Abstract
The great majority of human immunodeficiency virus type 1 (HIV-1) strains enter CD4+ target cells by interacting with one of two coreceptors, CCR5 or CXCR4. Here we describe a transmitted/founder (T/F) virus (ZP6248) that was profoundly impaired in its ability to utilize CCR5 and CXCR4 coreceptors on multiple CD4+ cell lines as well as primary human CD4+ T cells and macrophages in vitro yet replicated to very high titers (>80 million RNA copies/ml) in an acutely infected individual. Interestingly, the envelope (Env) glycoprotein of this clade B virus had a rare GPEK sequence in the crown of its third variable loop (V3) rather than the consensus GPGR sequence. Extensive sequencing of sequential plasma samples showed that the GPEK sequence was present in virtually all Envs, including those from the earliest time points after infection. The molecularly cloned (single) T/F virus was able to replicate, albeit poorly, in cells obtained from ccr5Δ32 homozygous donors. The ZP6248 T/F virus could also infect cell lines overexpressing the alternative coreceptors GPR15, APJ, and FPRL-1. A single mutation in the V3 crown sequence (GPEK->GPGK) of ZP6248 restored its infectivity in CCR5+ cells but reduced its ability to replicate in GPR15+ cells, indicating that the V3 crown motif played an important role in usage of this alternative coreceptor. These results suggest that the ZP6248 T/F virus established an acute in vivo infection by using coreceptor(s) other than CCR5 or CXCR4 or that the CCR5 coreceptor existed in an unusual conformation in this individual.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) enters target cells by first binding to the primary receptor CD4 and then to a coreceptor, generally one of the chemokine receptors CCR5 and CXCR4 (4). CD4 binding induces structural changes in the envelope (Env) glycoprotein that form and expose the coreceptor binding site. There are two main interactions between Env and coreceptor (13, 14, 25, 50, 51): the base of the third variable loop (V3) engages the N terminus of the coreceptor, while the crown of the V3 loop that includes the highly conserved GPGR/Q arch motif binds to the extracellular loops of the coreceptor, with the second extracellular loop of the coreceptor being particularly important (16, 25, 35, 48, 62). Although some HIV-1 strains are able to use a variety of different G protein-coupled receptors to gain entry into CD4+ cell lines, the great majority of these viruses use CCR5 and/or CXCR4 as coreceptors to infect primary cells (3, 4, 10, 23, 47, 66). CCR3, GPR15, APJ, and FPRL-1 are among the most frequently used alternative coreceptors when overexpressed on cell lines (11, 26, 43, 47, 57). Rare cases of HIV-1 strains that are able to use FPRL-1 and GPR1, but not CCR5 or CXCR4, have been reported (57); however, their in vivo relevance remains unknown.
To characterize the biological processes underlying HIV/simian immunodeficiency virus (SIV) transmission, we recently developed an experimental strategy that permits the identification, enumeration, and molecular cloning of transmitted/founder (T/F) viruses (28, 53). This strategy, which employs single-genome amplification (SGA) and direct amplicon sequencing of HIV/SIV RNA or DNA from the plasma or infected cells, makes it possible to infer the nucleotide sequence of the viral strain(s) that initiated productive infection weeks earlier (1, 28, 29, 37, 53, 58, 67). An important prediction of this approach has been that inferred T/F viruses are fully functional and encode all proteins necessary to establish a new infection. Indeed, this prediction has been borne out in numerous studies, which have shown that T/F viral genes as well as full-length genomes are biologically active. Sets of T/F Envs have been shown to mediate efficient virus entry in single-round infection assays, and they invariably use CCR5 as a coreceptor (28, 34). Similarly, T/F infectious molecular clones (IMCs) of HIV-1, SIVmac and SIVagm all produce replication competent virus that grow to high titers in primary CD4+ T cells (22, 38, 54).
To generate a comprehensive panel of T/F Env constructs for biological studies, we recently characterized more than 100 plasma samples from acute infection cases, including commercially available seroconversion panels of serial plasma donors (28). Sequences spanning the rev-vpu-env region of the HIV-1 genome were amplified using the SGA method and used to infer the T/F env sequences (28, 54). A subset of these were then cloned and subjected to functional studies. When a panel of subtype B env genes derived by this method was tested, all but one were highly functional in single-round infection assays. The exception was an Env construct from an acutely infected plasma source donor with a highly unusual GPEK V3 loop crown sequence that failed to utilize CCR5 or CXCR4 efficiently to infect cell lines or primary CD4+ cells, although function was restored when the V3 crown sequence was changed to GPER. The poor functionality of this unambiguously identified T/F Env was in stark contrast to the ability of the corresponding virus to replicate in vivo, as evidenced by high plasma viral load (>80 million RNA copies/ml) in the infected host. Here, we investigated the mechanisms underlying this highly unusual T/F virus phenotype.
MATERIALS AND METHODS
Amplification of the HIV-1 env gene.
Serial plasma samples collected from an acutely infected plasma donor, ZP6248, were purchased from ZeptoMetrix. A total of seven plasma samples were collected between 12 February and 9 March 1997, and viral loads (VLs) were determined by the COBAS Amplicor HIV-1 monitor test. Viral RNA was extracted from plasma collected on 26 February, 2 March, and 9 March using the PureLink viral RNA/DNA minikit (Invitrogen, Carlsbad, CA) and used for cDNA synthesis using Superscript III (Invitrogen, Carlsbad, CA) and primer Env3′ex (5′-TTGCTACTTGTGATTGCTCCATGT-3′) (nucleotides [nt] 8913 to 8936 in HXB2) in a 50-μl volume. Single-genome amplification (SGA) was used to obtain rev-vpu-env cassettes as previously described (28, 32). The first-round PCR was carried out with primers Env5′ex (5′-TAGAGCCCTGGAAGCATCCAGGAAG-3′) (nt 5853 to 5877) and Env3′ex, and the second-round PCR was done with primers Env1Atopo (5′-caccGGCTTAGGCATCTCCTATGGCAGGAAGAA-3′) (nt 5957 to 5983) and Env3′in (5′-GTCTCGAGATACTGCTCCCACCC-3′) (nt 8904 to 8882).
Sequence analysis.
All SGA amplicons were sequenced directly by cycle sequencing and dye terminator methods using an ABI 3730xl genetic analyzer (Applied Biosystems, Foster City, CA). Individual sequences were assembled and edited using the Sequencher program 4.7 (Gene Codes, Ann Arbor, MI). The env sequences were aligned using CLUSTAL W (60), and manual adjustment for optimal alignment was done using MASE (20). The env sequence of the T/F virus was inferred as previously described (28). A phylogenetic tree was constructed with complete env gene sequences using the neighbor-joining method (52) and the Kimura two-parameter model (30). Highlighter analysis was performed through the LANL database website (http://www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter.html).
Generation of V3 mutants.
Site-directed mutagenesis was carried out to introduce mutations into the V3 region of the env gene using the Quikchange site-directed mutagenesis kit (Stratagene, La Jolla, CA) as previously described (40). A single mutation (E312G) was introduced into the ZP6248.wt env gene to make the ZP6248.E312G mutant. The mutagenesis reaction was transformed into XL1-Blue supercompetent cells, and positive clones were confirmed by sequencing.
Construction of YU2-ZP4248 chimeras.
To conveniently insert a foreign env gene into the replication-competent YU2 proviral clone, we first generated a modified YU2 clone (pYU2-ΔenvMN), in which the partial vpu gene, complete env, and partial nef gene (nt 6186 to 8800) were replaced by a multiple cloning site fragment ACGCGTATCACAAGATTAGTGcGGCcgc (MluI-14bp spacer-NotI). The unique MluI and NotI sites (italic letters) that are rare among HIV-1 env genes were introduced. The first 2 bp and the last 8 bp (underlined) are from the vpu and nef coding regions, respectively. The small letters indicate those bases that are different from the original YU2 sequence.
To clone the ZP6248.wt env gene into pYU2-ΔenvMN, both ZP6248.wt and ZP6248.E312G were amplified with Platinum Taq High Fidelity polymerase. MluI and NotI restriction sites were introduced at the 5′ and 3′ ends of the amplified env genes, respectively, at the corresponding positions in the HIV-1 genome. The PCR products were purified, digested with MluI and NotI, and ligated into the pretreated pYU2-ΔenvMN vector. The positive clones from transformed JM109 cells were identified and confirmed by sequencing.
Single-round infection assay.
TZM-bl cells were infected with pseudovirions as previously described (31). Infectivity was determined by measuring luciferase activity in the cell lysates using the Bright-Glo luciferase assay system (Promega Corp., Madison, WI) on a Victor 2 luminometer (Perkin-Elmer Life Sciences, Shelton, CT).
Determination of coreceptor usage.
GHOST(3) indicator cell lines expressing CCR1, CCR2b, CCR3, CCR4, CCR8, CXCR4, Hi5, X4R5, GPR15/BOB, STRL33/Bonzo, and CX3CR1 were obtained from the NIH AIDS Research and Reference Reagent Program. Cells (3 × 105) were seeded in 12-well plates the day before infection. One hundered nanograms of p24 of each pseudovirion or infectious virus prepared from transfected 293T cells was added to each well. After absorption for 6 h at 37°C, complete medium was added, and the cell culture was maintained at 37°C with 5% CO2. The supernatant was collected daily, and virus replication was monitored by measuring the p24 antigen content using the Alliance HIV-1 p24 Antigen ELISA kit (PerkinElmer, Boston, MA). Coreceptor usage was also determined on NP-2 cells expressing CD4 along with either CCR5, CXCR4, or other G protein-coupled receptors with Env pseudovirions as previously described (43, 57).
Molecular cloning of the full-length ZP6248 T/F virus.
To obtain an infectious molecular clone (IMC) of the ZP6248 T/F virus, SGA methods were used to amplify overlapping 5′ and 3′ half genomes as well as the viral long terminal repeat (LTR) from plasma viral RNA. The full-length T/F sequence was inferred as described (22, 38, 54) and chemically synthesized as three subgenomic fragments. These fragments were then cloned using unique restriction enzyme sites into a low-copy-number plasmid (modified pBR322 vector), which has been shown to prevent construct instability (54). The integrity of the IMC was confirmed by nucleotide sequence analysis following large-scale plasmid purification.
Western blotting.
HIV-1 IMCs were transfected into 293T cells in a T75 flask using FuGENE6 transfection reagent (Roche Diagnostics; Indianapolis, IN). Briefly, 40 μg DNA was mixed with 120 μl of FuGENE6 in a total volume of 1.5 ml of serum-free DMEM, incubated for 30 min, and added to 293T cells (70% confluence) seeded 1 day earlier at 1.5 ×107 cells per flask. Forty-eight hours after transfection, the supernatants were harvested, and the viruses were pelleted through a 20% sucrose cushion at 27,000 rpm for 2 h using an AH-629 swinging bucket aluminum rotor. The pelleted virus particles and transfected 293T cells were lysed with 250 μl of lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 20 mM EDTA, 1% Triton X-100, 0.1% SDS, pH 7.4). Western blot analysis was performed as previously described (31). The same amount of each virus (based on the p24 contents: 300 ng of the cell lysate and 10 ng of pelleted virus particles) was loaded on a NuPAGE Novex 4 to 12% Bis-Tris gel (Invitrogen, Carlsbad, CA). After the samples were transferred to a nitrocellulose membrane, the membrane was blocked in phosphate-buffered saline (PBS) containing 1% casein and 0.01% NaN3 for 1 h. The blot was reacted with purified HIVIG (Quality Biological, Gaithersburg, MD) and the mouse monoclonal antibody (MAb) 13D5 (1 μg/ml) to the HIV-1 Env protein. Finally, the membrane was reacted with IRDye 700DX-conjugated affinity-purified anti-human IgG (Rockland Immunochemicals, Gilbertsville, PA) and Alexa-Fluor 680-conjugated goat anti-mouse IgG (Invitrogen; Carlsbad, CA). Fluorescence was detected on an Odyssey infrared imaging system (LiCor Biosciences, Lincoln, NE).
Infection of CD4+ T cells and macrophages.
Human CD4+ T cells were obtained from the Center for AIDS Research's Immunology Core at the University of Pennsylvania. All donors gave written informed consent. Homozygous cells (ccr5 wild type [wt] and ccr5Δ32) were stimulated with plate-bound anti-CD3 (eBiosciences, San Diego, CA) and anti-CD28 (BD Biosciences, San Jose, CA) antibodies for 3 days prior to infection. Cells were maintained in RPMI with 10% fetal bovine serum (FBS) and interleukin 2 (IL-2) (20 units/ml) throughout the experiment. Cells were then infected with HIV-1 pseudovirions expressing green fluorescent protein (GFP) in the presence or absence of 50 μM AMD3100 as previously described (46). Prototype R5 (JRFL) and X4 (LAI) pseudovirions were used as controls.
GFP pseudovirions were made by cotransfecting Env-pcDNA3.1 with pNL4-3deltaE-EGFP in 293T cells. Viruses were harvested 72 h posttransfection and pelleted through a 20% sucrose cushion. Cells were then spinoculated with HIV-GFP pseudovirions for 2 h at 1,200 × g. Three days postinfection, cells were analyzed by flow cytometry. Experiments were repeated in two different ccr5Δ32 homozygous donors, each time with a ccr5 wt control donor.
Infections of matched donor CD4+ T cells and macrophages with the ZP6248 T/F and control IMCs were performed as previously described (54). Briefly, virus stocks were prepared by transfecting a 10-cm dish 30% confluent with 293T cells with 6 μg of IMC DNA using FuGene (Roche, Indianapolis, IN). CD4+ cells were positively selected and plated in polystyrene tissue culture dishes for 2 h in Hanks balanced salt solution (HBSS) with 10 mM Ca2+ and Mg2+ to remove adherent monocytes and then stimulated for 48 h with 3 μg/ml staphylococcal enterotoxin B. CD4+ T cells (5 × 105) were inoculated overnight with 50,000 IU of the control viruses (or 100 ng p24 for ZP6248.wt). Monocyte-derived macrophages were derived by similarly plating peripheral blood mononuclear cells (PBMCs) in 24-well polystyrene plates to isolate monocytes, which were cultured with Dulbecco modified Eagle medium (DMEM) supplemented with 10% giant-cell-conditioned medium, 10% human AB serum, and 5 U/ml rhM-CSF (R&D Systems, Minneapolis, MN). After 6 days, macrophages were inoculated overnight with 100,000 IU of the control viruses (or 200 ng p24 for ZP6248.wt) and then washed three times. Virus replication was monitored by measuring p24 antigen concentration in the supernatant.
Flow cytometry analysis of HIV-1-infected CD4+ T cells.
CD4+ T cells (1 × 106 to 2 × 106) per condition were stained with live/dead Aqua (Invitrogen, Carlsbad, CA), anti-CD3 Qdot655 (Invitrogen, Carlsbad, CA), anti-CD4 Alexa 700 (BD Biosciences, San Jose, CA), anti-CD45RO PE Texas Red (Beckman Coulter, Miami, FL), anti-CCR7 IgM (BD Biosciences, San Jose, CA), and anti-IgM PE (Invitrogen, Carlsbad, CA). Approximately 106 events were collected per condition. The gating strategy was as follows: singlets (FSC-A by FSC-H), live cells (SSC-A by live/dead), lymphocytes (SSC-A by FSC-A), and T cells (SSC-A by CD3). To assess viral tropism, infected cells, defined as GFP+, were back-gated on memory markers CCR7 and CD45RO. Flow cytometry was performed on a BD LSR II, and data were analyzed with FlowJo 8.8.6 (TreeStar, Inc., Ashland, OR).
CD4- and CCR5-dependent infection determined by Affinofile analysis.
293T cells expressing CD4 and CCR5 under independent inducible promoters, called Affinofiles, were used to assess the CD4 and CCR5 utilization efficiency of ZP6248.wt and ZP6248.E312G as previously described (27). Briefly, cells were plated in 96-well plates 48 h prior to induction. An entire 96-well plate was used per virus, representing 48 different CD4 and CCR5 conditions each in duplicate. To induce cells, 2-fold serial dilutions of ponasterone A and minocycline (Invitrogen, Carlsbad, CA), starting at 2 μM and 5 ng/ml, respectively, were added and cells were incubated for 18 h prior to infection. To infect cells, approximately 25 ng p24 of HIV-NL43-luc+ vpr+ pseudovirions were added to each well in the 96-well plate. At 72 h postinfection, luciferase activity was assessed using the Bright-Glo luciferase assay system and vector angles and magnitude were quantified by VERSA (http://versa.biomath.ucla.edu/). Control viruses TA1 (35) and maraviroc-resistant R3 (61) were included in all experiments, which were repeated in duplicate in each of at least three independent experiments.
Detection of the minority variant by PASS.
The parallel allele-specific sequencing (PASS) assay was performed as previously described (7). Briefly, 20 μl of 6% acrylamide gel mix, containing 1 μM acrydite-modified primer 1H1 (5′Acr-CAATAATTGTCTGGCCTGTACCGTCA-3′) (nt 7834 to 6859), cDNA template, 0.3% diallyltartramide, 5% Rhinohide, 0.1% APS, 0.1% TEMED (N,N,N′,N′-tetramethylethylenediamine), 0.2% bovine serum albumin (BSA), and 17 μl cDNA were used to cast a gel on a bind-silane (Amersham Biosciences, Piscataway, NJ)-treated glass slide. The in-gel PCR amplification was then performed in a PTC-200 thermal cycler with a mix of 1 μM primer 1C1 (5′-GGATCAAAGCCTAAAGCCATGTGT-3′) (nt 6560 to 6583), 0.1% Tween 20, 0.2% BSA, 1× PCR buffer, 100 μM deoxynucleoside triphosphate (dNTP) mix, 3.3 units of Jumpstart Taq DNA polymerase (Sigma, St. Louis, MO), and H2O (up to 300 μl) under a sealed SecureSeal chamber (Grace Bio-Labs, Inc., Bend, OR). PCR was carried out as follows: 94°C for 3 min; 65 cycles of 94°C for 30 s, 56°C for 45 s, and 72°C for 1 min; and 72°C for 3 min. After PCR amplification, single-base extension (SBE) was performed with mutant and wt bases distinctively labeled with Cy3 and Cy5, respectively, using the sequencing primer 5′-AACAACACAAGAAAAGGTGTACATATAGGACCAG-3′ (nt 7125 to 7164) that annealed just upstream of the mutation site (GAA->GGA). The gel was scanned to obtain images with a GenePix 4000B microarray scanner (Molecular Devices, Sunnyvale, CA) and then analyzed with the Progenesis PG200 (Nonlinear Dynamics, Durham, NC) software. Wild-type and mutant viruses were determined by comparing their relative fluorescence intensities to normalized values.
Coreceptor expression on PBMCs.
A panel of antibodies was used to examine coreceptor surface expression on peripheral blood lymphocytes (PBLs), either phytohemagglutinin (PHA)-stimulated or unstimulated, using flow cytometry. Four labeling steps were carried out. (i) Cells (2.5 × 105) were incubated with 20 μg/ml of each of the following antibodies: anti-GPR15 (R&D Systems, Minneapolis, MN), anti-FPRL-1 (R&D Systems, Minneapolis, MN), anti-APJ (R&D Systems, Minneapolis, MN), anti-CXCR4 (BD Pharmingen, San Diego, CA), or anti-CCR5 (BD Pharmingen, San Diego, CA). (ii) Cells were incubated with 10 μg/ml of FITC-labeled anti-mouse IgG (H+L) antibody (KPL, Gaithersburg, MD). (iii) PE-Cy7-labled anti-CD4 antibody (BD Pharmingen, San Diego, CA) was incubated with the cells. (iv) The cells were finally incubated with Aqua Vital dye (Invitrogen, Carlsbad, CA). Each labeling step was carried out for 30 min at 4°C. Lymphocytes were washed with 3 ml of PBS containing 1% BSA between steps. Lymphocytes fixed in PBS containing 1% paraformaldehyde were analyzed on a BD LSRII.
GenBank accession numbers.
The sequences determined in this study were deposited in GenBank under the accession numbers EU575573-EU575592 (env) and JN400469-JN400492 (half genome).
RESULTS
Viral kinetics during acute HIV-1 infection in subject ZP6248.
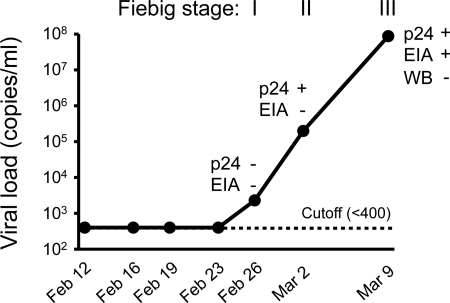
Serial samples from an acutely infected male plasma source donor, ZP6248, infected with a clade B HIV-1 were obtained before and after seroconversion, and virus load (VL), p24, and HIV-1 specific antibody levels were determined (Fig. 1). VLs were below the limit of detection (400 copies/ml) for the first four time points (between 12 and 23 February 1997) and then began to rise, reaching 87.7 million copies/ml within the next 11 days (Fig. 1). Subsequent samples were not available for analysis. p24 antigen was detected at the last two time points, and HIV-1 specific antibodies (Abs) were detected by enzyme immunoassay (EIA) but not by Western blot analysis at the final time point. According to the Fiebig staging system that is based on the sequential appearance of viral RNA, p24 antigen, and antibodies in plasma during acute infection (21), subject ZP6248 met the criteria for Fiebig stage I, Fiebig stage II, and Fiebig stage III at the indicated time points (Fig. 1). The time course from Fiebig stage I to Fiebig stage III was similar to those observed in other acute HIV-1 infections (37).
Fig. 1.
Laboratory staging of acute HIV-1 infection in subject ZP6248. Plasma viral load (RNA copies/ml) was determined using the COBAS Amplicor HIV-1 monitor test (the threshold of the assay is 400 copies/ml). p24 antigen was detected using the Alliance HIV-1 p24 Antigen ELISA Kit (PerkinElmer, Boston, MA). HIV-1 specific antibodies were detected using the GS HIV-1/HIV-2 plus O EIA kit and the GS HIV-1 Western blot kit (Bio-Rad, Hercules, CA). The temporal appearance of HIV-1 specific markers according to the classification by Fiebig (21) is shown.
Infection by a single T/F virus with an unusual V3 crown.
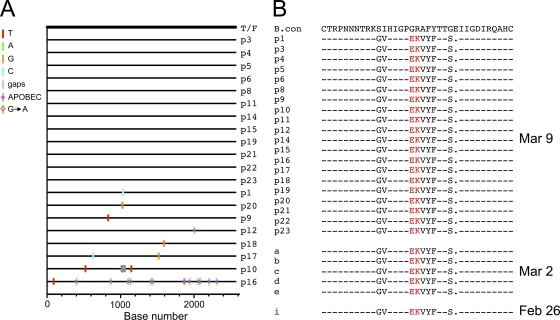
Twenty full-length env gene sequences were obtained from the 9 March plasma sample using the SGA method. Sequence analysis showed limited genetic diversity in the virus population, except for one env sequence which contained 13 G-to-A substitutions, characteristic of APOBEC-mediated hypermutation (Fig. 2A). A single T/F env gene sequence was inferred, which like most previously characterized T/F Envs was genotypically consistent with an R5 phenotype (28). Examination of the deduced amino acid sequences of 20 SGA amplicons revealed a highly unusual V3 sequence that contained a GPEK rather than the clade B consensus GPGR crown motif (Fig. 2B). Only 13 of more than 90,000 HIV-1 V3 sequences in GenBank had this same GPEK V3 crown motif, with 7 of these being from a single subject. As these are just V3 or gp120 sequences, their coreceptor usage patterns have not been determined. Six additional V3 sequences obtained from two earlier samples (26 February and 2 March) all had the identical GPEK crown sequences (Fig. 2B). In addition, the one amino acid deletion at the position 320 in the V3 is also uncommon. Thus, subject ZP6248 appears to have become productively infected with a single T/F virus that exhibited an unusual V3 crown.
Fig. 2.
Identification of a rare GPEK V3 crown sequence in the ZP6248 envelope glycoprotein. (A) Single genome amplification (SGA) was used to infer the transmitted founder (T/F) env sequence as described (28). A Highlighter plot denotes the location of nucleotide substitutions in each SGA-derived env sequence compared to the T/F virus. The position of these substitutions is indicated on the bottom (p16 contains G-to-A changes characteristic of APOBEC-mediated hypermutation). Nucleotide substitutions and gaps are color coded (all env sequences were derived from the 9 March time point). (B) Alignment of Env protein sequences in the V3 domain. Sequences from three time points (9 March, 2 March, and 26 February) are compared to the subtype B consensus sequence. The rare EK V3 crown motif found in all ZP6248 sequences is highlighted in red. Dashes in the alignment indicate sequence identity to the consensus; dots indicate deletions.
ZP6248 T/F Env fails to mediate virus entry into TZM-bl and GHOST cells.
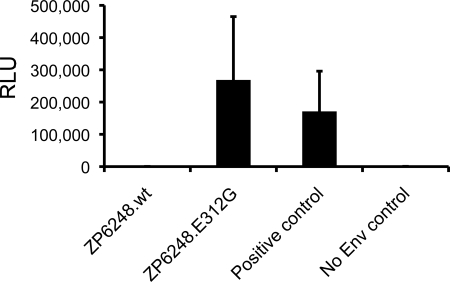
To examine its biological activity, the molecularly cloned ZP6248 T/F env gene was cotransfected with an env-deficient HIV-1 backbone (SG3Δenv). Infectivity of the resulting pseudovirions was assessed in TZM-bl cells, which express CD4 as well as CCR5 and CXCR4. Unlike all previously analyzed T/F Envs (28, 34, 38), the ZP6248 T/F Env construct failed to mediate infection in both TZM-bl cells (Fig. 3) and GHOST (3) cells expressing CD4 and either CCR5 or CXCR4 (data not shown). Thus, despite a very high virus load in the infected individual, the inferred T/F Env failed to mediate detectable virus infection in two commonly used reporter cell lines.
Fig. 3.
Infectivity of pseudoviruses containing ZP6248 Env glycoproteins. Viral pseudotypes bearing the indicated Env glycoproteins were used to infect TZM-bl cells stably expressing CD4, CXCR4, and CCR5. Infectivity was determined by measuring relative light units (RLU) in cell lysates 48 h after infection. ZP6248.wt represents the T/F Env, while ZP6248.E312G contained a single amino acid substitution in the V3 crown. A subtype C Env (ZM53 [32]) was used as a positive control, while Env-deficient pseudotypes (SG3Δenv) were used as a negative control.
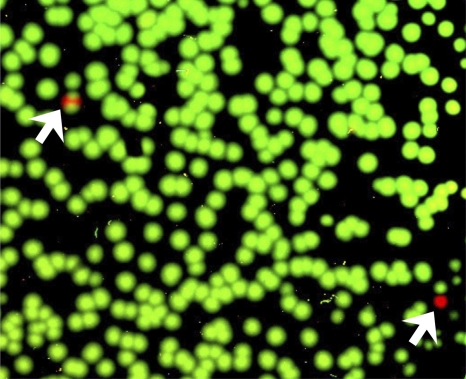
Since it is known that V3 plays an important role in coreceptor binding (13, 14, 25, 50, 51), we introduced a single nucleotide substitution (A->G) into the ZP6248.wt Env to generate a mutant, ZP6248.E312G, with a consensus glycine (G) rather than the aspartic acid (E) at position 312. Pseudovirions generated with the ZP6248.E312G Env mutant were fully infectious in TZM-bl cells expressing CCR5 and CXCR4 (Fig. 3). This finding thus identified the rare GPEK crown motif as the reason for the inability of the ZP6248 Env to mediate infection of TZM-bl cells via either of the two major HIV coreceptors and indicated that a single mutation in the V3 crown sequence restored its infectivity to levels comparable to other primary R5 Envs. Given this, we used a parallel allele-specific sequencing method (7) to determine if viral genomes bearing clade B consensus V3 loop sequences were present in this individual. We analyzed 1,390 viral genomes from the 9 March plasma sample and found only two that contained the E312G sequence (Fig. 4). Therefore, we conclude that the inferred T/F Env accurately reflects the dominant viral genotype in this individual, and that virions bearing the unusual GPEK motif were responsible for the primary infection.
Fig. 4.
Detection of the rare ZP6248.E321G mutation during acute infection. The presence of the ZP6248.E321G mutation (A to G) was examined by parallel allele-specific sequencing (PASS) analysis in plasma collected at the 9 March time point. In this assay, cDNA annealed to an acrydite-modified primer is immobilized in an acrylamide gel, after which in-gel PCR is performed, with the resulting products accumulating around the individual cDNA templates. Sequencing primers that anneal just upstream of the mutation site in V3 (GAA->GGA) can be used to distinguish wt and mutant bases at the same position by single-base extension using Cy3- and Cy5-labeled adenosine (wt) and guanosine (mutant), respectively. The gel was scanned to obtain images with a GenePix 4000B microarray scanner, and the spot number was counted using the Progenesis PG200 software. Green and red spots indicate wt and mutant bases, respectively, detected in individual viral genomes. Two mutants were identified by arrows. A partial image from one of the three experiments is shown.
ZP6248 T/F Env uses multiple alternative coreceptors for cell entry.
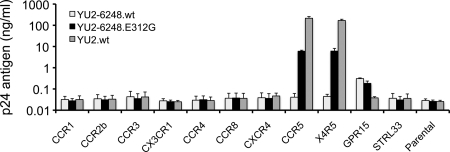
The fact that the T/F Env clone failed to mediate efficient virus entry, coupled with our finding that a single amino acid substitution in the V3 loop rendered the ZP6248 Env functional in the TZM-bl assay, raised the possibility that the T/F virus from which it was derived had an altered in vivo coreceptor preference. It was also possible that the ZP6248 T/F Env recognized CCR5 only when expressed in specific cell types or that it did not pseudotype efficiently. To differentiate between these possibilities, we generated replication-competent YU2 chimeras containing either wild-type (YU2-6248.wt) or mutant (YU2-6248.E312G) ZP6248 env genes and tested these constructs in a panel of GHOST (3) cells expressing CCR1, CCR2b, CCR3, CX3CR1, CCR4, CCR8, CXCR4, CCR5, CCR5/CXCR4, GPR15/BOB, and STRL33/Bonzo. Consistent with the results obtained with pseudovirions, YU2-6248.wt did not infect CCR5+, CXCR4+, or CCR5/CXCR4+ cells but replicated, albeit at very low titers, in GPR15+ cells (Fig. 5). Conversely, YU2-ZP6248.E312G replicated well in CCR5+ and CCR5/CXCR4+ cells but grew somewhat more poorly in GPR15+ cells.
Fig. 5.
Coreceptor usage of ZP6248 Env. GHOST (3) cell lines expressing CD4 and the indicated coreceptors were infected with two replication-competent YU2/ZP6248 chimeras (YU2-6248.wt and YU2-6248.E312G) as well as wild-type HIV-1 YU2.wt. Viruses normalized by p24 content (100 ng) were used to infect cells, and virus replication was monitored by measuring p24 antigen in culture supernatants 7 days after infection. The y axis shows the mean of p24 concentrations from three independent experiments. Error bars represent one standard error of the mean.
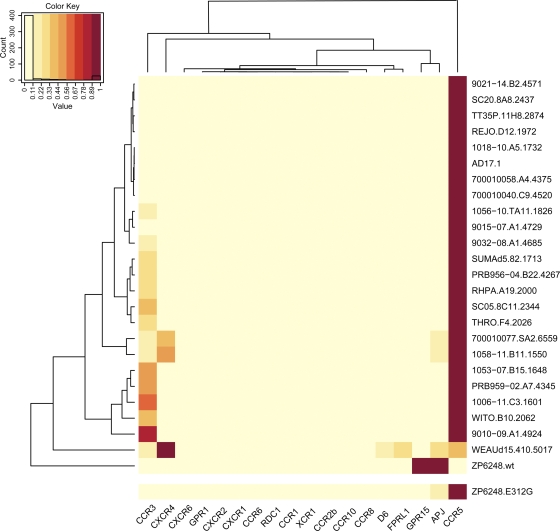
To further test the coreceptor preference of the ZP6248 Env glycoprotein in a different cellular context, we infected NP-2 cells expressing CD4 along with either CCR5, CXCR4, or other G protein-coupled receptors with Env pseudovirions (57). We found that ZP6248.wt mediated nearly equivalent levels of infection on cells expressing GPR15 or APJ and weakly infected cells expressing FPRL-1 or CCR5 (Table 1). Once again, the virus with the E312G mutant exhibited greatly enhanced infectivity in cells expressing CD4 and CCR5. When the GPEK motif was introduced into a different background (HxB2), use of GPR15 was not imparted (data not shown). Thus, the unusual GPEK motif is necessary but not sufficient for the utilization of GPR15. Finally, we compared the coreceptor usage of the ZP6248 Env to that of 24 clade B T/F pseudovirions using a panel of NP-2 cells expressing various G protein-coupled receptors. In stark contrast to the other Envs in this panel which used CCR5 or CXCR4 most efficiently, ZP6248.wt predominantly used GPR15 and APJ, while the use of other coreceptors was negligible (Fig. 6). Additionally, we noted that ZP6248 used GPR15 much better than any other Envs in this panel (data not shown). The ZP6248.E312G mutant, on the other hand, showed a pattern of entry into NP-2 cells indistinguishable from that of many other T/F Envs (Fig. 6). Thus, the ZP6248 T/F Env is clearly distinct from other T/F Envs we have analyzed and failed to mediate efficient infection of cell lines expressing CD4 and CCR5 unless residue 312 in the V3 crown sequence was changed to consensus clade B glycine.
Table 1.
Determination of coreceptor usage of ZP6248 in NP-2 cell linesa
| Env | Usage (RLU) of coreceptor |
||||
|---|---|---|---|---|---|
| GPR15 | APJ | FPRL-1 | CCR5 | CD4 only | |
| ZP6248.wt | 10,991 | 9,972 | 355 | 1,183 | 11 |
| ZP6248.E312G | 1,331 | 6,428 | 2,300 | 45,542 | 6 |
| NL4-3 (X4) | 82 | 9,433 | 6 | 220 | 23 |
| YU2 (R5) | 101 | 940 | 2,256 | 342,801 | 7 |
| NL4-3 without Env | 7 | 6 | 8 | 13 | 8 |
NP-2 cells expressing CD4 and the various potential coreceptors as noted were infected with NL4-3 luc+ pseudovirions. The ability to use the coreceptor was assessed by measuring relative light units (RLUs) in cell lysates and comparing the results to those for CXCR4- and CCR5-using control Envs.
Fig. 6.
Comparison of the ZP6248 Env tropism to that of other subtype B T/F Envs. Entry of ZP6248.wt and ZP6248.E312G pseudotypes into 18 different NP-2 cell lines stably expressing CD4 and the indicated seven-transmembrane domain receptors was compared to that of 24 clade B T/F Envs. The cell line that was infected most efficiently for each virus was normalized to one (dark red), while the entry into the other cell lines was expressed as a fraction of this value as indicated by the key within the figure. Dendrograms on the top and left of the heat map represent hierarchical clustering of the data that show coreceptors with similar patterns of usage by Envs (top) and Envs with similar tropism for coreceptor usage (left).
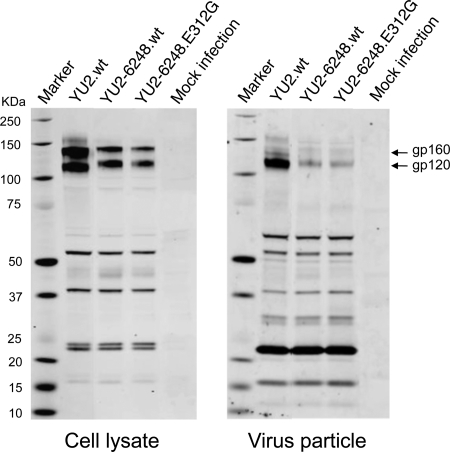
To determine if ZP6248.wt Env was expressed and incorporated into virions properly, we analyzed Env content in cell lysates and secreted virus particles from transfected 293T cells by Western blotting. Compared to YU2, both YU2-6248.wt and YU2-6248.E312G contained less Env, though their cleavage efficiency appeared normal (Fig. 7). However, since the YU2-6248.E312G virus was highly infectious on cells expressing CD4 and CCR5, it is evident that the amount of Env on YU2-6248.wt was not limiting for virus infection.
Fig. 7.
Protein expression from transfected cells. 293T cells were transfected with the proviral clones indicated, and cell lysates and viral supernatants were harvested 48 h posttransfection. Protein expression and processing was examined by Western blot analysis using an HIV-1-positive human serum sample and a mouse MAb (13D5) specific for the HIV-1 Env protein (31). The position of the Env precursor (gp160) and the extracellular domain (gp120) are indicated. Protein standards are indicated on the left.
Full-length ZP6248 T/F virus infects GPR15+ but not CCR5+ GHOST cells.
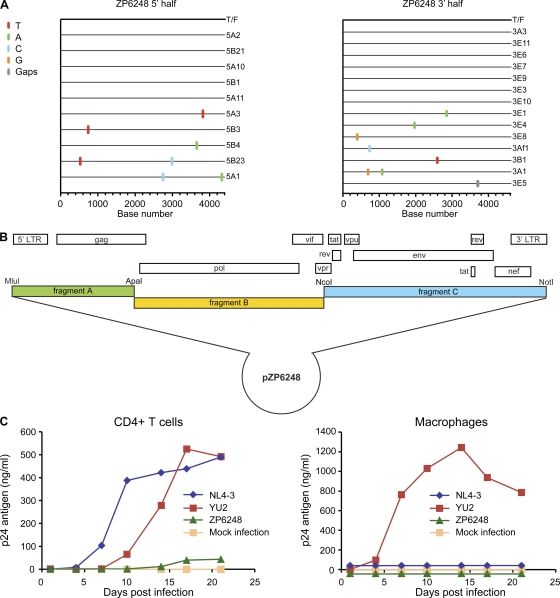
To determine whether the atypical function of the ZP6248 Env would be recapitulated in its regular genomic context, we inferred the sequence of the entire ZP6248 T/F provirus and generated a full-length IMC. Overlapping genomic halves were amplified from plasma viral RNA from the 9 March sample using the SGA method as previously described (54). As observed for the env gene, both half-genome sequences were homogenous and coalesced to an unambiguous consensus sequence representing the T/F virus (Fig. 8A), which was then synthesized and cloned using three subgenomic fragments (Fig. 8B). When virus derived from the molecular clone was used to infect a GHOST cell panel, only GPR15-expressing cells became weakly infected, demonstrating that the full-length ZP6248 T/F virus exhibited the same coreceptor preferences as the viral pseudotypes and the YU2-ZP6248 chimera (data not shown).
Fig. 8.
Molecular cloning and biological characterization of the T/F virus of subject ZP6248. (A) 5′ (4,377 bp) and 3′ (4,528 bp) half-genomes overlapping by 66 bp were amplified by SGA from plasma viral RNA (9 March time point) and used to infer the sequence of the single T/F virus infecting subject ZP6248 as previously described (22, 38, 54). The Highlighter plots denote the location of nucleotide substitutions compared to the inferred T/F sequence, with their position indicated on the bottom. Nucleotide substitutions and gaps are color coded. (B) The full-length ZP6248 genome was synthesized in three overlapping fragments and cloned into a modified pBR322 vector as described previously (59). (C) Viruses harvested from transfected 293T cells were examined for their ability to replicate in activated primary human CD4+ lymphocytes (left) and monocyte-derived macrophages (right) from the same donor. The x axis indicates days postinfection; the y axis denotes p24 antigen concentration (ng/ml) in the culture supernatant.
ZP6248 T/F virus infects CD4+ T cells from ccr5Δ32 homozygous donors.
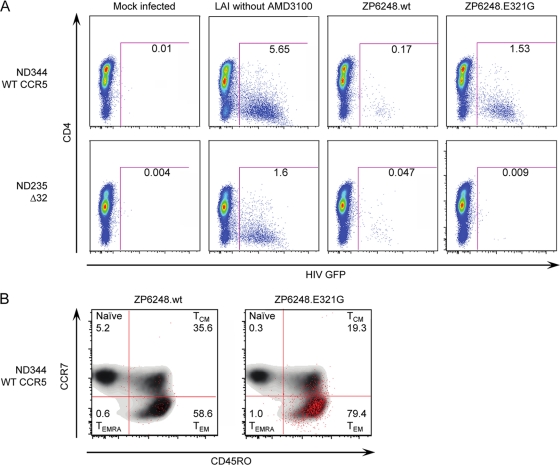
We next sought to determine if ZP6248 Env could mediate efficient infection of primary human CD4+ T cells and if so, to identify the receptors used for virus entry. Unfortunately, cells were not available from this donor, though we were able to determine that this individual had a wild-type ccr5 genotype (not shown). We obtained both ccr5 wt and ccr5Δ32 homozygous human CD4+ T cells from several donors and performed infection assays using ZP6248.wt and ZP6248.E312G Env pseudovirions expressing a GFP reporter. One of five representative experiments using cells from different ccr5 wt and ccr5Δ32 homozygous donors is shown in Fig. 9A. In the presence of the CXCR4 inhibitor AMD3100, ZP6248.wt pseudoviruses infected 0.17% of ccr5 wt cells and 0.047% of ccr5Δ32 homozygous cells compared to 1.53% and 0.009% for ZP6248.E312G pseudovirions, respectively. Similar results were obtained in the absence of AMD3100 using cells from four other ccr5 wt donors and one additional ccr5Δ32 homozygous donor. In addition, the replication-competent ZP6248 T/F virus replicated with severely delayed kinetics and to a much lower titer in CD4+ T cells, and it failed to replicate in macrophages (Fig. 8C). Interestingly, prolonged culture of the ZP6248 T/F virus in CD4+ T cells resulted in the selection of a variant with a GPGK V3 crown motif that conferred efficient CCR5 use (data not shown). Finally, the ZP6248 T/F virus replicated even more poorly and only at high inoculum counts in ccr5Δ32 homozygous CD4+ T cells (data not shown). Thus, Env pseudovirions and IMC-derived virus bearing an unusual V3 loop sequence could utilize one or more alternative coreceptors to mediate inefficient entry into ccr5Δ32 homozygous cells, although it also used CCR5 poorly.
Fig. 9.
Infection of human CD4+ T cells. Env pseudovirions expressing a GFP reporter were used to infect primary ccr5 Δ32/Δ32 or ccr5 wt CD4+ T cells in the presence of the CXCR4 antagonist AMD3100. Viral infection was assessed by flow cytometry after gating on live CD3+ cell singlets. (A) ccr5 Δ32/Δ32 or ccr5 wt CD4+ T cells were infected with ZP6248.wt and ZP6248.E312G in the presence of AMD3100. Mock-infected cells were used as a negative control, while the X4 HIV-1 strain LAI was used as a positive control in the absence of AMD3100. (B) Infected cells were back-gated onto memory markers CCR7 and CD45R0 (CCR7+ CD45RO−, naïve; CCR7+ CD45RO+, central memory; CCR7− CD45RO+ effector memory; CCR7− CD45RO−, effector memory RA) to evaluate memory subset tropism of ZP6248.wt and ZP6248.E312G on CCR5 wild-type cells (ND344). The uninfected cells are shown in a contour plot to better reveal the distribution of infected (GFP+ cells) shown as red dots. 174 and 1,742 GFP+ events were collected for PZ6248.wt and ZP6248.E312G infected cells, respectively. The data are shown from one of five independent experiments with cells from five ccr5 wt donors and two ccr5Δ32 homozygous donors.
Tropism analysis of the ZP6248 T/F virus.
The failure of the ZP6248 T/F Env to mediate efficient infection of primary CD4+ T cells raised the possibility that it was uniquely adapted to its host, which, in turn, could have exhibited unusual features, such as a typical CCR5 expression, posttranslational processing, or conformation. While cells were not available from this donor, we reasoned that a more detailed analysis of viral tropism might shed light on this issue. Therefore, we employed a highly sensitive CD4+ T cell subset tropism assay to determine if virions bearing the ZP6248.wt or ZP6248.E312G Envs exhibited any unusual properties. CD4+ T cells were obtained from several donors and infected with GFP-expressing pseudovirions. Cells from individual donors were stained with CCR7 and CD45RO to define naïve, central memory (TCM), effector memory (TEM), and effector memory RA cell subsets (TEMRA). HIV-1-infected cells were then back-gated to evaluate tropism among different CD4+ T cell subsets. 5.2% of ZP6248.wt pseudovirus-infected cells were naïve, 35.6% were TCM, 58.6% were TEM, and 0.6% were TEMRA, compared to 0.3%, 19.3%, 79.4%, and 1.0% for ZP6248.E312G, respectively (Fig. 9B). This distribution of infected cells among the various CD4+ T cell subsets is similar to what we have observed for other clade B viruses, indicating that the ZP6248.wt Env and ZP6248.E312G Envs did not exhibit unusual CD4+ T cell subset preference. Similar results were obtained when cells from four additional donors were analyzed (data not shown). There were no consistent trends in the tropism for naïve cells for ZP6248.wt versus ZP6248.E312G across five independent experiments.
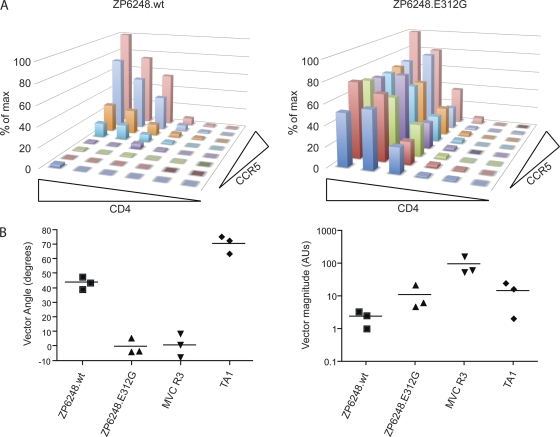
We also took a second approach to examine the tropism of ZP6248 Env pseudovirions, performing infection assays on Affinofile cells, which are a human 293T cell line and express CD4 and CCR5 under independent inducible promoters, making it possible to modulate receptor levels across a broad physiologic range (27). The Affinofile cells were infected under various CD4 and CCR5 expression conditions with pseudoviruses bearing the ZP6248.wt or ZP6248.E312G Envs. We also performed infection assays with two control viruses: TA1 requires high levels of CCR5 for efficient virus infection, while MVC R3 can infect cells expressing very low levels of CCR5 (44). We found that ZP6248.wt Env required relatively high levels of both CCR5 and CD4 for infectivity, while ZP6248.E312G Env could utilize very low levels of CCR5 at moderate CD4 expression levels (Fig. 10A). The background luciferase expression on NP-2 and Affinofile cells is 2 to 3 orders of magnitude lower than that of TZM-bl cells (64). This significantly increases the detection sensitivity. In the Affinofile cell system, the sensitivity to changes in CD4 and CCR5 use can be quantified by determining a vector angle (27). A low vector angle is consistent with very efficient CCR5 use, while a high vector angle is associated with inefficient CCR5 use (35, 61). The ZP6248.wt Env had a vector angle of 45°, consistent with its being equally dependent upon CCR5 and CD4 expression levels, while the ZP6248.E312G Env had a very low vector angle, consistent with very efficient CCR5 use at intermediate to high expression levels of CD4 (Fig. 10B). Of note, the infection level of ZP6248.wt pseudovirions was nearly one order of magnitude lower than that of ZP6248.E312G pseudovirions and TA1 and nearly two orders of magnitude lower than that of MVC R3 (Fig. 10B). Taken together, these findings are consistent with previous cell line and primary cell data, which show that ZP6248.wt Env can utilize CCR5 for entry but does so very inefficiently and only when CCR5 and CD4 expression levels are high.
Fig. 10.
ZP6248.wt inefficiently utilizes CCR5. (A) ZP6248.wt and ZP6248.E312G pseudovirions were used to infect affinofile cells which express CD4 and CCR5 with independent induction systems. ZP6248.wt requires high CCR5 expression for infection compared to ZP6248.E312G, which can utilize very low levels of CCR5. Both Envs require moderate CD4 expression for infection. (B) The sensitivity to changes in CD4 and CCR5 levels was quantified by viral entry receptor sensitivity analysis (VERSA), which yields a single vector angle. Lower vector angles correspond to Envs that utilize CCR5 very efficiently as seen with the maraviroc-resistant Env MVC R3 (1°), while higher vector angles are associated with inefficient CCR5 use as seen with the V3-loop truncated virus TA1 (71°). ZP6248.wt has a vector angle of 44° compared to 0° for ZP6248.E312G. In addition, the vector magnitude can be quantified to assess overall infectivity. ZP6248.wt is 5-fold less infectious than ZP6248.E312G and 40-fold less infectious than MVC R3, consistent with other cell lines and primary cell data.
Expression of coreceptors on the surface of CD4+ T cells.
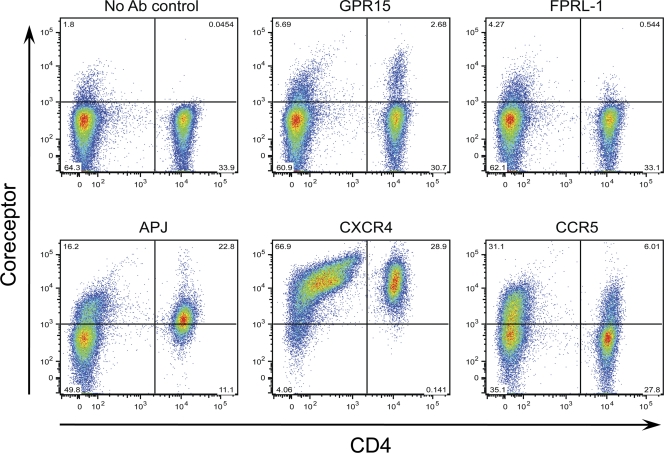
Since ZP6248 was able to infect CD4+ T cells without using CCR5 or CXCR4 coreceptors, albeit inefficiently, we asked whether alternative coreceptors, such as GPR15, APJ, and FPRL-1, were expressed on the surface of these target cells. We thus performed flow cytometry analysis of PBMCs from five normal healthy donors using antibodies specific for GPR15, APJ, and FPRL-1. GPR15 was detected on 7.4% of CD4+ T cells, while APJ was present on 53.4% of CD4+ T cells (Fig. 11). FPRL-1 was also detected but at a much lower frequency (1.1% of CD4+ T cells). As expected, CXCR4 was found on nearly all CD4+ T cells, while CCR5 was expressed on 12.5% CD4+ T cells. Similar results were obtained for GPR15, APJ, and FPRL-1 on both unstimulated and PHA-treated PBMCs. These results demonstrate that GPR15, APJ, and FPRL-1 are all expressed on the surface of CD4+ T cells and could thus serve as alternative coreceptors for ZP6248 in vivo.
Fig. 11.
Coreceptor expression on peripheral blood mononuclear cells. PHA-stimulated or nonstimulated cells were labeled with antibodies specific for GPR15, FPRL-1, APJ, CXCR4, and CCR5, and the percentage of positive cells was determined. Representative results from one of five blood donors are shown. The x axis indicates CD4+ T cells, while the y axis denotes cells expressing the tested coreceptors. No significant differences for expression of GPR15, FPRL-1, and APJ were noticed on PHA-stimulated and nonstimulated cells, and the results from nonstimulated cells are shown.
DISCUSSION
HIV-1 infection is primarily established by viruses that use CCR5 for entry (24, 28, 41, 44, 54). In this study, we identified and characterized a T/F virus, ZP6248, that replicated to a VL of 87.7 million copies/ml during acute infection in vivo, yet its Env glycoprotein failed to efficiently facilitate virus entry into CCR5 or CXCR4-bearing target cells in vitro. This finding was clearly at odds with previous results showing that T/F viruses are fully functional and invariably use CCR5 alone or, less commonly, in combination with CXCR4 to infect cells (28, 34). We sought to determine if the poor functionality of this T/F Env was due to methodological issues or whether this virus established infection in a naïve host by utilizing unusual viral and/or host factors that could provide insight into the mechanisms of HIV-1 transmission.
A trivial explanation that could account for the poor functionality of the ZP6248 T/F Env is that its sequence does not reflect that of the virus that established infection in this host. Because the SGA and bioinformatics approaches we have taken to identify multiple T/F Envs have invariably yielded fully functional Env proteins that utilize CCR5 (and sometimes CXCR4 as well) to infect cells, this explanation is highly unlikely. As is common in recently infected individuals, extensive sequencing of env shortly after this individual became infected showed very limited genetic diversity, and so our inference of the T/F env sequence is unambiguous. In addition, the ZP6248 T/F Env exhibited a highly unusual GPEK V3 crown sequence. All env genes sequenced at three early time points after infection contained this motif, as did all but 2 of 1,390 genomes analyzed by PASS. lt is important to note that this frequency (0.15%) is well above the level of background in this assay (39); therefore, a variant capable of using CCR5 efficiently in vitro was present in the acutely infected patient but was overshadowed in vivo by a variant with poorer in vitro CCR5 use. Importantly, all V3 sequences obtained from three different PCR primer sets (whole env gene, partial env gene, and half viral genome) were identical, except two GPGK variants identified by PASS. Thus, this individual was infected by a virus with a very unusual phenotype.
Another technical issue that could account for poor viral infectivity would be if the ZP6248 T/F Env is packaged into virions poorly. We think this possibility is unlikely, as we analyzed the ZP6248 T/F Env in four viral backgrounds: viral pseudotypes based on HIV-1 HxB2 and SG3 backbones, a replication-competent HIV-1 YU2 chimera, and the cognate T/F virus derived from subject ZP6248. In addition, immunoblots revealed levels of correctly processed ZP6248 Env in virus particles that were comparable to levels of the fully functional ZP6248.E312G mutant. Therefore, the low infection levels cannot be explained by poor expression, processing, or virion incorporation of Env. In addition, it is important to stress that the single E312G mutation in the V3 loop rendered the corresponding Env fully functional in virus infection assays, and it is difficult to envision how such a change could influence Env incorporation into virus particles.
Despite adequate levels of expression and processing, the ZP6248 Env exhibited poor functionality on two different, commonly used cell lines as well as on primary human CD4+ T cells from 14 different donors. Detailed tropism studies revealed no unusual properties with regard to T cell subset infection, though relatively high levels of CCR5 and CD4 were needed to support infection. In addition, the sequence of the ZP6248 Env has no unusual genetic features save for the highly atypical V3 loop crown. Based on its sequence, this Env is predicted to use CCR5. Might this virus have used CCR5 to mediate efficient virus infection in vivo, even though it failed to do so in vitro? If so, we would predict that there was something unusual about the manner in which CCR5 was presented on the surface of cells in this individual—otherwise we should have observed robust infection in our in vitro assays. Unfortunately, we cannot rule out this possibility, as cells from this individual are not available for study, although we were able to obtain enough cellular debris from plasma to determine that both CCR5 alleles had wild-type sequences in the ORF. In addition, the unusual V3 loop sequence in this individual provides some clues: it is thought that the tip of the V3 loop interacts with the second extracellular loop of the viral coreceptors (25, 65), and this region of CCR5 (and CXCR4) can exhibit conformational heterogeneity (5, 36). In fact, CCR5 inhibitors such as maraviroc alter the conformation of the extracellular loops of CCR5 thus blocking interactions with Env (17, 33, 55). One mechanism by which HIV-1 can become resistant to these allosteric inhibitors is through mutations in the V3 loop that enable it to interact with the altered receptor conformation (2, 6, 45, 46, 61, 63). In addition, several studies have shown via the use of panels of conformation-specific monoclonal antibodies that CCR5 exhibits conformational heterogeneity on the surface of human CD4+ T cells, though the mechanisms that account for this as well as the implications of altered conformation on viral infectivity are not known (5, 36). Thus, it is possible that CCR5 in this individual exhibited an unusual conformation that was not recapitulated by the cells used in our in vitro assays, and that the ZP6248 T/F Env with its unusual V3 sequence was well adapted to use this conformation.
Another possibility is that virus in this patient used a coreceptor other than CCR5 to initiate infection. If so, this would be unprecedented for HIV-1 but not for SIV; SIVrcm from red-capped mangabeys often uses CCR2 as a coreceptor, as genetic absence of CCR5 is common in this species (8). In addition, SIVsmm from sooty mangabeys can also use a coreceptor other than CCR5 to mediate infection in vivo: genetic absence of CCR5 is relatively common in sooty mangabeys, but CCR5-negative animals are still infected with SIVsmm, though their virus loads are, on average, one-half log less than what is typically seen in CCR5+ animals (49). The coreceptor used by SIVsmm to replicate in sooty mangabeys has not yet been identified, though in vitro and genetic studies appear to rule out the use of CXCR4. Therefore, in at least two nonhuman primate species, robust in vivo replication by SIV can be achieved by utilization of coreceptors other than CCR5 or CXCR4.
Several HIV-1 strains have been shown to employ coreceptors other than CCR5 or CXCR4 to infect cells (3, 4, 10, 23, 47, 66), with CCR3, GPR15, APJ, and FPRL-1 being among those most frequently used (11, 26, 43, 47, 57). However, these studies have typically employed cell lines that likely overexpress these coreceptors, so it is difficult to assess whether utilization of these coreceptors can lead to infection of primary human cell types. In fact, with few exceptions, HIV-1 strains fail to infect human T cells from individuals who lack CCR5 in the presence of the CXCR4 inhibitor AMD3100, arguing that in this context alternative coreceptors typically cannot mediate infection (11, 56). However, ZP6248 was able to use three alternative coreceptors (GPR15, APJ, and FPRL-1) to infect cell lines, using the former two much more efficiently than CCR5. This pattern is strikingly different from that of other clade B T/F Envs recently analyzed in NP-2 cells expressing various putative alternative coreceptors (64). Therefore, it is reasonable to ask if these receptors are expressed in vivo on cell types relevant to transmission. Previous studies have shown GPR-15 mRNA in human colonic tissue and CD4+ T lymphocytes (12, 15, 19), FPRL-1 in a large variety of cells and organs, including T and B lymphocytes (42), and APJ in the human brain (18) and in activated PBMCs (9). Using currently available antibodies, we have found that GPR15 and APJ are expressed on the surface of 7.7% and 56% of CD4+ T cells, respectively, while FPRL-1 can be found on only about 1% of CD4+ T cells. The presence of these coreceptors on the surface of CD4+ T cells suggests that they may be used by ZP6248 in vivo, with GPR-15 as the most likely candidate.
Almost all T/F HIV-1 Envs use CCR5 efficiently in vitro and presumably in vivo as well, given the resistance of individuals who lack CCR5 to virus infection. However, this study reinforces the idea that the Env glycoprotein is incredibly plastic, and it shows that our in vitro assays at times fail to recapitulate important virological properties that operate in vivo. Whether by exploiting an atypical CCR5 conformation or by using coreceptors not previously known to mediate transmission, the virus that infected subject ZP6248 did so without efficient use of CCR5 in its standard form in multiple assays. In light of this, while attempting to block CCR5 use as an option for target cell entry (e.g., by maraviroc-containing microbicides) will likely be an effective prevention strategy, our report cautions that atypical presentation of CCR5 or alternative coreceptors may sometimes be used by HIV to result in the establishment of a primary infection.
ACKNOWLEDGMENTS
We thank Hiroo Hoshino for NP-2 cell lines, Haili Zhang, Yan Zhou, and Robert Siliciano for bicyclam JM-2987 and pNL4-3-deltaE-EGFP, Brooke Walker for editorial assistance, and the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH for GHOST (3) cell lines.
This work was supported by NIH grants AI067854 (B.F.H., B.H.H., G.M.S., and F.G.), AI040880 (R.W.D.), Bill and Melinda Gates Foundation grants 37874, 38691, and 38643 (B.F.H., B.H.H., and G.M.S.), and the Duke and UAB Centers for AIDS Research (AI064518 and AI27767). C.B.W. and N.F.P. were supported by T32 AI007632 and T32 GM008361, respectively.
Footnotes
Published ahead of print on 10 August 2011.
REFERENCES
- 1. Abrahams M. R., et al. 2009. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J. Virol. 83:3556–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baba M., Miyake H., Wang X., Okamoto M., Takashima K. 2007. Isolation and characterization of human immunodeficiency virus type 1 resistant to the small-molecule CCR5 antagonist TAK-652. Antimicrob. Agents Chemother. 51:707–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Begaud E., et al. 2003. Broad spectrum of coreceptor usage and rapid disease progression in HIV-1-infected individuals from Central African Republic. AIDS Res. Hum. Retroviruses 19:551–560 [DOI] [PubMed] [Google Scholar]
- 4. Berger E. A., Murphy P. M., Farber J. M. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657–700 [DOI] [PubMed] [Google Scholar]
- 5. Berro R., et al. 2011. Multiple CCR5 conformations on the cell surface are used differentially by human immunodeficiency viruses resistant or sensitive to CCR5 inhibitors. J. Virol. 85:8227–8240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berro R., Sanders R. W., Lu M., Klasse P. J., Moore J. P. 2009. Two HIV-1 variants resistant to small molecule CCR5 inhibitors differ in how they use CCR5 for entry. PLoS Pathog. 5:e1000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cai F., et al. 2007. Detection of minor drug-resistant populations by parallel allele-specific sequencing. Nat. Methods 4:123–125 [DOI] [PubMed] [Google Scholar]
- 8. Chen Z., et al. 1998. Natural infection of a homozygous delta24 CCR5 red-capped mangabey with an R2b-tropic simian immunodeficiency virus. J. Exp. Med. 188:2057–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choe H., et al. 1998. The orphan seven-transmembrane receptor apj supports the entry of primary T-cell-line-tropic and dualtropic human immunodeficiency virus type 1. J. Virol. 72:6113–6118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cilliers T., et al. 2003. The CCR5 and CXCR4 coreceptors are both used by human immunodeficiency virus type 1 primary isolates from subtype C. J. Virol. 77:4449–4456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cilliers T., et al. 2005. Use of alternate coreceptors on primary cells by two HIV-1 isolates. Virology 339:136–144 [DOI] [PubMed] [Google Scholar]
- 12. Clayton F., et al. 2001. Gp120-induced Bob/GPR15 activation: a possible cause of human immunodeficiency virus enteropathy. Am. J. Pathol. 159:1933–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cormier E. G., Dragic T. 2002. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J. Virol. 76:8953–8957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cormier E. G., Tran D. N., Yukhayeva L., Olson W. C., Dragic T. 2001. Mapping the determinants of the CCR5 amino-terminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes. J. Virol. 75:5541–5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deng H. K., Unutmaz D., KewalRamani V. N., Littman D. R. 1997. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature 388:296–300 [DOI] [PubMed] [Google Scholar]
- 16. Doranz B. J., et al. 1997. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J. Virol. 71:6305–6314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dragic T., et al. 2000. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc. Natl. Acad. Sci. U. S. A. 97:5639–5644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edinger A. L., et al. 1998. An orphan seven-transmembrane domain receptor expressed widely in the brain functions as a coreceptor for human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol. 72:7934–7940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Farzan M., et al. 1997. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J. Exp. Med. 186:405–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Faulkner D. V., Jurka J. 1988. Multiple aligned sequence editor (MASE). Trends Biochem. Sci. 13:321–322 [DOI] [PubMed] [Google Scholar]
- 21. Fiebig E. W., et al. 2003. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS 17:1871–1879 [DOI] [PubMed] [Google Scholar]
- 22. Gnanadurai C. W., et al. 2010. Genetic identity and biological phenotype of a transmitted/founder virus representative of nonpathogenic simian immunodeficiency virus infection in African green monkeys. J. Virol. 84:12245–12254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goodenow M. M., Collman R. G. 2006. HIV-1 coreceptor preference is distinct from target cell tropism: a dual-parameter nomenclature to define viral phenotypes. J. Leukoc. Biol. 80:965–972 [DOI] [PubMed] [Google Scholar]
- 24. Gorry P. R., et al. 2002. Persistence of dual-tropic HIV-1 in an individual homozygous for the CCR5 Delta 32 allele. Lancet 359:1832–1834 [DOI] [PubMed] [Google Scholar]
- 25. Huang C. C., et al. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Isaacman-Beck J., et al. 2009. Heterosexual transmission of human immunodeficiency virus type 1 subtype C: macrophage tropism, alternative coreceptor use, and the molecular anatomy of CCR5 utilization. J. Virol. 83:8208–8220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Johnston S. H., et al. 2009. A quantitative affinity-profiling system that reveals distinct CD4/CCR5 usage patterns among human immunodeficiency virus type 1 and simian immunodeficiency virus strains. J. Virol. 83:11016–11026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Keele B. F., et al. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Keele B. F., et al. 2009. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J. Exp. Med. 206:1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111–120 [DOI] [PubMed] [Google Scholar]
- 31. Kirchherr J. L., et al. 2011. Identification of amino acid substitutions associated with neutralization phenotype in the human immunodeficiency virus type-1 subtype C gp120. Virology 409:163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kirchherr J. L., et al. 2007. High throughput functional analysis of HIV-1 env genes without cloning. J. Virol. Methods 143:104–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kondru R., et al. 2008. Molecular interactions of CCR5 with major classes of small-molecule anti-HIV CCR5 antagonists. Mol. Pharmacol. 73:789–800 [DOI] [PubMed] [Google Scholar]
- 34. Kraus M. H., et al. 2010. A rev1-vpu polymorphism unique to HIV-1 subtype A and C strains impairs envelope glycoprotein expression from rev-vpu-env cassettes and reduces virion infectivity in pseudotyping assays. Virology 397:346–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Laakso M. M., et al. 2007. V3 loop truncations in HIV-1 envelope impart resistance to coreceptor inhibitors and enhanced sensitivity to neutralizing antibodies. PLoS Pathog. 3:e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee B., et al. 1999. Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J. Biol. Chem. 274:9617–9626 [DOI] [PubMed] [Google Scholar]
- 37. Lee H. Y., et al. 2009. Modeling sequence evolution in acute HIV-1 infection. J. Theor. Biol. 261:341–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li H., et al. 2010. High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog. 6:e1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu J., et al. 2011. Analysis of low frequency mutations associated with drug-resistance to raltegravir before antiretroviral treatment. Antimicrob. Agents Chemother. 55:1114–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lu X., Hora B., Cai F., Gao F. 2010. Generation of random mutant libraries with multiple primers in a single reaction. J. Virol. Methods 167:146–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Michael N. L., et al. 1998. Exclusive and persistent use of the entry coreceptor CXCR4 by human immunodeficiency virus type 1 from a subject homozygous for CCR5 delta32. J. Virol. 72:6040–6047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Migeotte I., Communi D., Parmentier M. 2006. Formyl peptide receptors: a promiscuous subfamily of G protein-coupled receptors controlling immune responses. Cytokine Growth Factor Rev. 17:501–519 [DOI] [PubMed] [Google Scholar]
- 43. Nedellec R., et al. 2009. Virus entry via the alternative coreceptors CCR3 and FPRL1 differs by human immunodeficiency virus type 1 subtype. J. Virol. 83:8353–836319553323 [Google Scholar]
- 44. O'Brien T. R., et al. 1997. HIV-1 infection in a man homozygous for CCR5 delta 32. Lancet 349:1219. [DOI] [PubMed] [Google Scholar]
- 45. Ogert R. A., et al. 2008. Mapping resistance to the CCR5 co-receptor antagonist vicriviroc using heterologous chimeric HIV-1 envelope genes reveals key determinants in the C2-V5 domain of gp120. Virology 373:387–399 [DOI] [PubMed] [Google Scholar]
- 46. Pfaff J. M., et al. 2010. HIV-1 resistance to CCR5 antagonists associated with highly efficient use of CCR5 and altered tropism on primary CD4+ T cells. J. Virol. 84:6505–6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pohlmann S., Krumbiegel M., Kirchhoff F. 1999. Coreceptor usage of BOB/GPR15 and Bonzo/STRL33 by primary isolates of human immunodeficiency virus type 1. J. Gen. Virol. 80:1241–1251 [DOI] [PubMed] [Google Scholar]
- 48. Quinonez R., Sinha I., Singh I. R., Sutton R. E. 2003. Genetic footprinting of the HIV co-receptor CCR5: delineation of surface expression and viral entry determinants. Virology 307:98–115 [DOI] [PubMed] [Google Scholar]
- 49. Riddick N. E., et al. 2010. A novel CCR5 mutation common in sooty mangabeys reveals SIVsmm infection of CCR5-null natural hosts and efficient alternative coreceptor use in vivo. PLoS Pathog. 6:e1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rizzuto C., Sodroski J. 2000. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res. Hum. Retroviruses 16:741–749 [DOI] [PubMed] [Google Scholar]
- 51. Rizzuto C. D., et al. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949–1953 [DOI] [PubMed] [Google Scholar]
- 52. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 53. Salazar-Gonzalez J. F., et al. 2008. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 82:3952–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Salazar-Gonzalez J. F., et al. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med. 206:1273–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Seibert C., et al. 2006. Interaction of small molecule inhibitors of HIV-1 entry with CCR5. Virology 349:41–54 [DOI] [PubMed] [Google Scholar]
- 56. Sharron M., et al. 2000. Expression and coreceptor activity of STRL33/Bonzo on primary peripheral blood lymphocytes. Blood 96:41–49 [PubMed] [Google Scholar]
- 57. Shimizu N., et al. 2009. Broad usage spectrum of G protein-coupled receptors as coreceptors by primary isolates of HIV. AIDS 23:761–769 [DOI] [PubMed] [Google Scholar]
- 58. Stone M., et al. 2010. A limited number of SIVenv variants are transmitted to rhesus macaques vaginally inoculated with SIVmac251. J. Virol. 84:7083–7095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Takehisa J., et al. 2009. Origin and biology of simian immunodeficiency virus in wild-living western gorillas. J. Virol. 83:1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tilton J. C., et al. 2010. A maraviroc-resistant HIV-1 with narrow cross-resistance to other CCR5 antagonists depends on both N-terminal and extracellular loop domains of drug-bound CCR5. J. Virol. 84:10863–10876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Trkola A., et al. 2001. Potent, broad-spectrum inhibition of human immunodeficiency virus type 1 by the CCR5 monoclonal antibody PRO 140. J. Virol. 75:579–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tsibris A. M., et al. 2008. In vivo emergence of vicriviroc resistance in a human immunodeficiency virus type 1 subtype C-infected subject. J. Virol. 82:8210–8214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wilen C. B., et al. 2011. Phenotypic and immunologic comparison of clade B transmitted/founder and chronic HIV-1 envelope glycoproteins. J. Virol. 85:8514–8527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Xiang S. H., et al. 2005. Functional mimicry of a human immunodeficiency virus type 1 coreceptor by a neutralizing monoclonal antibody. J. Virol. 79:6068–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Xiao L., Rudolph D. L., Owen S. M., Spira T. J., Lal R. B. 1998. Adaptation to promiscuous usage of CC and CXC-chemokine coreceptors in vivo correlates with HIV-1 disease progression. AIDS 12:F137–143 [DOI] [PubMed] [Google Scholar]
- 67. Yeh W. W., et al. 2010. Autologous neutralizing antibodies to the transmitted/founder viruses emerge late after simian immunodeficiency virus SIVmac251 infection of rhesus monkeys. J. Virol. 84:6018–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]