Abstract
Viruses commonly use host cell survival mechanisms to their own advantage. We show that Akt, an important signaling kinase involved in cell survival, phosphorylates the RNA-dependent RNA polymerase (RdRp) from norovirus, the major cause of gastroenteritis outbreaks worldwide. The Akt phosphorylation of RdRp appears to be a feature unique to the more prevalent norovirus genotypes such as GII.4 and GII.b. This phosphorylation event occurs at a residue (Thr33) located at the interface where the RdRp finger and thumb domains interact and decreases de novo activity of the polymerase. This finding provides fresh insights into virus-host cell interactions.
TEXT
Norovirus (NoV) is the most common cause of acute gastroenteritis outbreaks, causing vomiting and diarrhea in affected individuals (14). Since 1996, five variants of the genogroup II-genotype 4 (GII.4) lineage have been associated with global pandemics of acute gastroenteritis of increased frequency (23). Overall, GII.4 variants are the cause of 62 to 80% of all NoV outbreaks globally (11, 23). There are a number of factors contributing to this higher epidemiological fitness, including host-cell receptor binding patterns and duration of herd immunity (reviewed in reference 7). Also, our group recently showed that the GII.4 viruses had a greater capacity to evolve than other NoV genotypes through higher rates of mutation (6). Therefore, the viral RNA-dependent RNA polymerase (RdRp) is likely to play an important role in driving viral evolution and fitness (6). Posttranslational modifications of viral proteins (15), including RdRps (17), are frequently found, and this led us to examine a potential posttranslational modification of such a polymerase from a representative pandemic NoV strain, GII.4 2006b (NSW696T/06/AU—GenBank EF684915). This variant was associated with a global pandemic in 2007 to 2008 (23) and caused three consecutive epidemics of gastroenteritis in Australia in 2006 (26), 2007, and 2008 (12).
Viruses take advantage of existing pathways and mechanisms of the host cell; among these is the phosphatidylinositol 3-kinase (PI3K)/Akt signaling pathway (5). Akt is a serine/threonine protein kinase that phosphorylates downstream targets, leading to numerous effects, with the net result of increasing cell growth, survival, and proliferation. Consequently, perturbations of the Akt signaling pathway have been implicated in the development of diseases, including many human cancers (2) and diabetes (28). With such a fundamental role in the regulation of cell growth and proliferation, it is perhaps not surprising that a number of viruses have been found to interact directly with the PI3K/Akt pathway to effect greater control of their replication within the host cell. In hepatitis C virus (HCV), expression of viral proteins, including NS3/4A, NS4B, and NS5A, results in the activation of the PI3K/Akt pathway (reviewed in reference 3), which is required for efficient virus replication (4). Similarly, influenza A virus activates the PI3K/Akt pathway through viral nonstructural protein 1 (13). This appears to suppress the induction of apoptosis, which may arrest the cell at a stage that supports viral replication. Alternatively, not all viruses activate the PI3K/Akt pathway. For example, with measles virus, infection leads to a downregulation of Akt that appears to play a role in immune suppression (8).
Given the common exploitation by viruses of Akt signaling, we sought to determine whether the NoV RdRp may be a substrate for Akt. Akt phosphorylates proteins at the minimal consensus sequence of RxRxx(S/T)h (where x is any amino acid and h is a bulky hydrophobic amino acid) (1). The findings regarding the minimal consensus motif have been supported by another peptide library screening approach (20). However, there is considerable variability with respect to this consensus in the sequences surrounding Akt phosphorylation sites (22). Therefore, to help determine the potential for phosphorylation on the NoV GII.4 2006b RdRp, we investigated the amino acid sequence by the use of Scansite software (21). This program predicts various phosphorylation sites and protein binding motifs on the basis of likelihood (stringency) using minimal consensus sequences. The Thr33 residue of the 2006b RdRp was identified as both a high-stringency Akt substrate site and a 14-3-3 zeta binding site. The RdRp Thr33 residue is mostly conserved among pandemic strains and almost absent in nonpandemic strains (Table 1) (χ2, P < 0.001).
Table 1.
RdRp strains and their predicted Akt phosphorylation sites and prevalences
| Genotype | Strain | GenBank accession no. | RdRp sequencea | Scansite scoreb | Prevalencec |
|---|---|---|---|---|---|
| GI.f | Otofuke | AB187514 | FWKSTPQ | None predicted | * |
| GI.1 | Norwalk | NC_001959 | FWRSSPE | None predicted | * |
| GI.2 | Southampton | L07418 | FWKSSPE | None predicted | * |
| GI.2 | WUG1 | AB081723 | FWKSSPE | None predicted | * |
| GI.4 | Chiba | AB042808 | FWRSTPE | None predicted | * |
| GI.5 | SzUG1 | AB039774 | FWRSTTE | 0.464 | * |
| GI.6 | Hesse DEU98 | AF093797 | FWRSTPE | None predicted | * |
| GII.b | Picton | AY919139 | FWRSSTT | 0.407 | * |
| GII.bd | C14 | AY845056 | FWRSSTT | 0.407 | *** |
| GII.c | Snow Mountain | AY134748 | FWRSSTV | 0.056 | * |
| GII.d | OsakaNI | DQ366347 | FWRSSNS | None predicted | * |
| GII.d | Yuri | AB083780 | FWRSSNA | None predicted | * |
| GII.g | St George | GQ845370 | FWRSSTA | 0.159 | *** |
| GII.4 | 2009-10 | GQ845367 | FWRSSTA | 0.159 | ***** |
| GII.4 | 2008 | AB445395 | FWRSSTA | 0.159 | *** |
| GII.4 | 2006a | EF187497 | FWRSSTA | 0.159 | *** |
| GII.4d | 2006b | EF684915 | FWRSSTA | 0.159 | ***** |
| GII.4 | Hunter | DQ078814 | FWRSSTA | 0.159 | ***** |
| GII.4 | Farmington | AY502023 | FWRSSTA | 0.159 | ***** |
| GII.4 | Lordsdale | X86557 | FWRSSTA | 0.159 | *** |
| GII.4 | Cairo | GQ845368 | FWRSSTA | 0.159 | *** |
| GII.4 | Mc37 | AY237415 | FWRSSTA | 0.159 | *** |
| GII.4 | Guangzhou | DQ369797 | FWRSSTA | 0.159 | *** |
| GII.4 | Osaka | AB541319 | FWRSSTT | 0.407 | *** |
| GII.4 | US-1995/96 | DQ078829 | FWRSSTT | 0.407 | ***** |
| GII.4 | MD145-12 | AY032605 | FWRSSTT | 0.407 | *** |
| GII.1 | Hawaii | U07611 | FWRSSTT | 0.407 | * |
| GII.3 | Saitama U18 | AB039781 | FWRSSNA | None predicted | * |
| GII.5 | Neustrelitz260 | AY772730 | FWRSSNT | None predicted | * |
| GII.6 | Saitama U17 | AB039779 | FWRSSPD | None predicted | * |
| GII.6 | Saitama U16 | AB039778 | FWRSSPD | None predicted | * |
| GII.7 | Saitama U4 | AB039777 | FWRSSPD | None predicted | * |
| GII.7 | Saitama U3 | AB039776 | FWRSSPD | None predicted | * |
| GII.7 | GIFU99 | AB084071 | FWRSSPD | None predicted | * |
| GII.7d | Mc17 | GQ849131 | FWRSSPD | None predicted | * |
| GII.8 | Saitama U25 | AB039780 | FWRSSPD | None predicted | * |
| GII.11 | Swine43 | AB126320 | FWRSSTA | 0.159 | * |
| GIII.1 | Jena | AJ011099 | FWRSSPA | None predicted | * |
| GIII.2 | Dumfries | AY126474 | FWRSSPA | None predicted | * |
| GIII.2 | Newbury2 | AF097917 | FWRSSPA | None predicted | * |
| GV.1 | Murine NoV-1 CW1 | DQ285629 | FWRTSPE | None predicted | * |
| GV.1 | Murine NoV-4 S18 | FJ446719 | FWRTSPE | None predicted | * |
| GV.1 | Murine NoV-1 CW3 | EF014462 | FWRTSPE | None predicted | * |
RdRp residues 28 to 34 relative to GII4 2006b variant NSW696T/06/AUS (GenBank accession no. EF684915). Residues shown in boldface characters represent the predicted phosphorylation site.
Values refer to Scansite stringency scores: <0.2, high stringency (indicated with gray shading and boldface characters); 0.2 to 1, medium stringency (indicated with gray shading).
Prevalence data are presented as follows: ***** indicates that the strain was associated with a global pandemic, *** indicates that the strain was associated with a regional epidemic, and * indicates that the strain was associated with a local outbreak or with sporadic disease in the natural host.
The strain was used in the experiments whose results are presented in Fig. 1.
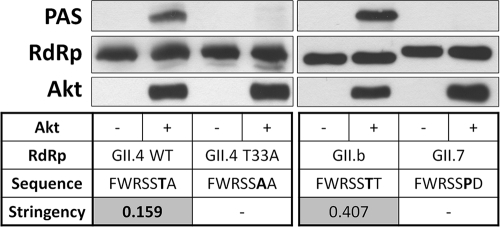
In light of the in silico predictions and the complementary expertise of the investigators (12, 22), we decided to test Akt-mediated phosphorylation of purified recombinant RdRp. Briefly, the coding region for NoV RdRp with a C-terminal hexahistidine tag was cloned into pGEX-4T-1 (GE Healthcare) and then expressed and purified as previously described (6). An N-terminal glutathione-S-transferase (GST) tag was included to increase the size of the RdRp to allow separation of the RdRp from the Akt protein by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). An in vitro Akt kinase assay (22) was employed to compare wild-type RdRp (GII.4 2006b) to a mutated version, Thr33Ala, in which the predicted phosphorylation site had been mutated to alanine. To detect phosphorylation, we made use of an antibody designed for identification of phosphorylated Akt substrates (PAS) (catalog. no. 9611; Cell Signaling Technology [29]). Our results showed clear Akt-dependent phosphorylation of the wild-type GII.4 2006b RdRp (Fig. 1), which was absent in the mutated version. A second antibody that is more specific for phosphorylated Akt substrates (catalog no. 9614; also from Cell Signaling Technology) also detected wild-type phosphorylated RdRp but not the mutated form (data not shown). In addition, we tested a representative RdRp with a medium-stringency site (GII.b C14) (C14/2002/AU—GenBank AY845056) and one with no predicted phosphorylation site (GII.7 Mc17) (Mc17/2001/TH—GenBank AY237413). The results indicated that the medium-stringency sequence was phosphorylated at Thr33 also but that the GII.7 RdRp with no corresponding Thr residue was not (Fig. 1).
Fig. 1.
Akt phosphorylates RdRp at Thr33. Wild-type (WT) or mutant (T33A) recombinant NoV GII.4 2006b, GII.b C14, and GII.7 Mc17 RdRp (1 μg) were incubated with or without recombinant Akt (400 ng) as indicated at 30°C for 30 min. Reactions were stopped by addition of loading buffer, and samples were subjected to SDS-PAGE and Western blotting with phosphorylated Akt substrate (PAS) antibody, followed by stripping and probing for RdRp and Akt. Data are representative of the results of three independent experiments. The residue shown in bold in each of the sequences represents the residue corresponding to Thr33 in WT GII.4 2006b RdRp. Stringency values refer to Scansite stringency (<0.2, high stringency; 0.2 to 1, medium stringency).
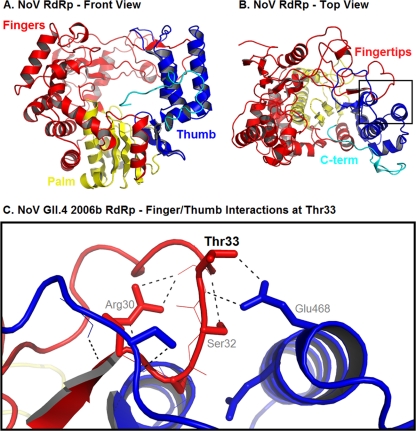
Akt's phosphorylation of NoV RdRp may provide additional insights into the evolutionary strategies by which viruses exploit the host for their own benefit. The Akt pathway is active when cells are proliferating rapidly, so it is possible that NoV takes advantage of this situation to increase its survival. How this phosphorylation event may influence the function of RdRp is unclear, since there is currently very little information linking the structure and function of the NoV RdRp, largely due to the lack of a cell culture system for the virus. Most of the important residues have been predicted from published crystal structures (19, 27) and by comparisons of structures to those of functionally defined residues in poliovirus and foot and mouth disease virus (see, e.g., reference 25). The phosphorylated Thr33 residue is located at the tip of the finger domain, which interacts with the top of the thumb and encloses the active site (Fig. 2A and B). The results of a study by Thompson et al. suggest that this structure, which is unique to RdRps, plays a role in stabilizing poliovirus RdRp (25). Those same residues may play a similar stabilizing role in NoV RdRp, allowing a conformation that facilitates RNA synthesis. Homology modeling predicts extensive interdomain interactions within this region of the RdRp, including two polar bridging interactions that link the finger and thumb domains at Thr33 in the GII.4 2006b RdRp (Fig. 2C). When the Thr33 residue is phosphorylated, the negative charge of the phosphate should repel the negatively charged side chain of Glu468, altering the dynamics of these interdomain finger/thumb interactions.
Fig. 2.
Three-dimensional modeling of NoV RdRp, showing finger-thumb domain interactions at Thr33. (A) Front view of NoV GII.4 2006b RdRp, highlighting the finger (red), thumb (blue), and palm (yellow) domains. (B) Top view, highlighting the fingertips (red) and the C-terminal (C-term) region (cyan). (C) Closeup view of the boxed region in panel B, showing the finger-thumb interactions at Thr33 for 2006b RdRp. Images were produced using PyMOL, version 1.3 (Schrödinger, LLC).
The HCV RdRp (NS5B) has been shown to be phosphorylated by protein kinase C-related kinase 2, and that phosphorylation event played a role in regulating HCV RNA replication (16). To explore the possibility that the phosphorylation of Thr33 is functionally significant, we used a de novo GTP incorporation assay as previously described (6) to compare the enzyme kinetics of the wild-type 2006b RdRp with that of a mutated form (Thr33Glu) that should mimic phosphorylation. The phosphomimetic mutant had a lower maximum enzyme velocity (Vmax = 100 fmol·min−1) than the wild-type RdRp (Vmax = 125 fmol·min−1), indicating a lower rate of RNA synthesis (Fig. 3). The phosphomimetic mutant also had a lower affinity for the GTP substrate, with a Km value of 10.4 μM compared to 4.8 μM for the wild-type RdRp. The differences in enzyme kinetics between the wild-type and phosphomimetic RdRp suggest that the phosphorylation of Thr33 may modulate the function of the polymerase. In HCV, it has been shown that disruptions to the interactions between the finger and thumb domains lead to a reduction in de novo RdRp activity whereas efficient primer extension activity is maintained (9, 10, 24). Since NoV utilizes both de novo and protein-primed RNA synthesis, it is possible that phosphorylation of NoV RdRp could alter the balance between these modes of RNA synthesis.
Fig. 3.
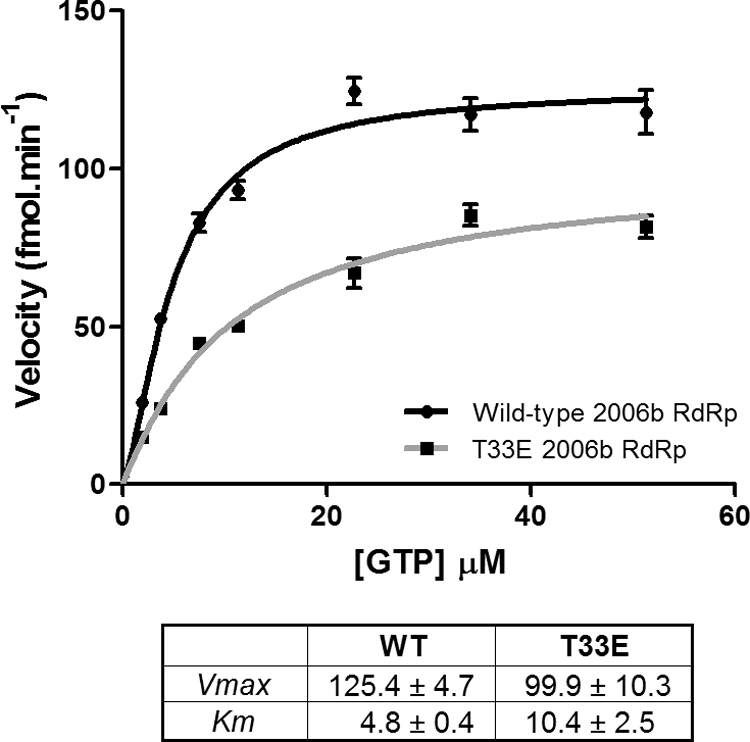
Comparison of enzyme kinetics of the wild-type 2006b RdRp with a Thr33Glu phosphomimetic mutant. The enzyme kinetics of the wild-type (WT) 2006b RdRp were compared with that of a Thr33Glu phosphomimetic mutant (T33E). Briefly, 50 ng of RdRp was added to initiate a reaction in which 3H-GTP was incorporated into homopolymeric C RNA. After 10 min of incubation, the reaction was stopped, the reaction mixture was harvested onto a filterplate, and then the incorporated nucleotides were detected via scintillation counting. Samples were tested in triplicate, and background values were subtracted to measure the levels of incorporated nucleotides (expressed as femtomoles per minute). The levels were measured across a range of increasing GTP substrate concentrations, reaching a final concentration of 51 μM. The data were analyzed using the Hill equation in GraphPad Prism, version 5.04. The kinetic parameters are presented with standard deviations.
In contrast to examples of viral exploitation of the Akt pathway (3, 8, 13), a decrease in polymerase activity could indicate that Akt phosphorylation of the NoV RdRp is an antiviral strategy protecting the infected cell. However, this idea may be hard to reconcile with our observation that the Akt phosphorylation event occurs in the most epidemiologically fit genotype (GII.4) and is generally absent from others. Further work would be needed to distinguish between these possibilities and to determine the viral outcomes of this phosphorylation event.
Phosphorylation may not just alter function directly but may act as a molecular signal for a binding partner. In line with this idea, a high-stringency 14-3-3 zeta binding site on the phosphorylated Thr33 residue was predicted by the use of Scansite. 14-3-3s are ubiquitous proteins that bind to phosphorylated residues; therefore, further alterations of protein function could occur (18). In this case, due to the high intrinsic disorder within this region of the polymerase, it is possible that 14-3-3 binding acts as a clamp that fixes the polymerase into a functionally significant conformation. The interaction of 14-3-3 zeta and NoV RdRp may also affect the localization within the cell to aid evasion of the host cell recognition system. The possibility that phosphorylation of NoV RdRp may affect intermolecular interactions represents another interesting avenue for further research.
Here, we have discovered a novel Akt substrate, a viral polymerase, and identified its phosphorylation site. This residue appears to be critical for the activity of the NoV RdRp, according to the results of our in vitro polymerase assay. Importantly, it is routinely present in pandemic strains but mostly lacking in nonpandemic genotypes. Future work should include confirming the NoV RdRp phosphorylation event in vivo and determining its impact on viral replication and whether this favors the host or the virus. In this respect, it would be worthwhile to engineer the phosphorylation site into the existing murine NoV infectious cell culture system. The current work provides a good basis for future efforts aimed at increasing our understanding of how phosphorylation of viral proteins may affect replication.
Footnotes
Published ahead of print on 17 August 2011.
REFERENCES
- 1. Alessi D. R., Caudwell F. B., Andjelkovic M., Hemmings B. A., Cohen P. 1996. Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett. 399:333–338 [DOI] [PubMed] [Google Scholar]
- 2. Altomare D. A., Testa J. R. 2005. Perturbations of the AKT signaling pathway in human cancer. Oncogene 24:7455–7464 [DOI] [PubMed] [Google Scholar]
- 3. Bode J. G., Brenndorfer E. D., Karthe J., Haussinger D. 2009. Interplay between host cell and hepatitis C virus in regulating viral replication. Biol. Chem. 390:1013–1032 [DOI] [PubMed] [Google Scholar]
- 4. Brenndörfer E. D., et al. 2009. Nonstructural 3/4A protease of hepatitis C virus activates epithelial growth factor-induced signal transduction by cleavage of the T-cell protein tyrosine phosphatase. Hepatology 49:1810–1820 [DOI] [PubMed] [Google Scholar]
- 5. Buchkovich N. J., Yu Y., Zampieri C. A., Alwine J. C. 2008. The TORrid affairs of viruses: effects of mammalian DNA viruses on the PI3K-Akt-mTOR signalling pathway. Nat. Rev. Microbiol. 6:266–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bull R. A., Eden J. S., Rawlinson W. D., White P. A. 2010. Rapid evolution of pandemic noroviruses of the GII.4 lineage. PLoS Pathog. 6:e1000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bull R. A., White P. A. 2011. Mechanisms of GII.4 norovirus evolution. Trends Microbiol. 19:233–240 [DOI] [PubMed] [Google Scholar]
- 8. Carsillo M., Kim D., Niewiesk S. 2010. Role of AKT kinase in measles virus replication. J. Virol. 84:2180–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chinnaswamy S., Murali A., Li P., Fujisaki K., Kao C. C. 2010. Regulation of de novo-initiated RNA synthesis in hepatitis C virus RNA-dependent RNA polymerase by intermolecular interactions. J. Virol. 84:5923–5935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chinnaswamy S., et al. 2008. A locking mechanism regulates RNA synthesis and host protein interaction by the hepatitis C virus polymerase. J. Biol. Chem. 283:20535–20546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Donaldson E. F., Lindesmith L. C., Lobue A. D., Baric R. S. 2010. Viral shape-shifting: norovirus evasion of the human immune system. Nat. Rev. Microbiol. 8:231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eden J. S., et al. 2010. Norovirus GII.4 variant 2006b caused epidemics of acute gastroenteritis in Australia during 2007 and 2008. J. Clin. Virol. 49:265–271 [DOI] [PubMed] [Google Scholar]
- 13. Ehrhardt C., et al. 2007. Influenza A virus NS1 protein activates the PI3K/Akt pathway to mediate antiapoptotic signaling responses. J. Virol. 81:3058–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Estes M. K., Prasad B. V., Atmar R. L. 2006. Noroviruses everywhere: has something changed? Curr. Opin. Infect. Dis. 19:467–474 [DOI] [PubMed] [Google Scholar]
- 15. Kadaveru K., Vyas J., Schiller M. R. 2008. Viral infection and human disease—insights from minimotifs. Front. Biosci. 13:6455–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim S. J., Kim J. H., Kim Y. G., Lim H. S., Oh J. W. 2004. Protein kinase C-related kinase 2 regulates hepatitis C virus RNA polymerase function by phosphorylation. J. Biol. Chem. 279:50031–50041 [DOI] [PubMed] [Google Scholar]
- 17. Mackenzie J. M., Kenney M. T., Westaway E. G. 2007. West Nile virus strain Kunjin NS5 polymerase is a phosphoprotein localized at the cytoplasmic site of viral RNA synthesis. J. Gen. Virol. 88:1163–1168 [DOI] [PubMed] [Google Scholar]
- 18. Muslin A. J., Tanner J. W., Allen P. M., Shaw A. S. 1996. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84:889–897 [DOI] [PubMed] [Google Scholar]
- 19. Ng K. K., et al. 2004. Crystal structure of Norwalk virus polymerase reveals the carboxyl terminus in the active site cleft. J. Biol. Chem. 279:16638–16645 [DOI] [PubMed] [Google Scholar]
- 20. Obata T., et al. 2000. Peptide and protein library screening defines optimal substrate motifs for AKT/PKB. J. Biol. Chem. 275:36108–36115 [DOI] [PubMed] [Google Scholar]
- 21. Obenauer J. C., Cantley L. C., Yaffe M. B. 2003. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 31:3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sharpe L. J., Luu W., Brown A. J. 2011. Akt phosphorylates Sec24: new clues into the regulation of ER-to-Golgi trafficking. Traffic 12:19–27 [DOI] [PubMed] [Google Scholar]
- 23. Siebenga J. J., et al. 2009. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001–2007. J. Infect. Dis. 200:802–812 [DOI] [PubMed] [Google Scholar]
- 24. Simister P., et al. 2009. Structural and functional analysis of hepatitis C virus strain JFH1 polymerase. J. Virol. 83:11926–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thompson A. A., Albertini R. A., Peersen O. B. 2007. Stabilization of poliovirus polymerase by NTP binding and fingers-thumb interactions. J. Mol. Biol. 366:1459–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tu E. T. V., et al. 2008. Epidemics of gastroenteritis during 2006 were associated with the spread of norovirus GII.4 variants 2006a and 2006b. Clin. Infect. Dis. 46:413–420 [DOI] [PubMed] [Google Scholar]
- 27. Zamyatkin D. F., Parra F., Machin A., Grochulski P., Ng K. K. 2009. Binding of 2′-amino-2′-deoxycytidine-5′-triphosphate to norovirus polymerase induces rearrangement of the active site. J. Mol. Biol. 390:10–16 [DOI] [PubMed] [Google Scholar]
- 28. Zdychová J., Komers R. 2005. Emerging role of Akt kinase/protein kinase B signaling in pathophysiology of diabetes and its complications. Physiol. Res. 54:1–16 [DOI] [PubMed] [Google Scholar]
- 29. Zhang H., et al. 2002. Phosphoprotein analysis using antibodies broadly reactive against phosphorylated motifs. J. Biol. Chem. 277:39379–39387 [DOI] [PubMed] [Google Scholar]