., Holmes E. C., Rocco C., Mangano A., Sen L. 2007. Extremely rapid spread of human immunodeficiency virus type 1 BF recombinants in Argentina. J. Virol. 81:427–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bártolo I., et al. 2009. Highly divergent subtypes and new recombinant forms prevail in the HIV/AIDS epidemic in Angola: new insights into the origins of the AIDS pandemic. Infect. Genet. Evol. 9:672–682 [DOI] [PubMed] [Google Scholar]
- 5. Bruselles A., Rozera G., Bartolini B., Prosperi M., del Nonno F. 2009. Use of massive parallel pyrosequencing for near full-length characterization of a unique HIV type 1 BF recombinant associated with a fatal primary infection. AIDS Res. Hum. Retroviruses 25:937–942 [DOI] [PubMed] [Google Scholar]
- 6. Buonaguro L., Tagliamonte M., Tornesello M. L., Buonaguro F. M. 2007. Genetic and phylogenetic evolution of HIV-1 in a low subtype heterogeneity epidemic: the Italian example. Retrovirology 4:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cuevas M. T., et al. 2009. Incidence of non-B subtypes of HIV-1 in Galicia, Spain: high frequency and diversity of HIV-1 among men who have sex with men. Euro Surveill. 14:19413. [DOI] [PubMed] [Google Scholar]
- 8. Delgado E., et al. 2010. Identification of a new HIV type 1 BF intersubtype circulating recombinant form (CRF44_BF) in Chile. AIDS Res. Hum. Retroviruses 26:821–826 [DOI] [PubMed] [Google Scholar]
- 9. Delgado E., et al. 2002. Identification of a newly characterized HIV-1 BG intersubtype circulating recombinant form in Galicia, Spain, which exhibits a pseudotype-like virion structure. J. Acquir. Immune Defic. Syndr. 29:536–543 [DOI] [PubMed] [Google Scholar]
- 10. de Oliveira T., Pillay D., Gifford R. J. and the U.K. Collaborative Group on HIV Drug Resistance 2010. The HIV-1 subtype C epidemic in South America is linked to the United Kingdom. PLoS One 5:e9311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Derache A., et al. 2008. Evolution of genetic diversity and drug resistance mutations in HIV-1 among untreated patients from Mali between 2005 and 2006. J. Antimicrob. Chemother. 62:456–463 [DOI] [PubMed] [Google Scholar]
- 12. Fernández-García A., et al. 2010. Identification of a new HIV type 1 circulating BF intersubtype recombinant form (CRF47_BF) in Spain. AIDS Res. Hum. Retroviruses 26:827–832 [DOI] [PubMed] [Google Scholar]
- 13. Gao F., et al. 1996. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J. Virol. 70:7013–7029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Geretti A. M. 2006. HIV-1 subtypes: epidemiology and significance for HIV management. Curr. Opin. Infect. Dis. 19:1–7 [DOI] [PubMed] [Google Scholar]
- 15. Gifford R. J., et al. 2007. Phylogenetic surveillance of viral genetic diversity and the evolving molecular epidemiology of human immunodeficiency virus type 1. J. Virol. 81:13050–13056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guimarães M. L., et al. 2009. Close phylogenetic relationship between Angolan and Romanian HIV-1 subtype F1 isolates. Retrovirology 6:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guindon S., Gascuel O. 2003. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 18. Hamers F. F., Downs A. M. 2004. The changing face of the HIV epidemic in Western Europe: what are the implications for public health policies? Lancet 364:83–94 [DOI] [PubMed] [Google Scholar]
- 19. Heath L., van der Walt E., Varsani A., Martin D. P. 2006. Recombination patterns in aphthoviruses mirror those found in other picornaviruses. J. Virol. 80:11827–11832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hemelaar J., Gouws E., Ghys P. D., Osmanov S. 2006. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS 20:W13–W23 [DOI] [PubMed] [Google Scholar]
- 21. Herm A. 2008. Recent migration trends: citizens of EU-27 member states become ever more mobile while EU remains attractive to non-EU citizens. EUROSTAT: Statistics in focus 98/2008. http://epp.eurostat.ec.europa.eu/cache/ity_offpub/ks-sf-08-098-en/ks-sf-08-098-en.pdf
- 22. Holguín A., Alvarez A., Soriano V. 2002. High prevalence of HIV-1 subtype G and natural polymorphisms at the protease gene among HIV-infected immigrants in Madrid. AIDS 16:1163–1170 [DOI] [PubMed] [Google Scholar]
- 23. Holguín A., Mulder M., Yebra G., López M., Soriano V. 2008. Increase of non-B subtypes and recombinants among newly diagnosed HIV-1 native Spaniards and immigrants in Spain. Curr. HIV Res. 6:327–334 [DOI] [PubMed] [Google Scholar]
- 24. Holguín A., Pena M. J., Troncoso F., Soriano V. 2007. Introduction of non-B subtypes among Spaniards newly diagnosed with HIV type 1 in the Canary Islands. AIDS Res. Hum. Retroviruses 23:498–502 [DOI] [PubMed] [Google Scholar]
- 25. Holmes E. C. 2007. When HIV spread afar. Proc. Natl. Acad. Sci. U. S. A. 104:18351–18352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jere A., Tripathy S., Agnihotri K., Jadhav S., Paranjape R. 2004. Genetic analysis of Indian HIV-1 nef: subtyping, variability and implications. Microbes Infect. 6:279–289 [DOI] [PubMed] [Google Scholar]
- 27. Jetzt A. E., et al. 2000. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J. Virol. 74:1234–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kalish M. L., et al. 2004. Recombinant viruses and early global HIV-1 epidemic. Emerg. Infect. Dis. 10:1227–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Konings F., et al. 2006. Genetic analysis of HIV-1 strains in rural Eastern Cameroon indicates the evolution of second-generation recombinants to circulating recombinant forms. J. Acquir. Immune Defic. Syndr. 42:331–341 [DOI] [PubMed] [Google Scholar]
- 30. Lewis F., Hughes G. J., Rambaut A., Pozniak A., Brown Leigh A. J. 2008. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Med. 5:e50 doi:10.1371/journal.pmed.0050050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mansky L. M., Temin H. M. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087–5094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McConnell M. J., et al. 2008. Molecular epidemiology of HIV type 1 in newly diagnosed patients in southern Spain. AIDS Res. Hum. Retroviruses 24:881–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moore C. B., et al. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439–1443 [DOI] [PubMed] [Google Scholar]
- 34. Muñoz-Nieto M., et al. 2008. HIV type 1 intersubtype recombinants during the evolution of a dual infection with subtypes B and G. AIDS Res. Hum. Retroviruses 24:337–343 [DOI] [PubMed] [Google Scholar]
- 35. Paraskevis D., et al. 2009. Tracing the HIV-1 subtype B mobility in Europe: a phylogeographic approach. Retrovirology 6:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Parczewski M., et al. 2010. Characteristics of HIV-1 non-B subtype infections in Northwest Poland. J. Med. Virol. 82:1306–1313 [DOI] [PubMed] [Google Scholar]
- 37. Perelson A. S., Neumann A. U., Markowitz M., Leonard J. M., Ho D. D. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time Science 271:1582–1586 [DOI] [PubMed] [Google Scholar]
- 38. Pérez-Alvarez L., et al. 2006. Isolation and biological characterization of HIV-1 BG intersubtype recombinants and other genetic forms circulating in Galicia, Spain. J. Med. Virol. 78:1520–1528 [DOI] [PubMed] [Google Scholar]
- 39. Perrin L., Kaiser L., Yerly S. 2003. Travel and the spread of HIV-1 genetic variants. Lancet Infect. Dis. 3:22–27 [DOI] [PubMed] [Google Scholar]
- 40. Plantier J. C., et al. 2009. A new human immunodeficiency virus derived from gorillas. Nat. Med. 15:871–872 [DOI] [PubMed] [Google Scholar]
- 41. Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25:1253–1256 [DOI] [PubMed] [Google Scholar]
- 42. Rambaut A., Posada D., Crandall K. A., Holmes E. C. 2004. The causes and consequences of HIV evolution. Nat. Rev. Genet. 5:52–61 [DOI] [PubMed] [Google Scholar]
- 43. Ruchansky D., Casado C., Russi J., Rabiza J., López-Galindez C. 2009. Identification of a new HIV-1 circulating recombinant form (CRF38_BF1) in Uruguay. AIDS Res. Hum. Retroviruses 25:351–356 [DOI] [PubMed] [Google Scholar]
- 44. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 45. Sax P. E., et al. 2005. Should resistance testing be performed for treatment-naive HIV-infected patients? A cost-effectiveness analysis. Clin. Infect. Dis. 41:1316–1323 [DOI] [PubMed] [Google Scholar]
- 46. Sierra M., et al. 2007. Identification of 3 phylogenetically related HIV-1 BG intersubtype circulating recombinant forms in Cuba. J. Acquir. Immune Defic. Syndr. 45:151–160 [DOI] [PubMed] [Google Scholar]
- 47. Soares E. A., et al. 2005. HIV-1 subtype C dissemination in southern Brazil. AIDS 19(Suppl. 4):S81–S86 [DOI] [PubMed] [Google Scholar]
- 48. Soares M. A., et al. 2003. A specific subtype C of human immunodeficiency virus type 1 circulates in Brazil. AIDS 17:11–21 [DOI] [PubMed] [Google Scholar]
- 49. Tatt I. D., Barlow K. L., Clewley J. P., Gill O. N., Parry J. V. 2004. Surveillance of HIV-1 subtypes among heterosexuals in England and Wales, 1997–2000. J. Acquir. Immune Defic. Syndr. 36:1092–1099 [DOI] [PubMed] [Google Scholar]
- 50. Taylor B. S., Sobieszczyk M. E., McCutchan F. E., Hammer S. M. 2008. The challenge of HIV-1 subtype diversity. N. Engl. J. Med. 358:1590–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. van der Kuyl A. C., Cornelissen M. 2007. Identifying HIV-1 dual infections. Retrovirology 4:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vasileva K. 2009. Citizens of European countries account for the majority of the foreign population in EU-27 in 2008. EUROSTAT: Statistics in focus 94/2009. http://epp.eurostat.ec.europa.eu/cache/ity_offpub/ks-sf-09-094/en/ks-sf-09-094-en.pdf
- 53. Vidal N., et al. 2000. Unprecedented degree of human immunodeficiency virus type 1 (HIV-1) group M genetic diversity in the Democratic Republic of Congo suggests that the HIV-1 pandemic originated in Central Africa. J. Virol. 74:10498–10507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wheeler W. H., et al. 2010. Prevalence of transmitted drug resistance associated mutations and HIV-1 subtypes in new HIV-1 diagnoses, U.S.-2006. AIDS 24:1203–1212 [DOI] [PubMed] [Google Scholar]
- 55. Worobey M., et al. 2008. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature 455:661–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yamaguchi J., et al. 2008. Identification of new CRF43_02G and CRF25_cpx in Saudi Arabia based on full genome sequence analysis of six HIV type 1 isolates. AIDS Res. Hum. Retroviruses 24:1327–1335 [DOI] [PubMed] [Google Scholar]
- 57. Yirrell D. L., et al. 2008. HIV subtypes in Scotland, 2000–2006. Epidemiol. Infect. 136:1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang M., et al. 2010. The role of recombination in the emergence of a complex and dynamic HIV epidemic. Retrovirology 7:25. [DOI] [PMC free article] [PubMed] [Google Scholar]