Abstract
Protein–protein interactions play crucial roles in the execution of various biological functions. Accordingly, their comprehensive description would contribute considerably to the functional interpretation of fully sequenced genomes, which are flooded with novel genes of unpredictable functions. We previously developed a system to examine two-hybrid interactions in all possible combinations between the ≈6,000 proteins of the budding yeast Saccharomyces cerevisiae. Here we have completed the comprehensive analysis using this system to identify 4,549 two-hybrid interactions among 3,278 proteins. Unexpectedly, these data do not largely overlap with those obtained by the other project [Uetz, P., et al. (2000) Nature (London) 403, 623–627] and hence have substantially expanded our knowledge on the protein interaction space or interactome of the yeast. Cumulative connection of these binary interactions generates a single huge network linking the vast majority of the proteins. Bioinformatics-aided selection of biologically relevant interactions highlights various intriguing subnetworks. They include, for instance, the one that had successfully foreseen the involvement of a novel protein in spindle pole body function as well as the one that may uncover a hitherto unidentified multiprotein complex potentially participating in the process of vesicular transport. Our data would thus significantly expand and improve the protein interaction map for the exploration of genome functions that eventually leads to thorough understanding of the cell as a molecular system.
Genome projects have revealed a number of novel genes from our genomes as well as those of various model organisms and a number of unique microorganisms (http://www.ncbi.nlm.nih.gov/Entrez/Genome/main_genomes.html). However, the vast majority of the genes revealed by genome sequencing lack any clue as to their specific functions. We have failed to predict functions for almost half of the genes even in the genomes of Escherichia coli and Saccharomyces cerevisiae, which have been extensively studied in molecular genetics (1, 2). It thus becomes evident that efforts other than structural analysis are necessary to fully exploit the genome data, and hence functional genomics was launched by using a variety of systematic approaches.
One of the most straightforward endeavors to reveal gene functions is their systematic disruption, which is achieved by homologous recombination in microorganisms like S. cerevisiae and Bacillus subtilis (3, 4) and, albeit functionally, by RNA interference in the nematoda Caenorhabditis elegans (5, 6). However, the genes identified for the first time by the genome projects in these traditional model organisms would be those having escaped a variety of phenotypic screens or those refractory to pursuit by genetic approaches. It is thus conceivable that only a fraction of such mutants display distinct cellular phenotypes unless novel examinations are introduced into the systematic screen.
In this context, more promising would be the comprehensive analysis of biomolecules such as mRNAs, proteins, and metabolites. Currently, the most powerful approach is the transcriptome analysis or expression profiling based on microarray or DNA chip technologies (7). Accumulation of gene expression data under various conditions has allowed one to classify genes into distinct classes, each of which shares a unique expression profile and is presumably under the same regulatory mechanism. The functions of well-characterized genes would give insight into those of novel ones in the same cluster.
Note that, albeit its power, expression profiling is essentially an indirect measure for biological process. Much finer information would be obtained by the analysis of proteins per se, which actually bear various biological functions (8). Here one should remember that any protein fails to execute its function unless it interacts with other biomolecules. In particular, interactions with other proteins are of extreme importance and can serve as highly informative hints for functional prediction: physical association between a novel protein and a well-characterized one readily indicates that the former has a function related to that of the latter. Comprehensive analysis of protein–protein interactions would thus comprise an integral part of functional genomics (8).
However, in contrast with nucleic acids, natures of proteins are so variable from one to another that genomewide exploring of their interactions cannot be fully accomplished by any single methodology. It is obvious that a variety of complementary approaches should be undertaken toward the completion of the protein interaction map. The plausible approaches can be divided into two categories: the top-down proteomic approach and the bottom-up genomic one. The former is represented by the mass spectrometric analysis of native protein complexes purified mainly by affinity capture (8). The latter are the approaches in which each protein encoded in the genome of interest is expressed for the examination of mutual interactions. These include the yeast two-hybrid system, phage display, protein chip, and so forth. Among these genomic approaches, the yeast two-hybrid system (9) is currently the only one that is so well-established to be used in a genomewide scale. For instance, we had launched a large-scale two-hybrid analysis of the budding yeast S. cerevisiae by developing a comprehensive screening system to examine interactions in all possible combinations between the ≈6,000 proteins encoded by its fully sequenced genome (10). The results of our pilot phase project as well as those by others (11) have clearly demonstrated the feasibility and power of the approach.
Here we have completed the systematic analysis to provide a two-hybrid dataset that, in conjunction with those by others, substantially expands our knowledge on the putative protein–protein interactions occurring in the budding yeast. Accumulation of these pair-wise or binary interactions reveals various intriguing and/or unexpected nexus of proteins, thereby providing testable hypotheses and useful hints for the functions of many novel proteins. Comparison between these two efforts also clarified the limitations inherent to large-scale two-hybrid protein interaction mapping, thereby giving invaluable lessons to similar projects involving other organisms.
Materials and Methods
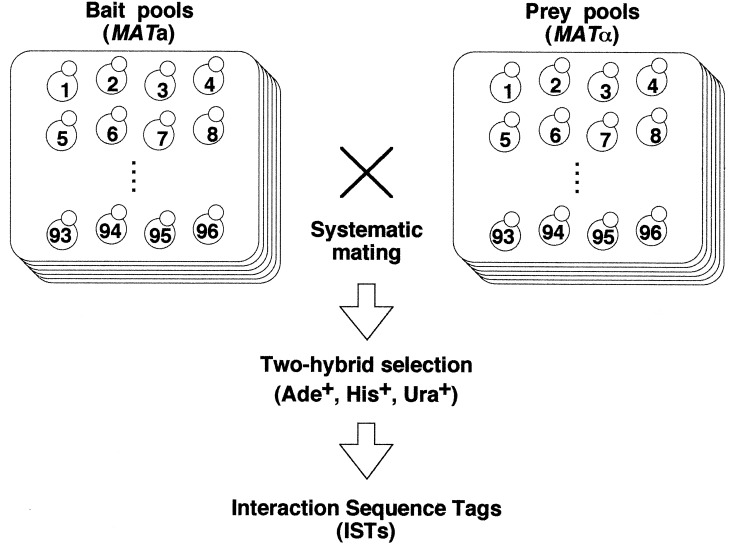
The comprehensive two-hybrid screening system has been described (10) and is summarized briefly below. We amplified each ORF by PCR using Pfu DNA polymerase and cloned into pGBK-RC, a Gal4 DNA-binding domain-based bait vector, and pGAD-RC, a Gal4 activation domain-based prey vector. We confirmed that all of these plasmids bear inserts of the expected sizes by using colony PCR followed by agarose gel electrophoresis. By our unique transformation procedure using 96-well plates, each bait plasmid was introduced into a MATa two-hybrid strain PJ69–2A bearing GAL2∷ADE2 and GAL1∷HIS3 reporter genes (12). Similarly, we transformed a MATα two-hybrid strain MaV204K, harboring SPAL10∷URA3 and UASGAL1∷HIS3 reporters (13), with individual prey plasmids. Each transformant was cultured in the well of flat-bottom 96-well plates filled with appropriate liquid media. After the removal of clones that activate reporter genes even in the absence of any interacting partners, those left in each plate were collected into a single tube to make a pool for screening: each pool is thus an equivolume mixture of cultures containing up to 96 independent clones. Because some ORFs were refractory to PCR amplification or cloning, we finally prepared 62 pools for both bait and prey, thereby covering ≈95% of the budding yeast ORFs.
To examine all possible combinations between the pools, we performed 3,844 mating reactions in total by using a multisample filtration apparatus (Millipore 1225 sampling manifold) to collect both bait and prey clones onto Millipore HA membrane, on which cells were allowed to mate. After the mating, diploid cells formed were selected for the activation of ADE2, HIS3, and URA3 reporter genes. Positive colonies were restreaked onto another selection plate that was supplemented with 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside to confirm the activation of the three reporter genes and endogenous MEL1 gene, another target of the transcription factor Gal4. The selected clones then were subjected to colony PCR with primers flanking the cloning sites of pGBK-RC and pGAD-RC. For the clones that are refractory to direct amplification from colonies, we isolated plasmid DNAs to use as the templates for PCR. Amplified inserts were directly read to obtain sequence tags, which subsequently were subjected to a blast search to decode interactions.
The description and various data for each protein were retrieved from the Yeast Proteome Database (YPD) (ref. 14, http://www.proteome.com/databases/index.html). The data for interactions between the orthologs and expression similarity were retrieved from the Database of Interacting Proteins (ref. 15, http://dip.doe-mbi.ucla.edu/) and the Biomolecular Relations in Information Transmission and Expression database (http://www.genome.ad.jp/brite/) of Kyoto Encyclopedia of Genes and Genomes (16), respectively. The software tool to support the modeling interaction networks has been described (17).
Results
Completion of the Comprehensive Two-Hybrid Screening.
We cloned almost all of the yeast ORFs individually as a DNA-binding domain fusion (bait) in a MATa strain and an activation domain fusion (prey) in a MATα strain and subdivided them into pools, each containing up to 96 clones (Fig. 1) (10). We finally prepared 62 pools for both baits and preys and mated them to each other in all possible 3,844 (= 62 × 62) combinations. The diploid cells formed by the mating, each bearing a pair of bait and prey, were plated onto the media lacking adenine, histidine, and uracil for the selection of clones simultaneously activating the three reporter genes, ADE2, HIS3, and URA3 (Fig. 1). The survivors of this primary selection then were transferred to the second media, which not only reconfirmed the activation of the three reporters but also examined the induction of endogenous MEL1, another gene regulated by Gal4. Because each of the four genes bears a unique Gal4-responsive promoter, false positive signals caused by fortuitous promoter-specific activation, which occasionally happens in two-hybrid screening (12), would be minimized. The growth and color on this plate were variable from clone to clone: some clones grew well but were pale (i.e., weak MEL1 induction) or reddish (i.e., weak ADE2 induction) whereas others displayed slow growth but dark blue colors. Such differential activation of reporter genes makes it difficult to estimate the strength of each interaction.
Figure 1.
Outline of the comprehensive two-hybrid analysis. We cloned almost all yeast ORFs individually as a DNA-binding domain fusion (bait) in a MATa strain and as an activation domain fusion (prey) in a MATα strain, and subsequently divided them into pools, each containing 96 clones. These bait and prey clone pools were systematically mated with each other, and the diploid cells formed were selected for the simultaneous activation of three reporter genes (ADE2, HIS3, andURA3) followed by sequence tagging to obtain ISTs.
Using the selection strategy described above, we finally obtained 15,523 positive clones. We next amplified the inserts of the plasmids cohabiting in each positive clone and subjected them to direct sequencing to obtain interaction sequence tags (ISTs), a pair of tag sequences for bait and prey (Fig. 1). Consequently, 13,754 ISTs were obtained from the 15,523 clones (Table 1). Removal of redundant and low-quality ISTs left 4,549 independent two-hybrid interactions among 3,278 proteins in total (Table 1). All of these data are available at http://genome.c.kanazawa-u.ac.jp/Y2H.
Table 1.
Summary of the comprehensive two-hybrid screening
| Mating reactions | 3,844 |
| Combinations to be examined | ∼3.5 × 107 |
| Positive colonies | 15,523 |
| ISTs | 13,754 |
| Independent two-hybrid interactions | 4,549 |
| More than 2 IST hits | 1,533 |
| More than 3 IST hits (core data) | 841 |
Comparison with the Other Large-Scale Two-Hybrid Project.
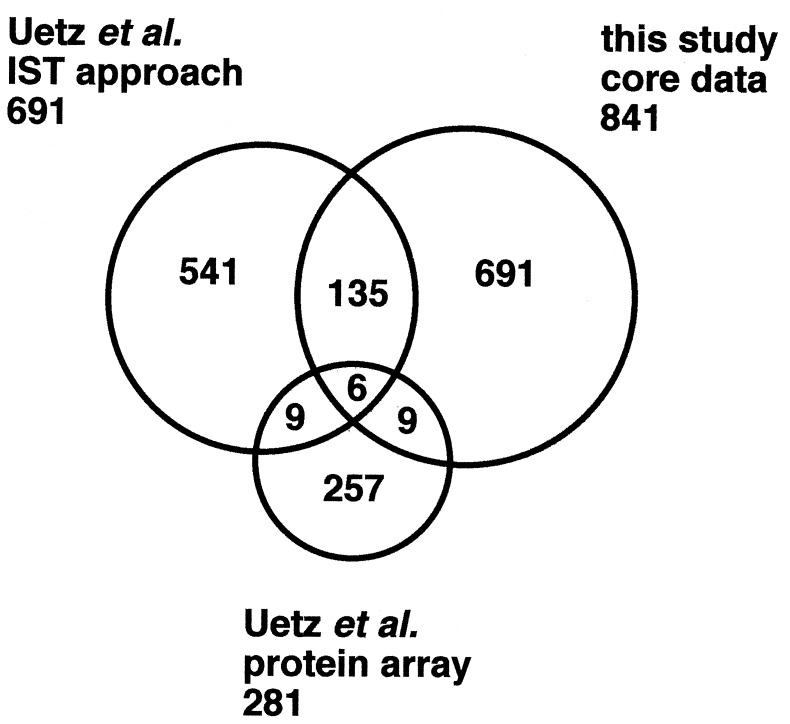
We compared our data with those by Uetz et al. (11), which also include interactions revealed by a high-throughput IST approach. First, we examined the numbers of known interactions recapitulated in each dataset, which may indicate the quality of screen. They obtained 691 pairs of proteins, 88 (i.e., 12.7%) of which are previously known to interact or to occur in the same complex according to the YPD (14) (Table 2). The subset of our data with more than three IST hits was found to have a similar rate for known interactions (i.e., 12.5%) but to be substantially larger than theirs. We designated this subset composed of 841 interactions involving 797 proteins as the core data.
Table 2.
Comparison between the two genomewide IST projects
We next examined how these two datasets overlap with each other (Fig. 2). Unexpectedly, the overlap was rather small; our core data and theirs, assumed to have similar quality (see above), share only 141 interactions, constituting 16.8% and 20.4% of the former and the latter, respectively. As expected, the fraction shared by the two projects shows a significantly higher rate for known interactions (i.e., 28.3%).
Figure 2.
Overlap among the results of large-scale two-hybrid projects. The Venn diagram indicates the overlap among the core data of our analysis and those obtained by the high-throughput IST analysis and the protein array approach by Uetz et al. (11).
A Genomewide Two-Hybrid Interaction Map.
To draw a genomewide two-hybrid interaction map, we connected 806 binary data, which were selected from the core data by removing redundancy caused by two-hybrid interactions detected in both orientations. Consequently a huge network composed of 417 proteins linked by 544 interactions and 132 much smaller networks involving 2–14 proteins were formed (Table 3). We were afraid that such a big network appeared mainly because of the noise in large-scale two-hybrid analyses. To rule out that possibility, we analyzed a dataset of 2,209 interactions among 1,858 proteins, which we extracted from the YPD (14) by excluding those revealed in systematic two-hybrid projects. These data thus would represent the interactions identified in conventional studies. Nevertheless, they also formed a big cluster that connected 1,003 proteins via 1,504 interactions (Table 3). Intriguingly, in both cases, the largest cluster includes the two-thirds of the interactions and half of the proteins. Thus, the emergence of a single huge network is not inherent to the systematic two-hybrid analysis: it may well reflect substantial crosstalks between the proteins actually occurring within a cell.
Table 3.
The largest network reconstructed from interaction data
| Dataset | Proteins in the largest network/total proteins (%) | Interactions in the largest network/total interactions (%) |
|---|---|---|
| Conventional studies* | 1,003/1,858 (54) | 1,504/2,209 (68) |
| This study† | ||
| Core data | 417/797 (52) | 544/806 (67) |
| All data | 2,838/3,278 (87) | 4,224/4,475 (94) |
This dataset was obtained by excluding interactions revealed by large-scale two-hybrid projects from those deposited in the YPD (14).
Synonymous interactions due to two-hybrid interactions detected in both orientations are counted as a single one.
However, the network does include some artifacts. For instance, common subunits shared by three RNA polymerases link up all of the components of RNA polymerases I, II, and III into a single cluster. It should be also noted that the proteins having numerous interaction partners, some of which are likely caused by noise in the two-hybrid screens, also accelerate the expansion of network. The use of all our data containing such proteins leads to a huge cluster, which involves the vast majority of proteins (87%) and interactions (94%) (Table 3). As expected, omission of the proteins with dozen of binding partners from the analysis substantially reduces the size of the largest cluster (data not shown).
Highlighting Biologically Relevant Subnetworks.
The observations described above indicate that any model for protein network should be extracted from the huge cluster with careful evaluation of each interaction. We thus attached additional features described below to each interaction in our data (http://genome.c.kanazawa-u.ac.jp/Y2H). We counted the co-occurrence of the interacting proteins in literature, because proteins appeared in the same paper are, in general, functionally related and their association is likely to be biologically relevant. We also retrieved data from YPD to allow users to readily know whether the interaction is independently confirmed by others, whether both of the two are known to occur in the same complex, whether the two genes are coexpressed, and whether the two show any genetic interaction such as synthetic lethality.
Furthermore, we referred to alternative interaction paths from bait to prey found in the previously known interactions, because their presence would suggest a functional linkage between the two proteins. (However, alternative paths with multiple intervening proteins would be of low value, because most proteins can be linked to form a single huge nexus as described above.) Such an alternative path forms a circular contig of interactions, which is termed an interaction cluster by others (18, 19) and may indicate a protein complex.
We also examined whether an interaction between their orthologs called interolog (18, 19) is reported in other organisms using the data deposited in the Database of Interacting Proteins (15). For instance, we found a novel nexus of Ufd1-Npl4-Cdc48, and a recent report, to our interest, identified a trimeric complex consisting of Ufd1, Npl4, and p97, each of which, respectively, represents the mammalian ortholog of Ufd1, Npl4, and Cdc48 (20). This finding suggests that the Ufd1-Npl4 complex links the AAA-ATPase Cdc48 to ubiquitin and nuclear transport pathways not only in mammalian cells but also in the budding yeast (20).
All of the features mentioned above would help one evaluate the biological relevance of each interaction to decide whether it should be integrated into the network model being constructed. It is obvious that the process for such modeling requires frequent referring to these items as well as many trials and errors in editing. We have thus developed a software tool to support such a task (17), which will be made available from our web site (http://genome.c.kanazawa-u.ac.jp/Y2H).
Examples of Biologically Intriguing Subnetworks.
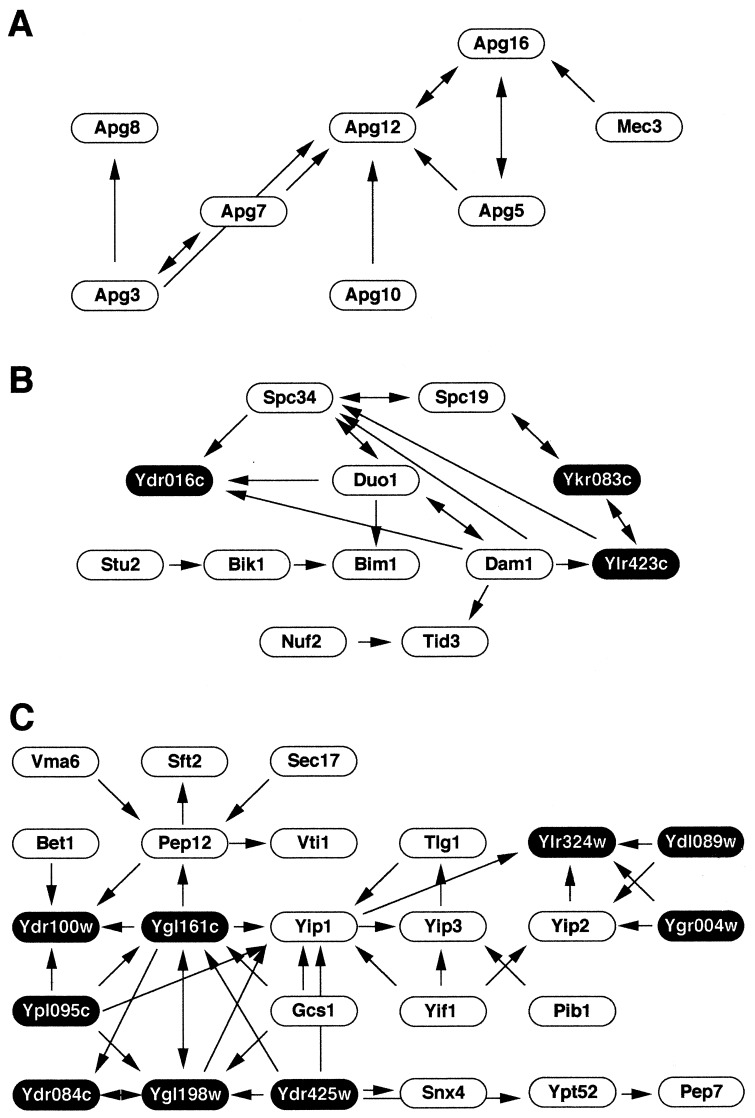
Using the data with caution, we could construct biologically intriguing models for protein interaction networks. For instance, the hypothetical network shown in Fig. 3A is composed of proteins known to function in the process of autophagy (21). The two proteins, Apg7 and Apg10, catalyze the conjugation of Apg12 to Apg5, and this conjugate is assumed to be further multimerized by the action of Apg16. It also partially reveals an alternative pathway including Apg3/Aut1 and Apg8/Aut7. Thus, this interaction network coincides well with the molecular mechanism underlying autophagy. In addition, we observed a two-hybrid interaction with multiple IST hits between Apg16 and Mec3, a checkpoint protein for G2 arrest after DNA damage, which also was detected in the other large-scale project (11). This interaction might be indicative of an unexpected link between autophagy and cell cycle control.
Figure 3.
Biologically intriguing interaction networks. Three subnetworks consisting of proteins involved in autophagy (A), spindle pole body function (B), and vesicular transport (C) are shown. The arrows indicate the orientation of each two-hybrid interaction, beginning from the bait to the prey. Hypothetical proteins of unknown functions are indicated by black ovals with white letters.
A network, which contains the components of spindle pole body and the proteins affecting its function, is depicted in Fig. 3B, a part of which had already emerged in our pilot phase (10). Based on the interaction scheme, we had pointed out the potential involvement of a hypothetical protein Ydr016c in spindle-pole body function (10). Indeed, it was recently reported that the protein localizes to intranuclear spindles and spindle pole bodies and that a temperature-sensitive mutant for this essential gene arrests with large buds and a short mitotic spindle (http://genome-www4.stanford.edu/cgi-bin/SGD/locus.pl?locus = DAD1). The gene is now designated as DAD1 for Duo1 and Dam1 Interacting. This case provides another example demonstrating the power of interaction analysis in the prediction of gene function: a protein with multiple interacting partners sharing a distinct function is certainly involved in the same biological process. Notably, this network also contains the product of another essential gene, YKR083C, currently lacking any clue as to its function. It is tempting to examine the defects of spindle pole body in mutants for this novel gene.
The large network in Fig. 3C involves many proteins known to function in the membrane fusion step of vesicular transport process. This network had been partly uncovered in our pilot phase, including the interactions between Yip1 and Yif1 (10). Indeed, these two proteins were recently demonstrated to form a tight complex on Golgi membrane and play an essential role that precedes vesicular docking and fusion (22). The completion of the two-hybrid analysis has substantially enlarged the network by incorporating additional proteins in similar functional categories as well as those of unknown function. Note that a complex crossing over of binary interaction arrows is observed among these novel proteins (Fig. 3C), which might indicate the presence of a hitherto unidentified multiprotein complex presumably playing a role in vesicular transport process.
Discussion
The large-scale two-hybrid analysis is currently the only feasible approach for comprehensive interaction mapping (19, 25, 26). Its power has been well demonstrated in this and other studies (10, 11, 18, 23, 24). Systematic interaction mapping provides various hypothetical networks that are biologically intriguing and/or unexpected, as exemplified by those extracted from our data (Fig. 3). These networks provide testable hypotheses, which eventually would improve our understanding of the cell as molecular machinery. Such hypothetical networks or protein complexes also would serve as the most appropriate targets for proteomics-based analysis (8).
On the other hand, this study demonstrates limitation of the large-scale two-hybrid approach as well. The data of two independent projects fail to largely overlap (Fig. 2). The reasons for this small overlap are not clear, but there are a number of plausible explanations. First, both projects used PCR-amplified ORFs and some of them would inevitably bear mutations abolishing interactions. Second, each project used unique plasmid construct, which may significantly affect the folding of hybrid proteins: some are folded correctly in our collection but not in that of Uetz et al. (11) and vice versa. However, it is impossible to predict or examine the folding for each of the ≈12,000 hybrid proteins. Third, strategy and stringency of the selection was different between the two projects. While we used three reporter genes and multicopy two-hybrid plasmids, Uetz et al. used a single reporter and low-copy vectors. Fourth, both screenings were obviously not saturated. We observed that some interactions, which had escaped the large-scale screen, could be detected when assayed in a one-to-one manner using our constructs. Finally, stochastic activation of reporter genes, more or less inherent to two-hybrid system, generates false signals, which would appear as low-hit ISTs unique to each project.
These results indicate that any single IST project is difficult to complete. For the exploration of protein interactome, it would be better to have several independent IST projects using different constructs and to combine their results. It is also important to integrate the results of an alternative approach for large-scale two-hybrid analysis, the protein array-based approach, which is sensitive but rather slow (11).
These large-scale approaches have, of course, various limitations inherent to the two-hybrid method itself. The most serious problem would be its reliability: the two-hybrid system is generally claimed to show many false positive or biologically meaningless signals. How are our comprehensive data reliable? To provide a rough estimate for the reliability, we analyzed our core data composed of 841 interactions (Table 1), which contain 415 interactions occurring between two “named” proteins from this dataset. Because most of such named proteins are associated with, at least, the minimum functional description, interactions between them would allow us to evaluate relevance of the interactions. Of the 415 interactions, 103 are previously shown to interact or to occur in the same complex by the conventional studies, according to YPD. We thus inspected each of the remaining 312 interactions and found that 85 are likely to occur, judging from the function of the proteins. In addition, this dataset contains 24 novel homotypic or oligomeric interactions. We thus assume that more than half of the interactions in the core dataset are of some biological relevance.
On the other hand, a number of false negatives are evident. Our analysis of two-hybrid interaction data deposited in the YPD indicates that, while providing a huge number of novel interactions, systematic two-hybrid projects have failed to recapitulate as much as ≈90% of the two-hybrid interactions that are identified in conventional studies. Note that this low coverage is not only due to the insufficient depth of the screening and potential misfolding or mutations abolishing the interactions, but also due to the use of full-length proteins, which often obscure a sizable fraction of interactions (10, 27). It becomes increasingly evident that, in many cases, interaction surfaces are usually masked and become exposed only when activation signals, such as phosphorylation or allosteric effector binding, induce conformational change in either or both of the proteins.
A plausible way to unmask such interactions is to screen libraries of fragmented preys using the baits that are preselected on the basis of specific function or structure. For instance, an exhaustive two-hybrid screening of a fragmented prey library with baits involved in RNA splicing has revealed a number of novel interactions which the whole-genome approaches with full-length ORFs have failed to detect (23, 24). Similarly, a computationally directed screen aiming at the comprehensive analysis of the association between coiled-coils also has uncovered many interactions, all but one of which escaped the detection by the genomewide approach (28). We also performed a directed screening of a high-quality genomic library (12) using all of the yeast Src homology 3 domains and WW domains as baits to identify many novel interactions (T.I., unpublished results).
However, the success of such screenings totally depends on the design of baits: some can reproduce all of the previously reported interactions whereas others not at all. It is, unfortunately, impossible to tell which boundaries should be used to ensure correct folding of each domain. Ideally, one should use both bait and prey in variously truncated forms to maximize the chance of correct folding and hence successful interactions, although the combinations to be examined will obviously explode in such screens. Thus, in practice, both genomewide screening with full-length ORFs and a variety of directed approaches using conventional fragmented libraries should be conducted as parts of a continued effort toward the exploration the protein interactome.
To uncover as many types of interactions as possible, it may be also effective to use other two-hybrid methods including the bacterial systems, those based on the reconstitution of transcription by RNA polymerase III, Ras signaling pathway, and ubiquitin function (see ref. 29 for a review). Emerging technologies such as protein chips (30) or a high-throughput mass spectrometric analysis of protein complex (8) also would considerably expand our knowledge on protein interactome.
These large-scale projects have been drastically increasing the number of putative protein–protein interactions in the database. Integration of these data leads to a huge network involving most proteins, which may well indicate the crosstalks between proteins, but is too big to interpret (Table 3). Others also noted the emergence of such a huge network, although they used a combined dataset containing interactions revealed by both conventional studies and large-scale screens (31, 32). While our analysis indicates that the huge cluster is not inherent to large-scale analysis, it may be artificially overextended by the incorporation of low-quality two-hybrid data (Table 3). Thus, careful evaluation of the network by the aid of bioinformatics is necessary. Various approaches would be plausible to draw useful information from such a huge network. For instance, one may color each node based on the assigned functional category. Alternatively, only the proteins preselected for particular functions are used for network construction, although such restriction may well miss a chance of totally unexpected discovery. Careful editing of network models is vital to draw hypotheses worth further pursuit from the huge network. Accordingly, bioinformatic tools to support such a process would become increasingly important.
One also should bear in mind that the current network models are totally lacking spatial and temporal resolution. Several distinct complexes sharing a common protein component may be artificially linked with each other in silico. To avoid this, we have to collect data on the architecture of each native protein complex as well as its spatiotemporal occurrence. It is also important to know quantitative aspects of interactions: what fraction of each protein is actually participating in the complex formation? Such interaction profiling would follow the cataloging phase in the protein interactome research. Furthermore, to learn the biology of each interaction, one has to examine the effect of its disruption (i.e., interaction targeting). The cataloging, profiling, and targeting of interactions eventually will allow one to draw a truly functional and dynamic map of protein–protein interactions that would undoubtedly lead us one step forward to comprehensive understanding of the cell as a molecular system.
Acknowledgments
We thank Y. Shibagaki, H. Shimano, and C. Kawagoe for help in data analysis. We are grateful to H. Sumimoto for stimulating discussion on protein–protein interactions and encouragement. This work was supported by research grants from the Ministry of Education, Science, Sports and Culture (MESSC) and Science and Technology Agency (STA). The development of the supporting software was performed as a part of the research and development project of Industrial Science and Technology Program supported by New Energy and Industrial Technology Development Organization (NEDO).
Abbreviations
- YPD
Yeast Proteome Database
- IST
interaction sequence tag
Footnotes
See commentary on page 4277.
References
- 1.Blattner F R, Plunkett G, 3rd, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 2.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel J D, Jacq C, Johnston M, et al. Science. 1996;274:563–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 3.Winzeler E A, Shoemaker D D, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke J D, Bussey H, et al. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 4.Ogasawara N. Res Microbiol. 2000;151:129–134. doi: 10.1016/s0923-2508(00)00118-2. [DOI] [PubMed] [Google Scholar]
- 5.Fraser A G, Kamath R S, Zipperien P, Martinez-Campos M, Sohrmann M, Ahringer J. Nature (London) 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 6.Gonczy P, Echeverri C, Oegema K, Coulson A, Jones S J M, Copley R R, Duperon J, Oegema J, Brehm M, Cassin E, et al. Nature (London) 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- 7.Lockhart D J, Winzeler E A. Nature (London) 2000;405:827–836. doi: 10.1038/35015701. [DOI] [PubMed] [Google Scholar]
- 8.Pandey A, Mann M. Nature (London) 2000;405:837–846. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- 9.Fields S, Song O K. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 10.Ito T, Tashiro K, Muta S, Ozawa R, Chiba T, Nishizawa M, Yamamoto K, Kuhara S, Sakaki Y. Proc Natl Acad Sci USA. 2000;97:1143–1147. doi: 10.1073/pnas.97.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uetz P, Giot L, Cagney G, Mansfield T A, Judson R S, Knight J R, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. Nature (London) 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 12.James P, Halladay J, Craig E A. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vidal M, Brachmann R K, Fattaey A, Harlow E, Boeke J. Proc Natl Acad Sci USA. 1996;93:10315–10320. doi: 10.1073/pnas.93.19.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costanzo M C, Hogan J D, Cusick M E, Davis B P, Fancher A M, Hodges P E, Kondu P, Lengieza C, Lew-Smith J E, Lingner C, et al. Nucleic Acids Res. 2000;28:73–76. doi: 10.1093/nar/28.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xenarios I, Rice D W, Salwinski L, Baron M K, Marcotte E M, Eisenberg D. Nucleic Acids Res. 2000;28:289–291. doi: 10.1093/nar/28.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Goto S. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshida M, Fukagawa H, Shibagaki Y, Shimano H, Mizuno T. Genome Inform Ser Workshop Genome Inform. 2000;11:364–365. [Google Scholar]
- 18.Walhout A J, Sordella R, Lu X, Hartley J L, Temple G F, Brasch M A, Thierry-Mieg N, Vidal M. Science. 2000;287:116–122. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- 19.Walhout A J, Boulton S J, Vidal M. Yeast. 2000;17:88–94. doi: 10.1002/1097-0061(20000630)17:2<88::AID-YEA20>3.0.CO;2-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer H H, Shorter J G, Seemann J, Pappin D, Warren G. EMBO J. 2000;19:2181–2192. doi: 10.1093/emboj/19.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klionsky D J, Ohsumi Y. Annu Rev Cell Dev Biol. 2000;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- 22.Matern H, Yang X, Andrulis E, Sternglanz R, Trepte H-H, Gallwitz D. EMBO J. 2000;19:4485–4492. doi: 10.1093/emboj/19.17.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fromont-Racine M, Rain J C, Legrain P. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 24.Fromont-Racine M, Mayes A E, Brunet-Simon A, Rain J C, Colley A, Dix I, Decourty L, Joly N, Ricard F, Beggs J D, Legrain P. Yeast. 2000;17:95–110. doi: 10.1002/1097-0061(20000630)17:2<95::AID-YEA16>3.0.CO;2-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Legrain P, Selig L. FEBS Lett. 2000;480:32–36. doi: 10.1016/s0014-5793(00)01774-9. [DOI] [PubMed] [Google Scholar]
- 26.Uetz P, Hughes R E. Curr Opin Microbiol. 2000;3:303–308. doi: 10.1016/s1369-5274(00)00094-1. [DOI] [PubMed] [Google Scholar]
- 27.Hu J C. Proc Natl Acad Sci USA. 2000;97:12935–12936. doi: 10.1073/pnas.97.24.12935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newman J R S, Wolf E, Kim P S. Proc Natl Acad Sci USA. 2000;97:13203–13208. doi: 10.1073/pnas.97.24.13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fashena S J, Serebriiskii I, Golemis E A. Gene. 2000;250:1–14. doi: 10.1016/s0378-1119(00)00182-7. [DOI] [PubMed] [Google Scholar]
- 30.MacBeath G, Schreiber S L. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 31.Fellenberg M, Albermann K, Zollner A, Mewes H M, Hani J. Intell Syst Mol Biol. 2000;8:152–161. [PubMed] [Google Scholar]
- 32.Schwikowski B, Uetz P, Fields S. Nat Biotechnol. 2000;18:1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]