Abstract
The MA domain of the retroviral Gag protein mediates interactions with the plasma membrane, which is the site of productive virus release. HIV-1 MA has a phosphatidylinositol-(4,5)-bisphosphate [PI(4,5)P2] binding pocket; depletion of this phospholipid from the plasma membrane compromises Gag membrane association and virus budding. We used multiple methods to examine the possible role of PI(4,5)P2 in Gag-membrane interaction of the alpharetrovirus Rous sarcoma virus (RSV). In contrast to HIV-1, which was tested in parallel, neither membrane localization of RSV Gag-GFP nor release of virus-like particles was affected by phosphatase-mediated depletion of PI(4,5)P2 in transfected avian cells. In liposome flotation experiments, RSV Gag required acidic lipids for binding but showed no specificity for PI(4,5)P2. Mono-, di-, and triphosphorylated phosphatidylinositol phosphate (PIP) species as well as high concentrations of phosphatidylserine (PS) supported similar levels of flotation. A mutation that increases the overall charge of RSV MA also enhanced Gag membrane binding. Contrary to previous reports, we found that high concentrations of PS, in the absence of PIPs, also strongly promoted HIV-1 Gag flotation. Taken together, we interpret these results to mean that RSV Gag membrane association is driven by electrostatic interactions and not by any specific association with PI(4,5)P2.
INTRODUCTION
Assembly and budding of retrovirus particles are complex processes mediated by the viral structural protein Gag. Several thousand Gag molecules along with two copies of the RNA genome and the viral glycoprotein Env are transported to the assembly site where Gag-lipid, Gag-Gag, and Gag-RNA interactions drive the formation of a virus particle. The assembly site is determined largely by the membrane-binding domain (MBD) at the N terminus of the Gag protein, which mediates membrane targeting and membrane binding (25, 43, 58, 59, 64, 68). For most retroviruses, productive viral assembly occurs at the plasma membrane (PM) (21, 30).
Across retroviral genera, sequence similarity among retroviral MBDs is limited; however, all previously studied retroviral MBDs fold into a small, globular domain with an alpha-helical core (40). The MBD usually contains two membrane-binding signals, an N-terminal myristate, which inserts into the hydrophobic interior of lipid membranes, and a surface patch of basic residues, which interacts with acidic phospholipids. Several retroviral MBDs are not myristoylated, including those of equine infectious anemia virus (EIAV) (10, 26) and Rous sarcoma virus (RSV) (38). In contrast, the basic patch is highly conserved, suggesting that electrostatic interactions are universally important in membrane binding of Gag (40). Depending on the type of retrovirus and the severity of the changes, mutations in the basic patch can shift Gag localization from the plasma membrane to intracellular membranes (22, 43, 60), promote promiscuous binding to cellular membranes (55), or abolish membrane binding entirely (6, 58). Mutations that increase the positive charge of the basic patch can rescue Gag localization to the PM or enhance the release of virus particles (5, 6).
Acidic phospholipids, especially phosphatidylserine (PS) and phosphatidylinositol phosphates (PIPs), are important cellular factors in mediating protein-membrane interactions (27, 35, 39, 67). PS has a single, net-negative charge, while PIPs have multiple negative charges due to phosphorylation of the inositol ring at positions 3, 4, and/or 5. The location and degree of phosphorylation are determined by spatial regulation of kinases and phosphatases, which results in the enrichment of specific species of PIPs at different cellular membranes (reviewed in reference 33). PS and PI(4,5)P2 are found primarily on the inner leaflet of the PM in mammalian cells, where they account for 25 to 35% and 0.5 to 1.0% of the phospholipids, respectively (2, 9, 36, 49). Recruitment of cellular MBDs (e.g., pleckstrin homology [PH] domains [16, 19, 63], C2 domains [37], and epsin N-terminal homology domains/AP180 N-terminal homology [ENTH/ANTH] domains [28]) to the PM is dependent on direct interactions with PS and/or PI(4,5)P2. However, the quantitative contribution of each of these acidic lipids to PM binding of proteins is uncertain since different studies have yielded conflicting results (27, 67).
As purified proteins, some retroviral MBDs (e.g., that of HIV-1 and HIV-2) bind specifically to versions of PI(4,5)P2 that have shortened fatty acid chains required for solubility (51, 54). Mutation of the residues involved in PI(4,5)P2 interaction reduces PM affinity in vivo and also binding to artificial liposomes in vitro (3, 10, 25, 51, 54, 57). Consistent with the inferred role for this lipid in virus assembly at the PM, the membrane surrounding HIV-1 and murine leukemia virus (MLV) virions is enriched in PI(4,5)P2 (9) as well as PS (2, 4, 47). Furthermore, overexpression of inositol polyphosphate-5-phosphatase E (5-phosphatase IV here referred to as 5ptase), which depletes cellular levels of PI(4,5)P2 (32), results in a decrease in Gag localization at the PM and a reduction in virus release (25, 42, 51, 60). In the case of HIV-1, binding to PI(4,5)P2 leads to exposure of the myristate, thereby enhancing the affinity of the MBD for the PM (53, 54).
The RSV MBD is not myristoylated, nor does it contain a linear sequence of basic residues as do EIAV and MLV. However, the three-dimensional structure of the protein reveals that multiple basic residues come together to form a surface patch (38). The RSV MBD has a net-positive charge of +5 (5). Small deletions or neutralization of two or more basic residues in the surface patch abolish PM localization and reduce virus release (5, 6, 41, 44, 64). Compensatory mutations that restore the net-positive charge of the MBD also restore PM localization, whereas some mutations that increase the overall charge alter the intracellular trafficking of RSV Gag, enhance Gag localization at the PM, and accelerate the budding kinetics of virus-like particles (VLPs) (6). These results were interpreted to imply that RSV Gag-membrane binding is driven chiefly by electrostatic interactions (5, 6).
Using computational modeling and liposome flotation assays with purified recombinant proteins, we previously reported that RSV and HIV-1 MA membrane binding in vitro is driven by electrostatic interactions with negatively charged lipids (13, 14). Preliminary data also suggested that PI(4,5)P2 at 1% of total phospholipid does not significantly enhance MA-liposome binding in the presence of physiological levels of PS. We have now extended these studies to the RSV Gag protein, in comparison with the HIV-1 Gag protein, which has been studied by others (1, 11, 12, 20, 29). By fluorescence imaging and virus release in cells and by liposome flotation of protein synthesized in a reticulocyte lysate, RSV Gag appears to have no specific requirement for PI(4,5)P2 in membrane targeting and viral assembly in vivo or for liposome binding in vitro. Rather, RSV Gag membrane association relies only on electrostatic interactions.
MATERIALS AND METHODS
DNA vectors.
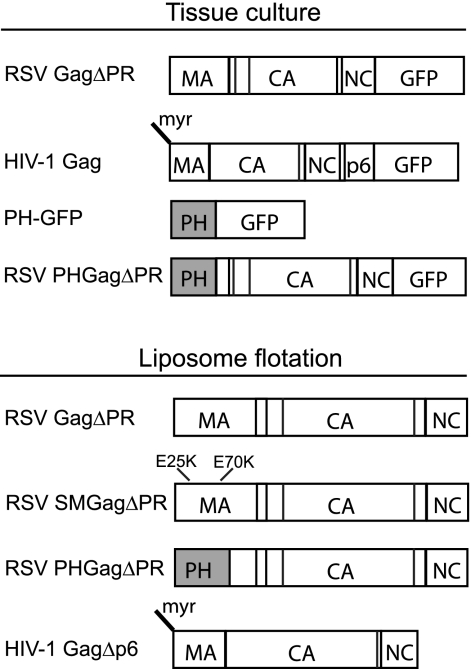
RSV and HIV Gag constructs used in this study are shown schematically in Fig. 1. All DNA constructs were generated with common subcloning techniques and were propagated in DH5α cells. The 5ptase expression vector, pcDNA4TO/Myc5ptaseIV (11), was a gift from Akira Ono. Plasmid PH-GFP, a gift from Barbara Baird, encodes the pleckstrin homology (PH) domain from human phospholipase C δ1 fused to enhanced green fluorescent protein (eGFP) under the control of the cytomegalovirus immediate early promoter. Fluorescent Gag proteins were created by cloning RSV GagΔPR derived from the Prague C strain or HIV-1 Gag derived from the BH-10 strain into the pEGFP-N1 vector (Clontech). Plasmid PHGag-GFP was constructed by subcloning the PH domain from pPH-GFP into RSV GagΔPR-GFP between the SacI site (nucleotide [nt] 255) in the leader region and the XhoI site in MA (nt 630), which has traditionally been used to define the C-terminal end of the RSV MBD (38).
Fig. 1.
Schematic representations of proteins. RSV and HIV-1 GFP-tagged proteins expressed in transfected cells are shown at the top along with the PH-GFP protein. Proteins translated in vitro and submitted to flotation analysis are shown at the bottom. Vertical lines in the boxes stand for protease cleavage sites.
The previously described plasmid RSV GagΔPR (8) was used to generate SuperM GagΔPR (SMGagΔPR) and PHGagΔPR. The RSV SMGag construct (6) was a gift from John Wills. The region of MA containing the SM mutations E25K and E70K was PCR amplified and cloned into pET3xc RSV GagΔPR using sites XbaI (in the backbone of the vector and added upstream of Gag by PCR) and XhoI (nt 630). RSV PHGagΔPR was constructed by PCR amplifying the PH domain from PH Gag-GFP and cloning the product into RSV GagΔPR using sites XbaI and XhoI. The plasmid expressing HIV-1 GagΔp6 (strain BH10) was a gift from Alan Rein. For simplicity, in the Results and Discussion sections of this paper, RSV GagΔPR and its derivatives as well as HIV-1 GagΔp6 are referred to as RSV Gag and HIV-1 Gag, respectively. Neither of these deleted domains affects the membrane-targeting properties of Gag or the first steps in assembly.
Cells and transfection.
Cell cultures and transfections were performed as previously described (14). DF1 (chicken) and QT6 (quail) fibroblasts were maintained in Dulbecco modified Eagle medium supplemented with 5% fetal bovine serum, 5% NuSerum (BD Biosciences), 1% heat-inactivated chick serum, standard vitamins, l-glutamine, penicillin, and streptomycin.
DF1 and QT6 cells were seeded onto glass coverslips for imaging or six-well plates for virus release assays 24 h prior to transfection. DF1 cells at 60% confluence were transfected with 2 μg of total DNA at a 1:1 ratio of fluorescent Gag to the 5ptase expression plasmid or to the control plasmid pBluescript SK(+) (Stratagene). QT6 cells at 30% confluence were transfected with 3 μg of total DNA at a 1:2 ratio of fluorescent Gag to 5ptase or control plasmid. Transient transfections were performed using FuGENE HD (Roche) according to the manufacturer's instructions.
Virus release and immunoblotting.
Medium and cells were collected 24 h posttransfection. The medium was centrifuged at 5,000 × g for 5 min to remove cellular debris. Virus-like particles (VLPs) were isolated by layering the cleared medium onto 0.50 ml virus pelleting buffer (15% sucrose in 20 mM Tris HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA) and centrifuging at 90,000 rpm for 45 min in a Beckman TLA 100.4 rotor. Western blotting was performed on cellular and VLP-associated Gag by using rabbit anti-RSV capsid (αCA) serum diluted 1:1,000 or rabbit anti-HIV-1 p24 (NIH AIDS Research and Reference Reagent Program, Reagent 4250) diluted 1:5,000, and then blots were probed with anti-rabbit IgG-alkaline phosphatase (Sigma) diluted 1:30,000. Following incubation in the ECF reagent (Sigma), blots were imaged on a Storm scanner (Molecular Dynamics). Bands were quantified using ImageQuant software.
Liposome binding assay.
Liposomes were prepared as previously described with some modifications (13). The lipid concentrations were chosen largely based on previously published studies, to facilitate comparisons. Chloroform solutions of purified natural l-α-phosphatidylcholine [egg PC], l-α-phosphatidylserine [brain PS], l-α-phosphatidylinositol-4,5-bisphosphate [brain PI(4,5)P2], l-α-phosphatidylinositol-4-phosphate [brain PI(4)P], and dry unsaturated 1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol-3′-phosphate) [18:1 PI(3)P], 1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol-3′,5′-bisphosphate) [18:1 PI(3,5) P2], and 1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol-3′,4′,5′-trisphosphate) [18:1 PI(3,4,5)P3] were purchased from Avanti Polar Lipids. Dry lipids were resuspended in chloroform per the manufacturer's directions. Chloroform solutions of PC and PS (67%:33%, 50%:50%, or 33%:67%), or PC, PS, and PIPs (61.75%:31%:7.25%) were mixed at the stated ratios and then dried under a stream of nitrogen and resuspended (1 h to overnight at 4°C with occasional mixing) in 20 mM HEPES (pH 7.0) to a concentration of 10 mg/ml under nitrogen gas. The resuspended lipids were passed at least 60 times through a 100-nm polycarbonate filter in an Avanti extruder to yield uniform liposomes. Liposomes were stored at 4°C under nitrogen and used no later than 10 days after preparation.
In vitro translation and liposome binding reactions were performed as previously described with modifications (11). Briefly, in vitro translation of the Gag constructs was performed using the T7-coupled TNT reticulocyte lysate system (Promega). Fifty-microliter reticulocyte reaction mixtures were prepared according to the manufacturer's instructions with [35S]methionine-cysteine (ExPRE35S35 protein labeling mix; Perkin Elmer) for protein labeling and incubated for 90 min at 30°C. Then, 100 μg of liposomes was added to the reaction mixture, for a final liposome concentration of 1.7 mg/ml, and incubated for an additional 30 min at 30°C.
In preparation for flotation, the reticulocyte reaction mixture with liposomes was diluted to a final volume of 0.25 ml with 20 mM HEPES (pH 7.0), mixed with 0.75 ml 67% sucrose, and then transferred to an ultracentrifuge tube. The mixture was overlaid with 1.6 ml 40% sucrose followed by 0.40 ml 4% sucrose. All sucrose solutions were prepared by dissolving sucrose (wt/wt) in 20 mM HEPES (pH 7.0). The sucrose gradients were centrifuged at 90,000 rpm at 4°C for 180 min in a Beckman TLA 100.4 rotor. Four 0.75-ml fractions were collected. The top one or two fractions represented liposome-bound Gag, and the bottom two fractions represented non-liposome-bound Gag. Unless otherwise noted, 30 μl of each fraction was resolved by SDS-PAGE. The gels were incubated for 30 min in 1 M sodium salicylate, dried, and placed on film at −80°C for 16 to 36 h. The resulting autoradiograms were scanned and quantified using ImageQuant software.
Many attempts were made to resolve the persistent high-molecular-weight band observed on the fluorograms of RSV Gag (and derivatives of Gag) flotations with liposomes containing PIPs. These included increasing the concentration of SDS in loading buffer, adding urea to 8 M, incubating with NaOH, and varying the time, pH, and temperature of samples incubated prior to gel loading. None of these treatments had a significant effect.
Immunofluorescence and confocal microscopy.
Monoclonal anti-myc antibody (Covance) was diluted 1:1,000 in phosphate-buffered saline (PBS) (pH 7.4). Tetramethyl rhodamine isothiocyanate-conjugated anti-mouse antibody was diluted 1:500 in PBS with 1% nonfat milk carrier. DF1 and QT6 cells were collected 12 to 16 h or 20 to 24 h posttransfection, respectively, and fixed with 3.7% formaldehyde in PBS for 15 min, permeabilized with 0.1% Tween 20 in PBS for 15 min, and then blocked with 4% bovine serum albumin for 30 min. Cells were stained with primary and secondary antibodies for 30 min each at 37°C and then mounted on glass slides with Fluoro-Gel (Electron Microscopy Sciences) for viewing on an Ultraview spinning disc confocal microscope (Perkin-Elmer) with a Nikon 100x Plan-Apochromat oil objective lens (numerical aperture [NA] 1.4). Image analysis and confocal stacks were generated with ImageJ software (v1.44d).
RESULTS
Effect of 5ptase overexpression on PI(4,5)P2 levels at the PM.
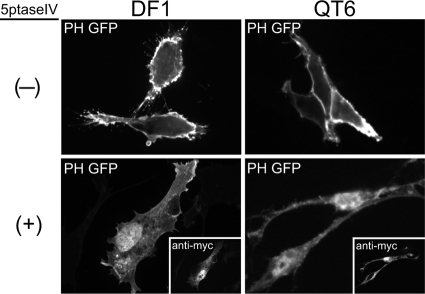
The enzyme 5ptase removes the phosphate at position 5 of the inositol ring on PIPs, converting PI(4,5)P2 and PI(3,4,5)P3 to PI(4)P and PI(3,4)P2, respectively (32). Previous studies using human 5ptase or the yeast homolog (Inp54p) reported that overexpression of this enzyme led to a significant decrease in PI(4,5)P2 levels at the PM (42, 65). We first sought to determine if human 5ptase had a similar effect on PI(4,5)P2 levels in avian cells. DF1 chicken fibroblasts and QT6 quail fibroblasts were cotransfected with DNAs encoding 5ptase and the PH domain of human phospholipase C δ1 (PLCδ1) fused to GFP. This well-studied PH domain binds to PI(4,5)P2 with high specificity and is commonly used as a reporter for the presence of PI(4,5)P2 in cells (16, 19, 63). Both cell types were initially imaged at 24 h posttransfection, but DF1 cells proved to be highly sensitive to 5ptase-induced apoptosis, displaying extensive vacuolation and blebbing at the PM (data not shown). Therefore, DF1 cells were imaged at 12 to 16 h posttransfection when the cells were still healthy. QT6 cells were more resistant to the toxic effects of PI(4,5)P2 depletion, appearing normal after 24 h, and consequently were imaged at 20 to 24 h posttransfection.
In the absence of 5ptase, PH-GFP localized primarily to the PM in both DF1 and QT6 cells (Fig. 2, top), especially to membrane ruffles and cellular protrusions expected to be enriched in PI(4,5)P2. Cotransfection of the myc-tagged 5ptase abolished PM localization of PH-GFP in both cell types (Fig. 2, bottom), resulting in a diffuse signal throughout the cytoplasm. Overexpression of 5ptase also led to a nuclear accumulation of PH-GFP. PLCδ1 has been shown to shuttle between the cytoplasm and the nucleus (66), suggesting that PI(4,5)P2-dependent association with membranes competes with transport into the nucleus. Our observations indicate that human 5ptase significantly depletes PI(4,5)P2 at the PM in these avian cell types.
Fig. 2.
Effect of 5ptase on PM localization of PH-GFP in avian cells. DF1 (left) or QT6 (right) cells were cotransfected with DNAs encoding PH-GFP plus a control plasmid (indicated by a minus sign, top) or myc-tagged 5ptase (indicated by a plus sign, bottom). Expression of 5ptase was detected by a monoclonal mouse anti-myc antibody (insets showing the same cells). Images are single, 0.1-μm confocal sections through the midbody of the cell and are representative for each condition (n > 30 cells).
PM localization of RSV Gag after PI(4,5)P2 depletion.
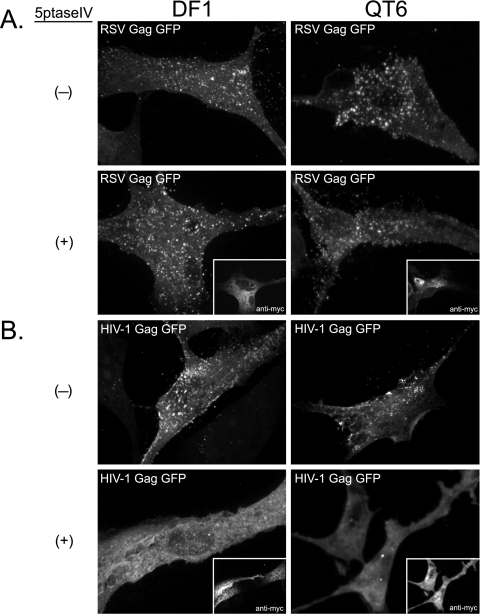
We next wanted to determine if depletion of PI(4,5)P2 affects recruitment of RSV Gag to the PM. DF1 and QT6 cells were cotransfected with DNAs expressing RSV Gag-GFP and myc-tagged 5ptase or RSV Gag-GFP and a control plasmid (Fig. 3A). A second set of transfections was performed in parallel using HIV-1 Gag-GFP as a positive control (Fig. 3B). By themselves, HIV-1 Gag-GFP and RSV Gag-GFP were concentrated in bright puncta at the PM, commonly interpreted to be assembling or budding virions, in both DF1 and QT6 cells (Fig. 3A and 3B, top). Consistent with previous reports on mammalian cells (11, 29), expression of 5ptase prevented HIV-1 Gag-GFP from forming puncta at the PM; instead, the fluorescence signal was diffuse and primarily cytoplasmic (Fig. 3B, bottom). Occasionally, the HIV-1 protein was seen in cytoplasmic accumulations, which may correspond to intracellular vesicles such as late endosomes (42). In contrast, in both types of avian cells, expression of 5ptase had no detectable effect on the punctate fluorescence signal of RSV Gag-GFP at the PM (Fig. 3A, bottom). We interpret these results to mean that, unlike HIV-1 Gag, RSV Gag does not depend on PI(4,5)P2 for PM binding.
Fig. 3.
Effect of 5ptase on PM localization of RSV Gag-GFP and HIV-1 Gag-GFP. DF1 (left) or QT6 (right) cells were cotransfected with DNAs encoding either RSV Gag-GFP (A) or HIV-1 Gag-GFP (B) and a control plasmid (indicated by a minus sign) or a plasmid encoding myc-tagged 5ptase (indicated by a plus sign; myc signal shown in insets). Images are projections of 10 to 15 confocal sections from each cell compiled by average fluorescence intensity and indicate total cellular fluorescence. Images are representative for each condition (n > 30).
To address if the RSV MBD is responsible for the lack of 5ptase effect on PM localization of RSV Gag, we replaced the viral MBD with the PH domain from PLC δ1, a cellular PI(4,5)P2-specific binding protein. The localization of this chimeric protein was examined in DF1 cells and QT6 cells as described above. In both cell types, in the absence of 5ptase, PHGag-GFP was observed primarily at the cell surface and in small vesicular structures near the cell periphery (see Fig. S1A in the supplemental material). Cellular and heterologous MBDs have been shown previously to support budding in the HIV-1 system (30, 56). In DF1 cells upon coexpression of RSV PHGag-GFP with 5ptase, the fluorescence signal at the PM was significantly reduced. Instead, fluorescence accumulated in large, cytoplasmic compartments (see Fig. S1A, white arrows), similar to the late endosomal localization of HIV-1 Gag in PI(4,5)P2-depleted cells (42). However, in some cells a residual fluorescence signal could be detected at the PM (see Fig. S1A, asterisks), suggesting that depletion of this lipid was not uniform across the cell population. Unexpectedly, in QT6 cells, 5ptase caused little loss of PM fluorescence when coexpressed with RSV PHGag-GFP (see Fig. S1A).
Why might phosphatase expression lead to nearly complete removal of PH-GFP and HIV-1 Gag-GFP from the PM but only modestly affect RSV PHGag-GFP at the PM? The difference between PH-GFP and PHGag-GFP might be accounted for by Gag multimerization in the latter, which would increase membrane affinity; the difference between RSV PHGag-GFP and HIV-1 Gag-GFP might be accounted for by the higher affinity of this PH domain for PI(4,5)P2 (Kd [dissociation constant] ∼ 1.7 μM [34, 62]) than of the HIV-1 MA domain for this phosphoinositide (Kd ∼ 150 μM [54]), although direct comparison of these values is not possible because of the different assays used and different fatty acid lengths of the lipid molecules. According to this model, after 5ptase expression, the residual low PI(4,5)P2 levels at the PM would be sufficient to bind the PH domain in PHGag, given the avidity effects due to multimerization, but not sufficient to bind the PH domains of PH-GFP, which does not multimerize, or the MA domain of HIV-1 Gag, which has a lower affinity for PI(4,5)P2. Given the differences observed in the two cell types, it is likely that increasing the ratio of 5ptase to Gag in transfections or specifically targeting 5ptase to the PM would be sufficient to prevent PHGag-GFP from binding the PM in QT6 cells.
Effect of PI(4,5)P2 depletion on virus release.
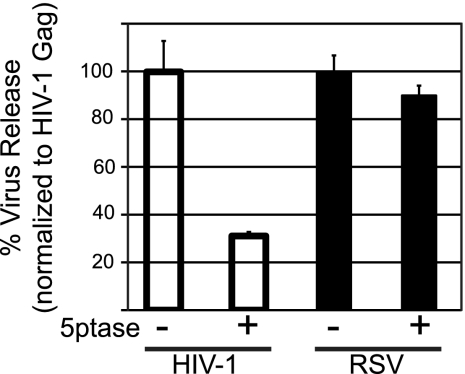
As a second approach to address the role of PI(4,5)P2 in RSV Gag PM binding, we measured the effect of 5ptase expression on the production of Gag-GFP VLPs. HIV-1 budding was previously shown to decrease by 3- to 5-fold upon PI(4,5)P2 depletion in mammalian cells (20, 42). QT6 cells were used for these experiments because DF1 cells did not tolerate the toxic effects of 5ptase long enough for interpretable data to be obtained. VLPs were collected by centrifugation of the medium 24 h posttransfection and quantified by Western blotting. While HIV-1 VLP release was decreased approximately 3-fold as a result of 5ptase expression, the production of RSV VLPs was not affected (Fig. 4), which is consistent with the fluorescence imaging results. We also measured VLP release for the chimeric RSV PHGag-GFP protein in QT6 cells. Phosphatase expression had no effect (see Fig. S1B in the supplemental material), consistent with the imaging results for this cell type. Overall, we interpret the RSV VLP release data to support the observations from fluorescence imaging and conclude that PI(4,5)P2 is not essential for PM targeting of RSV Gag.
Fig. 4.
Effect of 5ptase on virus release from QT6 cells. QT6 cells were cotransfected with DNAs encoding either HIV-1 Gag-GFP or RSV Gag-GFP plus a control plasmid (indicated by a minus sign) or a plasmid encoding myc-tagged 5ptase (indicated by a plus sign). Twenty-four hours posttransfection, virus-like particles (VLPs) were collected by centrifugation from the medium and the cells were lysed. The amounts of Gag-GFP in the lysate and in the VLP fractions were measured by Western blot analysis. Virus release was calculated as the amount of extracellular Gag as a fraction of total Gag. Error bars represent standard deviations from the mean from three independent experiments.
Role of PIPs in binding of RSV Gag to liposomes.
In our previously published protein flotation studies, we found that recombinant, purified RSV MA and myristoylated HIV-1 MA bound to liposomes containing physiologically relevant levels of the acidic lipid PS (13, 14). In preliminary experiments, the addition of 1% PI(4,5)P2 to standard PC/PS (2:1) liposomes did not lead to a significant increase in the amount of purified protein that floated. Based on these results and on computational modeling, we concluded that electrostatic interactions are the major factor in the binding of RSV and HIV-1 MA to membranes, rather than specific binding to PI(4,5)P2. More recent work from other laboratories has focused on liposome interaction of the whole HIV-1 Gag protein, synthesized in vitro in a reticulocyte system (11, 12, 29). These studies showed that high levels (>5%) of PIPs, including but not limited to PI(4,5)P2 and PI(3,4,5)P3, strongly augment the interaction of HIV-1 Gag with PC/PS (2:1) liposomes.
To further investigate how PIPs influence RSV Gag interaction with membranes, we performed liposome flotation reactions using a truncated version of RSV Gag that is missing the C-terminal protease domain (GagΔPR, hereafter referred to as RSV Gag). This 35S-labeled protein was synthesized using a commercial reticulocyte lysate, and the crude mixture was incubated with extruded, 100-nm PC/PS (2:1) liposomes with or without added PIPs. After centrifugation in a sucrose step gradient, the percent of liposome-associated protein was quantified by SDS-PAGE and fluorography. As a control, flotation reactions were carried out in parallel with HIV-1 Gag (11, 12, 29).
The presence of a high-molecular-weight band in the membrane fraction of RSV Gag was observed when the proteins were resolved by SDS-PAGE, which complicated the analysis of the flotation data. This band, at an apparent molecular mass greater than 250 kDa, was observed predominantly in the presence of PIPs. We infer that it corresponds to an aggregate or an assembled oligomer of RSV Gag, since the majority of total protein-associated radioactivity was in Gag, as evident when the reticulocyte reaction was analyzed in the absence of added liposomes (data not shown). All attempts to break apart this aggregate failed (see Materials and Methods). Therefore, we included the high-molecular-weight species in the quantification (Fig. 5B, filled bars).
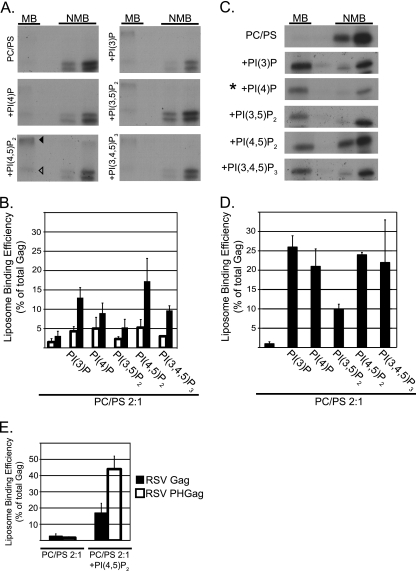
Fig. 5.
Flotation analysis of RSV Gag and HIV-1 Gag binding to PC/PS and PC/PS/PIP liposomes. All liposomes were PC/PS (2:1) without or with (+) the addition of 7.25% PIPs. (A and C) Representative flotation results for RSV Gag and HIV-1 Gag, respectively. MB, membrane bound (floated liposomes); NMB, not membrane bound. The RSV autoradiograms show the Gag band (open triangle) and a higher-molecular-weight band (filled triangle), which was consistently observed in the presence of liposomes containing PIPs. In C, the asterisk indicates that one-fourth of the total was used for the NMB samples. (B and D) Quantification of three or more flotation reactions for each liposome composition. The RSV quantification includes values for the Gag band only (open bars) or the Gag band plus higher-molecular-weight bands (filled bars). (E) Quantification of RSV PHGag and RSV Gag (data taken from Fig. 5D). Error bars represent standard deviations from the mean.
Compared with what was seen in previous studies using purified RSV MA, RSV Gag synthesized in the reticulocyte lysate interacted weakly with PC/PS (2:1) liposomes, being barely detectable in the floated fraction (Fig. 5A and B). At a concentration of 7.25% of total lipids, diverse PIPs increased the fraction of protein that floated but with little evidence of specificity; mono-, di-, and triphosphorylated PIPs all augmented membrane binding (Fig. 5A and B). Although PI(4,5)P2 appeared most effective, PI(3)P, PI(4)P, and PI(3,4,5)P3 also increased the amount of Gag protein in the membrane fraction by at least 3-fold. From these results, we conclude that the enhanced liposome interaction promoted by high concentrations of PIPs is due to electrostatics and not to specific recognition of the inositol head group by the RSV Gag protein.
To address if RSV Gag could be converted into a specific PI(4,5)P2 binding protein, we created the chimeric protein described above, in which the MA domain is replaced by a PH domain. RSV PHGag was found to associate with PI(4,5)P2-containing liposomes much more strongly than RSV Gag itself, while binding to PC/PS liposomes was unchanged (Fig. 5E). This result suggests that protein folding or other features of RSV Gag do not mask potential PIP interaction sites.
Several flotation studies reported that while HIV-1 Gag binds very poorly to PC/PS (2:1) liposomes, this interaction is strongly enhanced by inclusion of at least 5% PIPs in the artificial membranes (11, 12, 54). While PI(4,5)P2 and PI(3,4,5)P3 were most effective, other PIP species also augmented binding (11). We tested the ability of a collection of PIPs, all at 7.25% of total lipids in PC/PS (2:1) liposomes, to promote HIV-1 Gag flotation (Fig. 5C and D). The results confirm the weak interaction of Gag with PC/PS (2:1) and the strong enhancing effect of relatively high concentrations of PI(4,5)P2 and PI(3,4.5)P3. However, the monophosphorylated species PI(3)P and PI(4)P, which have not been tested previously, were as potent in enhancing membrane binding as PI(4,5)P2. Thus, our flotation data for HIV-1 do not support the prevailing model that HIV-1 Gag-membrane interaction has a specific requirement for PI(4,5)P2, as has been inferred from the properties of the phosphoinositide binding pocket in the MA domain reported in nuclear magnetic resonance (NMR) studies of MA with short-chain PIPs (54). Rather, the data are more consistent with a model in which the high negative charge of the PIP head group acts electrostatically to attract Gag to the PM or acts directly or indirectly to induce myristate exposure, which then enhances membrane binding through hydrophobic interactions.
In our hands, the fraction of HIV-1 Gag associated with PIP-containing liposomes was always at least 2-fold greater than for RSV Gag. Previously, we reported that RSV MA and myristoylated HIV-1 MA have similar dissociation constants governing the binding to PC/PS (2:1) liposomes and that forced dimerization of these MA proteins strengthens liposome association (13, 14), as expected on theoretical grounds. HIV-1 CA and Gag dimerize with a Kd of about 10−5 M (15, 24, 31, 50), an interaction that is mediated by the C-terminal domain of CA. Under similar conditions, purified RSV CA does not dimerize (31). We speculate that the strong liposome interaction of HIV-1 Gag compared with that of RSV Gag is due at least in part to dimerization of the former.
Electrostatic nature of RSV Gag binding to liposomes.
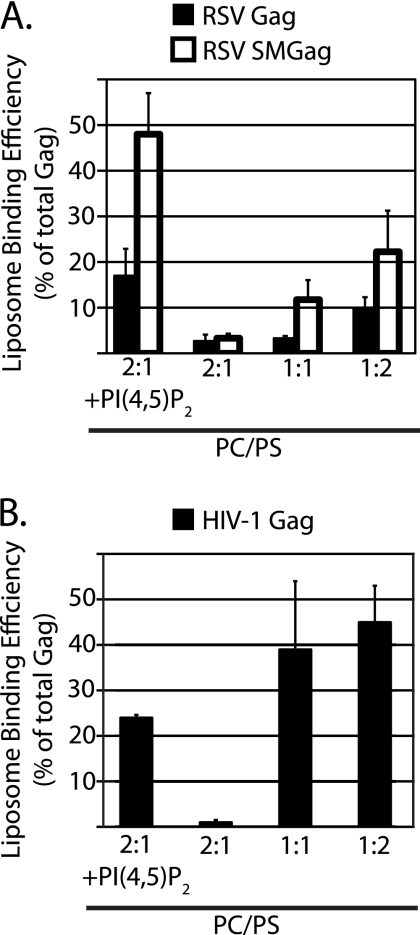
Purified RSV MA and HIV-1 MA interact more strongly with liposomes as the mole fraction of PS is increased (13, 14). To examine if the corresponding Gag proteins in reticulocyte lysates respond in a similar manner, we performed flotation reactions at different PS concentrations. As the fraction of PS increased from 33% to 67%, the amount of RSV Gag that floated increased by 3-fold (Fig. 6A, filled bars). We also examined a mutant RSV Gag protein called SuperM (SMGag) (6). In this mutant, two acidic residues in the RSV MBD are replaced by two basic residues, leading to a net increase of +4 in the overall charge of the MBD. In vivo, SMGag buds more rapidly than wild-type Gag and, unlike wild-type Gag, does not traffic through the nucleus before localizing to the PM (5). These early in vivo studies led to the initial conclusion that RSV Gag-membrane interaction is primarily electrostatic in nature. In the present flotation experiments, SMGag and wild-type Gag bound PC/PS (2:1) liposomes similarly. However, the mutant protein responded even more strongly to increased PS concentrations than the wild-type protein, with a more-than-6-fold increase (Fig. 6A, open bars). SMGag also responded more strongly to the presence of PI(4,5)P2 than did wild-type Gag.
Fig. 6.
Effect of PS concentration on binding of RSV and HIV Gag to PC/PS liposomes. The leftmost bar or bars show liposome association in the presence of PI(4,5)P2 as a reference point, with the data taken from Fig. 5. The other bars show liposome association at different PC:PS ratios without added PIPs. (A) RSV Gag (filled bars) and RSV SMGag (open bars). (B) HIV-1 Gag.
HIV-1 Gag liposome binding also rose dramatically at higher PS concentrations, with more than a 20-fold increase from 33% to 67% PS (Fig. 6B). These very high PS concentrations promoted even stronger HIV-1 Gag-liposome interaction than 7.25% PI(4,5)P2. We initially considered this result to be surprising, but it was invariably observed with different liposome batches and translation reactions. These results stand in contrast to results of previously reported experiments from the Ono laboratory, in which high PS concentrations could not supplant PI(4,5)P2 (11, 29).
Taken altogether, we interpret the data from the liposome flotation assays to support the model that for RSV Gag and HIV-1 Gag, membrane binding in vitro is driven primarily by charge interactions rather than by recognition of a specific phospholipid.
DISCUSSION
We have used in vivo and biochemical assays to show that RSV Gag does not have a specific requirement for PI(4,5)P2 in order to associate with membranes. Cellular depletion of PI(4,5)P2 by overexpression of a 5-phosphatase did not reduce RSV Gag-GFP localization to the PM nor did it compromise the release of VLPs. In contrast, in parallel experiments, HIV-1 Gag-GFP localization to the PM was nearly ablated and VLP production was inhibited by PI(4,5)P2 depletion, as reported previously by others (42). After synthesis in a reticulocyte lysate, RSV Gag protein bound weakly to standard PC/PS (2:1) liposomes. While this binding was augmented by inclusion of 7.25% PI(4,5)P2, other PIP species were equally effective in promoting binding. High levels of PS, in the absence of added PIPs, enhanced binding similarly. All of these results are consistent with our liposome flotation analysis of the purified RSV MA domain and confirm our conclusion that Gag-membrane binding is driven primarily by electrostatic interactions in RSV. Therefore, we infer that even after in vivo depletion of PI(4,5)P2 by 5ptase, the PM apparently retains enough negatively charged lipids to allow RSV Gag-PM targeting and binding.
While HIV-1 was intended primarily as a positive control, our results differ in some respects from those published previously. On the one hand, PI(4,5)P2 depletion abrogated HIV-1 Gag-GFP PM localization, as first found in the pioneering study by Ono et al. (42) and later confirmed when HIV-1 was used as a control for studies on equine infectious anemia virus (EIAV) (20), Mason-Pfizer monkey virus (MPMV) (60), and human T-cell lymphotropic virus (HTLV) (29), as well as in our own in vivo experiments. On the other hand, our biochemical results from liposome flotation assays diverged from those reported earlier (11, 29). We found no evidence of significant specificity of HIV-1 Gag for PI(4,5)P2. Other PIP species as well as high levels of PS promoted similar binding to liposomes. It is important to note that the lack of PIP specificity in vitro does not necessarily contradict the observation that phosphatase-mediated depletion of PI(4,5)P2 leads to the loss of Gag-GFP at the PM. Without a detailed understanding of the long-term effects of 5ptase on the pools of each phosphoinositide in different membrane compartments, it is not possible to directly connect biochemical data on flotation with fluorescence imaging of Gag-GFP in cells. In addition, other factors may functionally distinguish liposome binding of reticulocyte-translated proteins in vitro from PM binding in vivo, for example membrane curvature, the presence of other phospholipids and cholesterol, or the presence of rabbit reticulocyte proteins capable of binding to negatively charged lipids or to Gag.
What is the origin of the discrepancies between the flotation results for HIV-1 Gag reported here and the results published previously from the Ono lab (11, 29)? Our observation that PIP species other than PI(4,5)P2 also strongly promote liposome binding is perhaps less significant, since for those PIP species tested by both labs [PI(4,5)P2, PI(3,5)P2, and PI(3,4,5)P3], the raw data are not very different (11). However, our finding that high PS promotes HIV-1 Gag binding at least as strongly as 7% PI(4,5)P2 is in striking contrast to published results that showed only marginal Gag binding to liposomes with 60% PS (11, 12, 29). We tested and eliminated several possible explanations for this discrepancy, including different HIV-1 strain background (BH10 versus NL4-3), different origins of lipids (brain derived versus synthetic), and different Gag structures (lacking or including the p6 domain). Instead, the explanation appears to lie in the methods of liposome preparation and possibly the method of flotation. The previously reported experiments were carried out with liposomes that had been made by sonication in plastic tubes immersed in a water bath, conditions that are unlikely to effectively break up large multilamellar vesicles (G. Feigenson, personal communication). Our liposomes were made by repeated extrusion through a 100-nm filter, which is expected to produce small unilamellar vesicles and consequently a much larger available surface area. We traded liposome preparations with the Ono lab and in preliminary experiments confirmed the published results that for these sonicated liposomes, HIV-1 Gag floated much more extensively at 2:1 PC:PS plus 7% PI(4,5)P2 than at 1:1 PC: PS. In parallel, for our extruded liposomes, HIV-1 Gag bound more extensively at 1:1 PC:PS than at 2:1 PC:PS plus 7% PI(4,5)P2. The Ono lab observed a similar trend, although with quantitatively less flotation for the extruded than for the sonicated liposomes. We speculate that the differences in accessible membrane surface area, and perhaps also differences in liposome size and buoyant density as well as differences in centrifugation conditions, lead to the very different measurements of ability of HIV-1 Gag in the crude reticulocyte lysate to attach to artificial membranes. For example, basic reticulocyte proteins, which might be in vast excess over the translated radioactive HIV-1 Gag, might cover the smaller membrane surface of multilamellar liposomes and thereby prevent the weak binding of Gag to PS.
The role of PI(4,5)P2 in retroviral Gag-membrane interaction has been addressed for several retroviruses but with divergent results for different experimental settings and different viruses. In HIV-1, early work on in vitro assembly of purified Gag protein, in the absence of membranes, first suggested the importance of phosphoinositides. Assembly of properly sized VLPs required inositol hexakisphosphate or pentakisphosphate (7), which were interpreted to mimic a phosphoinositide in cells, possibly PI(3,4,5)P3. Some of the side chains interacting with these highly charged molecules have been mapped (57). In the original study by Ono et al. (42), phosphatase-mediated depletion of PI(4,5)P2 was found to redirect Gag-GFP away from the PM. A binding pocket for short-chain PI(4,5)P2 (54), which was later also identified for HIV-2 (51), suggested that this lipid species is responsible for HIV-1 Gag PM localization. More recently, PI(4,5)P2 was found to be significantly enriched in HIV-1 virions (9).
As measured by NMR, EIAV MA also was found to bind short-chain PIPs, but in this case several PIP species had affinities higher than that of PI(4,5)P2 (20). Early experiments were interpreted to mean that EIAV MA can bind to neutral liposomes but that membrane association is strongly enhanced by acidic lipids (48). However, in a more recent report, PM fluorescence of EIAV Gag-GFP was not affected by expression of 5ptase, although a possibly more potent PIP phosphatase induced major changes in subcellular localization (20).
In the MLV system, the expression of 5ptase reduced MLV VLP release (25), as in the case of HIV-1. Also, PI(4,5)P2 was found to be enriched in virions (9). In vitro, PIPs in PC liposomes were found to be required for binding of MA (25), but the MA was unmyristoylated and thus the biological significance is uncertain. This stimulatory effect was quantitatively similar for diverse PIPs, and little or no binding was observed to liposomes containing only PS. However, in the presence of 20% PS, PI(4,5)P2 specifically stimulated MA binding with an apparent Kd 4-fold lower than those of other PIPs, suggesting a synergism between these two negatively charged lipids.
Finally, HTLV-1 Gag and chimeric Gag proteins bearing the HTLV-1 MA domain did not show a specific requirement for PI(4,5)P2, either for PM localization in cells or for liposome binding from reticulocyte lysates. However, acidic phospholipids, including PIPs, stimulated liposome binding, similar to what we have observed for RSV Gag. Altogether, these diverse results for different retrovirus Gag proteins suggest that if a head group-specific PI(4,5)P2 interaction is biologically important, it may be limited to HIV-1 Gag and its close relatives.
In overview, at least five principles underlie PM targeting of retroviral Gag proteins. First, vesicular trafficking appears to play a central role, as demonstrated by the interaction of the HIV-1 MA domain with clathrin adapter protein AP3 and the effects of knocking out this binding (17). But understanding of the trafficking pathway by which any Gag protein reaches the PM remains rudimentary and controversial, despite attempts to follow this kinetically (46).
Second, as described above, a PI(4,5)P2 head group-specific binding pocket has been identified at the atomic level by NMR for HIV-1 and HIV-2 (51, 54). According to the myristoyl switch model proposed by these studies (23, 52–54), one of the fatty acid chains of PI(4,5)P2 is extracted from the lipid bilayer and is bound to a hydrophobic groove in MA, just as the N-terminal myristate is extracted from its pocket in MA and becomes inserted into the membrane. In this model, it remains uncertain how the formidable energy barrier would be overcome to promote the movement of a fatty acid chain from the lipid bilayer into a hydrophobic groove in a protein. The biochemical experiments on which the model is based, as well as the NMR experiments measuring the affinities of EIAV MA protein for purified phosphoinositides (20), were necessarily carried out with short-chain PIPs to maintain their solubility. A recent study using surface plasmon resonance to measure PIP binding to HIV-1 Gag concluded that the lengths of the PIP acyl chains are the major contributing factor in determining the PIP binding affinity of HIV-1 Gag, not the phosphorylation state of the inositol head group (3). Thus, the biological relevance of studies with short-chain PIPs remains to be established. Finally, interpreting the effects of 5ptase-mediated depletion of PI(4,5)P2 on Gag PM localization may be more difficult than commonly assumed. To date, all retrovirus studies on 5ptase-depletion, including ours, have been long-term, with the endpoints representing a cumulative effect of the phosphatase over many hours. Secondary effects of perturbing PIP pools are almost certain to occur and thus limit the interpretations of fluorescence imaging. All eukaryotic cells maintain tight control of the concentrations and localization of PIPs, which probably explains why overexpression of 5ptase can trigger apoptosis in at least some cell types.
Third, hydrophobic interactions are critical in mediating membrane interactions for those Gag proteins modified by an N-terminal myristate, like HIV-1 Gag and MLV Gag. It is widely accepted that insertion of a single fatty acid into a lipid bilayer is insufficient to lock a protein into the membrane. In the absence of other membrane-binding features, a subpopulation of proteins will be cytosolic as the fatty acid moiety samples the aqueous space. In the case of retroviral Gag proteins, several factors, including multimerization of Gag (61), binding of the MA domain to PIPs (1, 51, 54), or membrane proximity, can tip this balance toward membrane attachment. By bringing the myristate into close proximity to the PM, electrostatic interactions between the MBD and acidic phospholipids favor insertion of the myristate into the lipid bilayer where it is more stable. The quantitative contributions of these factors in promoting myristate insertion are unknown. Because of the complexity in understanding the behavior of myristate and protein binding grooves for it, the nature of retroviral Gag-membrane interaction may be easiest to work out for viruses like RSV, which rely primarily on electrostatic interactions with membranes.
Fourth, multimerization of any membrane-binding protein increases its avidity for the membrane, and this property is intrinsic to Gag. Multimerization is expected to be concentration dependent, and hence expression levels also should affect membrane binding, as found experimentally for HIV-1 MA (45). To date, the effects of multimerization on membrane binding have been modeled experimentally and computationally only by dimerization, using RSV MA and HIV MA proteins that were induced to dimerize artificially. The behavior of higher-order multimers, which might be formed in the cytoplasm or assembled on the membrane, has not been studied. Once a Gag lattice has grown beyond a minimal size, the lattice probably would remain firmly attached to the PM even after depletion of PI(4,5)P2. The role of kinetics in PM binding, which is likely to be critical, has not been addressed in any study. Future work on the effects of multimerization and of PI(4,5)P2 depletion in Gag PM binding may require truncated or mutated Gag proteins that do not multimerize (18).
Fifth, electrostatic interactions appear to be universal in Gag membrane targeting, perhaps providing the major force directing Gag to the PM. All MA proteins have basic patches on the membrane-proximal surface (40), and the inner leaflet of the PM is unique in its high concentration of acidic lipids. This negative charge results mainly from PS and PI(4,5)P2, but the relative contributions of each of these lipids are not well understood and may depend on cell type (27, 67). In addition, clustering of lipids in membranes may result in formation of microdomains with different charges. Fluorescent protein sensors have been developed to probe for the overall negative potential of the inner leaflet. These probes are based on a positively charged segment of polypeptide plus a lipid anchor that by itself is insufficient for steady-state PM localization (67). Such probes, together with PH-GFP and fluorescently marked Gag proteins, could help elucidate if the electrostatic interactions between MA domains and the PM are powered primarily by PI(4,5)P2, by PS, or by a combination of the two. Different retroviruses probably lie on a gamut, with some relying more on PI(4,5)P2 and others relying more on PS. Gag proteins that rely more on PI(4,5)P2 may do so in part because of a specific binding pocket for the head group or because of a local clustering of basic residues that is preferentially attracted to the multiple negative charges of this lipid. The latter effect could explain the frequently reported strong effects of PIPs in flotation analyses but without specificity for PI(4,5)P2. In summary, we propose as a unified, working model that not only RSV Gag but all retroviral Gag proteins rely primarily on electrostatic interactions to drive at least the first steps in PM binding.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by USPHS grant CA20081 to V.M.V.
We thank David Holowka, Akira Ono, and Alan Rein for DNA clones and Gerald Feigenson for advice on handling lipids. We are particularly appreciative of Akira Ono's willingness to trade liposomes to help resolve the discrepancies in HIV-1 Gag flotation results obtained by our two laboratories.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 3 August 2011.
REFERENCES
- 1. Alfadhli A., Still A., Barklis E. 2009. Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. J. Virol. 83:12196–12203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aloia R. C., Tian H., Jensen F. C. 1993. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc. Natl. Acad. Sci. U. S. A. 90:5181–5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anraku K., et al. 2010. Highly sensitive analysis of the interaction between HIV-1 Gag and phosphoinositide derivatives based on surface plasmon resonance. Biochemistry 49:5109–5116 [DOI] [PubMed] [Google Scholar]
- 4. Brugger B., et al. 2006. The HIV lipidome: a raft with an unusual composition. Proc. Natl. Acad. Sci. U. S. A. 103:2641–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Callahan E., Wills J. 2003. Link between genome packaging and rate of budding for Rous sarcoma virus. J. Virol. 77:9388–9398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Callahan E., Wills J. 2000. Repositioning basic residues in the M domain of the Rous sarcoma virus gag protein. J. Virol. 74:11222–11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Campbell S., et al. 2001. Modulation of HIV-like particle assembly in vitro by inositol phosphates. Proc. Natl. Acad. Sci. U. S. A. 98:10875–10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Campbell S., Vogt V. M. 1997. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J. Virol. 71:4425–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chan R., et al. 2008. Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J. Virol. 82:11228–11238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen K., et al. 2008. Solution NMR characterizations of oligomerization and dynamics of equine infectious anemia virus matrix protein and its interaction with PIP2. Biochemistry 47:1928–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chukkapalli V., Hogue I., Boyko V., Hu W., Ono A. 2008. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient Gag membrane binding. J. Virol. 82:2405–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chukkapalli V., Oh S. J., Ono A. 2010. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc. Natl. Acad. Sci. U. S. A. 107:1600–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dalton A., Murray P., Murray D., Vogt V. 2005. Biochemical characterization of Rous sarcoma virus MA protein interaction with membranes. J. Virol. 79:6227–6238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dalton A. K., Ako-Adjei D., Murray P. S., Murray D., Vogt V. M. 2007. Electrostatic interactions drive membrane association of the human immunodeficiency virus type 1 Gag MA domain. J. Virol. 81:6434–6445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. del Alamo M., Neira J. L., Mateu M. G. 2003. Thermodynamic dissection of a low affinity protein-protein interface involved in human immunodeficiency virus assembly. J. Biol. Chem. 278:27923–27929 [DOI] [PubMed] [Google Scholar]
- 16. DiNitto J., Lambright D. 2006. Membrane and juxtamembrane targeting by PH and PTB domains. Biochim. Biophys. Acta 1761:850–867 [DOI] [PubMed] [Google Scholar]
- 17. Dong X., et al. 2005. AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell 120:663–674 [DOI] [PubMed] [Google Scholar]
- 18. Dou J., et al. 2009. Characterization of a myristoylated, monomeric HIV Gag protein. Virology 387:341–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ferguson K., Lemmon M., Schlessinger J., Sigler P. 1995. Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell 83:1037–1046 [DOI] [PubMed] [Google Scholar]
- 20. Fernandes F., et al. 2011. Phosphoinositides direct equine infectious anemia virus gag trafficking and release. Traffic 12:438–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Finzi A., Orthwein A., Mercier J., Cohen E. A. 2007. Productive human immunodeficiency virus type 1 assembly takes place at the plasma membrane. J. Virol. 81:7476–7490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Freed E., Orenstein J., Buckler-White A., Martin M. 1994. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J. Virol. 68:5311–5320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Freed E. O. 2006. HIV-1 Gag: flipped out for PI(4,5)P(2). Proc. Natl. Acad. Sci. U. S. A. 103:11101–11102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gamble T. R., et al. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849–853 [DOI] [PubMed] [Google Scholar]
- 25. Hamard-Peron E., et al. 2010. Targeting of murine leukemia virus gag to the plasma membrane is mediated by PI(4,5)P2/PS and a polybasic region in the matrix. J. Virol. 84:503–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hatanaka H., et al. 2002. Structure of equine infectious anemia virus matrix protein. J. Virol. 76:1876–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heo W. D., et al. 2006. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science 314:1458–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hom R. A., et al. 2007. pH-dependent binding of the Epsin ENTH domain and the AP180 ANTH domain to PI(4,5)P2-containing bilayers. J. Mol. Biol. 373:412–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Inlora J., Chukkapalli V., Derse D., Ono A. 2011. Gag localization and virus-like particle release mediated by the matrix domain of human T-lymphotropic virus type-1 Gag are less dependent on phosphatidylinositol-(4,5)-bisphosphate than those mediated by the matrix domain of human immunodeficiency virus type-1 Gag. J. Virol. 85:3802–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jouvenet N., et al. 2006. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 4:e435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kingston R. L., et al. 2000. Structure and self-association of the Rous sarcoma virus capsid protein. Structure 8:617–628 [DOI] [PubMed] [Google Scholar]
- 32. Kisseleva M. V., Wilson M. P., Majerus P. W. 2000. The isolation and characterization of a cDNA encoding phospholipid-specific inositol polyphosphate 5-phosphatase. J. Biol. Chem. 275:20110–20116 [DOI] [PubMed] [Google Scholar]
- 33. Kutateladze T. 2010. Translation of the phosphoinositide code by PI effectors. Nat. Chem. Biol. 6:507–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lemmon M. A., Ferguson K. M., O'Brien R., Sigler P. B., Schlessinger J. 1995. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc. Natl. Acad. Sci. U. S. A. 92:10472–10476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Leventis P. A., Grinstein S. 2010. The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys. 39:407–427 [DOI] [PubMed] [Google Scholar]
- 36. Maldonado R., Blough H. 1980. A comparative study of the lipids of plasma membranes of normal cells and those infected and transformed by Rous sarcoma virus. Virology 102:62–70 [DOI] [PubMed] [Google Scholar]
- 37. Manna D., et al. 2008. Differential roles of phosphatidylserine, PtdIns(4,5)P2, and PtdIns(3,4,5)P3 in plasma membrane targeting of C2 domains. Molecular dynamics simulation, membrane binding, and cell translocation studies of the PKCalpha C2 domain. J. Biol. Chem. 283:26047–26058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McDonnell J. M., et al. 1998. Solution structure and dynamics of the bioactive retroviral M domain from Rous sarcoma virus. J. Mol. Biol. 279:921–928 [DOI] [PubMed] [Google Scholar]
- 39. McLaughlin S., Murray D. 2005. Plasma membrane phosphoinositide organization by protein electrostatics. Nature 438:605–611 [DOI] [PubMed] [Google Scholar]
- 40. Murray P., et al. 2005. Retroviral matrix domains share electrostatic homology: models for membrane binding function throughout the viral life cycle. Structure 13:1521–1531 [DOI] [PubMed] [Google Scholar]
- 41. Nelle T., Wills J. 1996. A large region within the Rous sarcoma virus matrix protein is dispensable for budding and infectivity. J. Virol. 70:2269–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ono A., Ablan S., Lockett S., Nagashima K., Freed E. 2004. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl. Acad. Sci. U. S. A. 101:14889–14894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ono A., Orenstein J., Freed E. 2000. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J. Virol. 74:2855–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Parent L., Wilson C., Resh M., Wills J. 1996. Evidence for a second function of the MA sequence in the Rous sarcoma virus Gag protein. J. Virol. 70:1016–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Perez-Caballero D., Hatziioannou T., Martin-Serrano J., Bieniasz P. 2004. Human immunodeficiency virus type 1 matrix inhibits and confers cooperativity on gag precursor-membrane interactions. J. Virol. 78:9560–9563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Perlman M., Resh M. 2006. Identification of an intracellular trafficking and assembly pathway for HIV-1 gag. Traffic 7:731–745 [DOI] [PubMed] [Google Scholar]
- 47. Pessin J. E., Glaser M. 1980. Budding of Rous sarcoma virus and vesicular stomatitis virus from localized lipid regions in the plasma membrane of chicken embryo fibroblasts. J. Biol. Chem. 255:9044–9050 [PubMed] [Google Scholar]
- 48. Provitera P., Bouamr F., Murray D., Carter C., Scarlata S. 2000. Binding of equine infectious anemia virus matrix protein to membrane bilayers involves multiple interactions. J. Mol. Biol. 296:887–898 [DOI] [PubMed] [Google Scholar]
- 49. Quigley J. P., Rifkin D. B., Reich E. 1971. Phospholipid composition of Rous sarcoma virus, host cell membranes and other enveloped RNA viruses. Virology 46:106–116 [DOI] [PubMed] [Google Scholar]
- 50. Rose S., et al. 1992. Characterization of HIV-1 p24 self-association using analytical affinity chromatography. Proteins 13:112–119 [DOI] [PubMed] [Google Scholar]
- 51. Saad J., et al. 2008. Structure of the myristylated human immunodeficiency virus type 2 matrix protein and the role of phosphatidylinositol-(4,5)-bisphosphate in membrane targeting. J. Mol. Biol. 382:434–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saad J., et al. 2007. Mutations that mimic phosphorylation of the HIV-1 matrix protein do not perturb the myristyl switch. Protein Sci. 16:1793–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Saad J., et al. 2007. Point mutations in the HIV-1 matrix protein turn off the myristyl switch. J. Mol. Biol. 366:574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Saad J., et al. 2006. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. U. S. A. 103:11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Scheifele L., Rhoads J., Parent L. 2003. Specificity of plasma membrane targeting by the Rous sarcoma virus gag protein. J. Virol. 77:470–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Scholz I., et al. 2008. Analysis of human immunodeficiency virus matrix domain replacements. Virology 371:322–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shkriabai N., et al. 2006. Interactions of HIV-1 Gag with assembly cofactors. Biochemistry 45:4077–4083 [DOI] [PubMed] [Google Scholar]
- 58. Soneoka Y., Kingsman S., Kingsman A. 1997. Mutagenesis analysis of the murine leukemia virus matrix protein: identification of regions important for membrane localization and intracellular transport. J. Virol. 71:5549–5559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Spearman P., Wang J. J., Vander Heyden N., Ratner L. 1994. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J. Virol. 68:3232–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stansell E., et al. 2007. Basic residues in the Mason-Pfizer monkey virus gag matrix domain regulate intracellular trafficking and capsid-membrane interactions. J. Virol. 81:8977–8988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tang C., et al. 2004. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc. Natl. Acad. Sci. U. S. A. 101:517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Uekama N., Sugita T., Okada M., Yagisawa H., Tuzi S. 2007. Phosphatidylserine induces functional and structural alterations of the membrane-associated pleckstrin homology domain of phospholipase C-delta1. FEBS J. 274:177–187 [DOI] [PubMed] [Google Scholar]
- 63. Varnai P., Balla T. 1998. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell Biol. 143:501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wills J., Craven R., Weldon R. J., Nelle T., Erdie C. 1991. Suppression of retroviral MA deletions by the amino-terminal membrane-binding domain of p60src. J. Virol. 65:3804–3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wiradjaja F., et al. 2007. Inactivation of the phosphoinositide phosphatases Sac1p and Inp54p leads to accumulation of phosphatidylinositol 4,5-bisphosphate on vacuole membranes and vacuolar fusion defects. J. Biol. Chem. 282:16295–16307 [DOI] [PubMed] [Google Scholar]
- 66. Yagisawa H. 2006. Nucleocytoplasmic shuttling of phospholipase C-delta1: a link to Ca2+. J. Cell. Biochem. 97:233–243 [DOI] [PubMed] [Google Scholar]
- 67. Yeung T., et al. 2008. Membrane phosphatidylserine regulates surface charge and protein localization. Science 319:210–213 [DOI] [PubMed] [Google Scholar]
- 68. Zhou W., Parent L. J., Wills J. W., Resh M. D. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 68:2556–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.