Abstract
A series of potently neutralizing monoclonal antibodies (MAbs) that target quaternary epitopes on the native Env trimer have recently been described. A common feature shared by these antibodies is the critical involvement of sites in both the V2 and V3 variable domains in antibody recognition. In this study the gp120 variable-region determinants were mapped for eight rhesus macaque monoclonal antibodies (RhMAbs) possessing potently neutralizing activity specific for a quaternary target in SF162 Env and compared to those originally identified for human MAb 2909. These studies showed that determinants for the epitopes defined by the RhMAbs differed in both the V2 (positions 160, 167, and 169) and V3 (positions 313 and 315) regions from 2909, and in a number of cases, from each other. Attempts to reconstitute expression of these epitopes on the cell surface by cotransfecting Envs containing either the V2 or the V3 determinant of the epitope were not successful, suggesting that these epitopes were expressed on individual protomers in a trimer-dependent manner. Several of the V2 positions found to be critical for expression of these quaternary epitopes also significantly affected exposure and neutralization sensitivity of targets in the V3 and CD4-binding domains. These results demonstrated a considerable diversity in the fine structure of this class of epitopes and further suggested a potentially important relationship between the expression of such quaternary epitopes and V1/V2-mediated masking of immunodominant epitopes.
INTRODUCTION
Human immunodeficiency virus (HIV) entry is mediated by oligomeric Env complexes present on the surface of virions, and recent evidence shows that epitopes that are dependent on quaternary structures specific for the native Env complexes can be particularly sensitive neutralization targets. This includes studies showing that a novel class of antibodies directed against broadly conserved quaternary neutralization epitopes (QNEs) that are expressed on native Env trimers but not soluble Env proteins possess broad and potent neutralizing activities (40) and are major components of broadly neutralizing activities found in some HIV-immune sera (41). Antibodies of this type generally bind poorly, if at all, to soluble Env proteins, and identification of monoclonal antibodies (MAbs) specific for quaternary epitopes required direct screening for virus-neutralizing activity, explaining why similar antibodies were not identified in earlier studies that utilized binding to purified proteins or peptides as screening assays.
The presence of quaternary neutralization epitopes in HIV type 1 (HIV-1) was first demonstrated in 2005 with the isolation of 2909, a MAb with an unusually potent neutralizing activity against the SF162 strain (10). 2909 was identified by screening Epstein-Barr virus (EBV)-transformed B-cell colonies from an infected human for neutralizing activity against virus pseudotyped with the SF162 Env. The binding activity of 2909 was specific for intact SF162 virions, and this MAb did not bind to soluble SF162 gp120 or gp140. Binding of the MAb to virions required the presence of both the V2 and V3 domains in gp120 and was abrogated by mutations in the HR1 region of TM that prevented the formation of trimeric Env complexes (14). The crystal structure of 2909 resembled that of the broadly neutralizing QNE-specific MAb, PG16, in the presence of a long protruding CDR3 loop that included several sulfated tyrosine residues (3, 25, 26).
Although 2909 activity was originally reported to be highly specific for the SF162 strain, we and others have shown that a single amino acid substitution in the V2 domain (Lys at position 160 in place of the more common Asn residue) was sufficient to introduce this epitope into a number of primary Envs (12, 43). This substitution resulted in the loss of the conserved N-linked glycosylation site that is an essential component of the broadly conserved quaternary epitopes PG9 and PG16 (40). Other than this rare substitution, the 2909 epitope was relatively conserved, leading us to hypothesize that more broadly expressed “2909-like” quaternary neutralization epitopes were likely to exist (27), an hypothesis confirmed by the isolation of PG9 and PG16.
The 2909 MAb was isolated from a donor infected with a unique HIV-1 isolate that contained Lys at position 160 (M. K. Gorny, unpublished data), thereby accounting for its cross-reactivity with SF162 Env and allowing its fortuitous identification by screening for neutralizing activity against SF162 pseudotypes. A similar neutralization screen performed with B cells from macaques infected with simian/human immunodeficiency virus SHIVSF162P4, which has an Env very similar in sequence to that of SF162, resulted in the isolation of a number of additional potently neutralizing MAbs with 2909-like specificities (34). That study established the relationship between the epitopes recognized by 2909 and the rhesus macaque MAbs (RhMAbs) and identified differences in the glycan requirements of 2909 and several of the RhMAbs at several positions in V2. A more recent study showed that 2909, but not the RhMAbs, was able to neutralize slightly more than one-third of primary Envs in which Asn160 was mutated to Lys, suggesting that these epitopes represented alternative antigenic variants of a related structure recognized by the broadly neutralizing PG9 and PG16 MAbs (43). In view of the dependence of these quaternary epitopes on oligomeric structure, specific carbohydrates, and elements in flexible variable loops, obtaining direct structural information of these epitopes will be very challenging. Therefore, other approaches are needed to determine the molecular characteristics of these targets.
This study provides a more detailed analysis of the epitopes recognized by eight of these RhMAbs and shows that these differed in both their V2 and V3 determinants from 2909, and in a number of cases, from each other. Key V2 determinants of these epitopes were also found to affect the masking activity of the V1/V2 domain for targets in both the V3 and CD4-binding domains (CD4-bd), supporting a role for these quaternary structures in epitope masking. Flow cytometry analyses of cells that coexpressed Envs containing either the V2 or V3 components of these epitopes suggested that these epitopes were localized on single gp120 protomers and not formed across neighboring protomers. The insights into the sequence requirements and structural diversity of these QNEs provided by these studies should help in understanding the nature of this class of epitope and in the design of immunogens capable of inducing such antibody specificities.
MATERIALS AND METHODS
Monoclonal antibodies.
The isolation of RhMAbs from macaques infected with SHIVSF162P4 was previously described (34). The Env of this SHIV was almost identical in sequence to the original HIV-1 SF162 Env. These antibodies were purified from EBV-transformed cultures on Protein A affinity columns. MAb 2909 was isolated from an HIV-1-infected individual by screening for the neutralization of HIV-1 pseudotyped with SF162 Env (10). 2F5 and 4E10 (33, 38), directed against epitopes in the ectodomain of gp41, were contributed by Hermann Katinger and obtained from the NIH AIDS Research and Reference Reagent Program. V3-specific MAbs 447-52D (9), B4e8 (2), 4117C (39), and 5145A (29), directed against an epitope overlapping the CD4-bd, were previously described.
Generation of chimeric and variant forms of Env.
SF162 Env variants containing modified V3 sequences were previously described (12, 16). Variants of the JR-FL Env (28) containing modified V2 residues were generated by sequentially introducing the necessary modifications by site-directed mutagenesis using the QuikChange kit, as described by the manufacturer (Stratagene, Inc.). The sequences of all mutant Envs were confirmed by sequencing the complete gene (Genewiz, Inc.). Amino acid numbering in Env is based on the HXB2 sequence and numbering of residues in the V3 domain start from the N-terminal cysteine.
Viral neutralization assays.
Neutralization activity was determined as previously described (17) with a single-cycle infectivity assay using virions generated from the Env-defective luciferase-expressing pNL4-3.Luc.R−E− genome (4) pseudotyped with the molecularly cloned HIV Env of interest. In brief, pseudotyped virions were incubated with serial dilutions of MAbs for 1 h at 37°C and were then added to U87-T4-CCR5 target cells plated out in 96-well plates in the presence of Polybrene (10 μg/ml). The amount of virus used in each neutralization assay was normalized to produce a standard level of luciferase activity (generally 25,000 to 50,000 units). After 24 h, cells were refed with RPMI medium containing 10% fetal bovine serum and Polybrene and incubated for an additional 24 to 48 h. Luciferase activity was determined 48 to 72 h postinfection with a microtiter plate luminometer (HARTA, Inc.), using assay reagents from Promega, Inc. Fifty-percent (IC50) and 90% (IC90) inhibitory concentrations presented in the tables were determined by interpolation from neutralization curves and are averages of at least three independent assays.
Analysis of epitope complementation by flow cytometric binding assays.
The ability of the quaternary epitopes analyzed in this study to assemble in trans upon coexpression of two nonreactive Envs was examined by flow cytometry. Envs which contained either the V2 or the V3 component of the epitope were cotransfected into 293 cells by using the TransIT-LT1 transfection reagent and protocol (Mirus Bio LLC, Madison, WI). One day later, cells were refed with fresh media containing 10% serum and incubated an additional 2 days. Cells were removed from plates for analysis by rinsing with 2 mM EDTA–phosphate-buffered saline (PBS) and stained with Env-specific MAbs conjugated to either fluorescein isothiocyanate (FITC) or phycoerythrin (PE). After incubation with the fluorescently labeled antibodies for 30 min at 4°C, the cells were washed, fixed in 4% paraformaldehyde, and analyzed for binding of MAbs specific for control or quaternary epitopes by using an Accuri C6 flow cytometer and C Flow software (Accuri Cytometers Inc., Ann Arbor, MI). Live cells were pregated and the extent of binding of 2909 and the RhMAbs was measured to determine the formation of quaternary epitopes. In one experiment, cells transfected with antigenically marked Envs were stained with MAbs specific for unique determinants present on the two Envs, to confirm that both Envs were efficiently coexpressed on the surface of individual cells.
RESULTS
RhMAbs possess unprecedented neutralization potency for SF162.
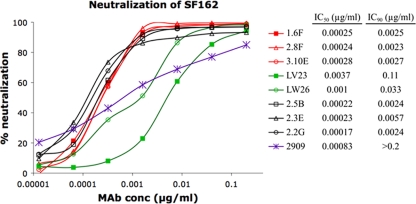
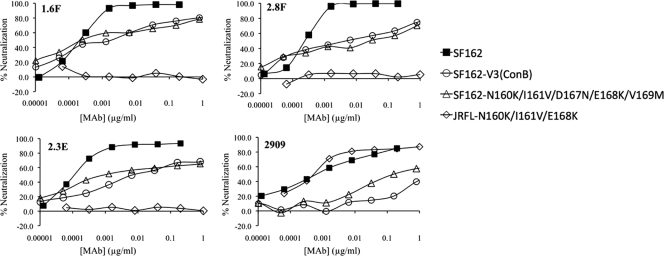
The SF162-specific RhMAbs described in this study were isolated by screening for neutralizing activity against SF162 viral pseudotypes (34), and the efficient identification of these MAbs was due in part to their remarkable neutralizing potency for this virus. Neutralization curves for eight RhMAbs and human MAb 2909 against the SF162 pseudotype are shown in Fig. 1. The human MAb 2909 is directed against a related quaternary epitope in SF162 Env that also is dependent on sites in the V2 and V3 domains (12). Most of the RhMAbs were considerably more potent than 2909 against the SF162 pseudotypes, as manifested both by their lower IC50s and by the shape of their neutralization curves (Fig. 1). The IC50s for six of the eight RhMAbs were in the range of 0.0002 to 0.0003 μg/ml; these activities are orders of magnitude more potent than those reported for antibodies against other sensitive epitopes in the V3 and CD4-binding domains of this Env (28). This remarkable potency was also seen when IC90 values are calculated, which were in the 0.002- to 0.006-μg/ml range for the six most potent MAbs, compared to >0.2 for 2909. The low IC90 value for 2909 is related to the shape of the neutralization curve for this antibody against SF162, which has a flat slope and does not plateau even at high concentrations (Fig. 1). This neutralization pattern was previously shown to be related to a reduced avidity for the V2 component of the epitope in SF162 Env (12). In contrast, the RhMAbs gave standard sigmoidal neutralization curves for the SF162 pseudotype, with steep slopes and pronounced plateaus at or close to 100% neutralization.
Fig. 1.
Neutralization curves for eight RhMAbs and human MAb 2909 against HIV-1 pseudotyped with wt SF162 Env. IC50 and IC90 values (μg/ml) for each MAb calculated from these curves are indicated. These curves and neutralization endpoints are representative of multiple assays.
Differences in V3 determinants of 2909 and QNE-specific RhMAbs.
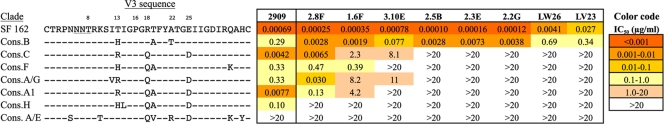
The QNE-specific RhMAbs resemble 2909 in that their ability to bind to virions required the presence of V2 and V3 but not V1 domains (34). The breadth of the V3 specificity of 2909 was previously shown by demonstrating its neutralizing activity for a series of chimeric SF162 Envs in which the V3 domain was replaced by various clade consensus (Cons) V3 sequences (12). While 2909 preferred the SF162 V3 sequence, it strongly neutralized Env chimeras with the clade C and A1 consensus V3 sequences and had weaker neutralizing activity against the consensus B, F, A/G, and H V3 chimeras. The V3 sequence requirements for the QNE-specific RhMAbs was determined by performing a similar analysis with the same set of V3 chimeric SF162 Envs (Table 1).
Table 1.
Sensitivity of QNE-specific MAbs to SF162 Env containing substitutions of various clade consensus V3 sequencesa,b
The sequences of the V3 domain of each of these proteins is compared to the endogenous SF162 sequence.
IC50s (μg/ml) for each of the MAbs is shown. The values shown in this and the following tables are averages of at least three independent experiments. The color code indicated is used for all of the tables in this paper.
The RhMAbs differed in the breadth of their reactivities with the consensus V3 sequences. Whereas two of the antibodies (2.8F and 1.6F) neutralized five of the seven clade consensus sequences, the others were considerably narrower, with five recognizing only the clade B consensus sequence. The fine specificities of the two broadest RhMAbs, 2.8F and 1.6F, differed from 2909 and from each other. Whereas 2909 strongly preferred the Cons C and A1 sequences to the Cons B sequence, 2.8F strongly neutralized the Cons B and Cons C sequences but reacted only weakly with the Cons A1 sequence, while 1.6F neutralized the Cons B sequence strongly and recognized the other consensus sequences only weakly.
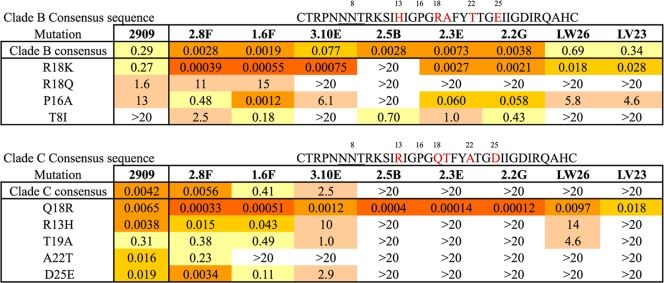
The dependence of these MAbs on specific V3 residues was determined by examining their neutralization potencies for point mutants introduced into the SF162 Cons B or Cons C V3 chimeric Envs. A key characteristic of the clade B V3 loop is the presence of an Arg at position 18 as part of the GPGR motif at the tip of the V3 loop (position 315 in Env), in place of Gln usually found at that position (GPGQ) for other clades (8). In contrast to 2909, which was not sensitive to substitutions at the tip of the loop, the RhMAbs were highly sensitive to mutations in this region and in most cases were strictly dependent on the basic residues Arg or Lys at position 18 (Table 2). Conversion of Gln18 in the consensus clade C sequence to Arg resulted in strong neutralization by all of the RhMAbs, including those that were nonreactive with the clade C consensus sequence, while the Arg-to-Gln conversion in the consensus clade B sequence resulted in loss of reactivity for those MAbs and a dramatic reduction in potency for the RhMAbs with cross-clade reactivity. The second most common residue at position 18 in clade B sequences is Lys (present at ∼10% frequency), and substitution of this residue affected the RhMAbs differently. The Arg18Lys substitution in the clade B sequence resulted in complete loss of reactivity for one of the RhMAbs (2.5B), had only minor effects for the reactive MAbs for several of the RhMAbs (2.3E and 2.2G), and caused significant increases in potency for the other five RhMAbs. Substitutions into the Cons C chimera at the other positions differing between the consensus B and C sequences resulted in only minor effects for the reactive MAbs and usually decreased neutralization sensitivity, with the greatest effect observed for the A22T mutation. These results demonstrated a dominant role for the tip of the V3 loop in recognition of the quaternary epitopes by the RhMAbs.
Table 2.
Effect of point mutations in the clade B and clade C consensus sequences on neutralization potency of QNE-specific MAbsa
IC50s (μg/ml) for each of the MAbs are shown. Color code is as in Table 1.
Differential effects were also observed for 2909 and the RhMAbs for point substitutions at different positions in the Cons B V3 region. The Pro at position 16 of the V3 loop is a highly conserved residue critical for maintaining the β-turn present at the tip of the V3 loop, and molecular dynamic simulations predicted that a Pro→Ala substitution at this position should result in a conformational change with loss of the β-hairpin structure that typically characterizes the V3 crown structure (13). The Pro16Ala mutation in the clade B V3 sequence increased the IC50 of 2909 by 44-fold and also affected sensitivity to most of the RhMAbs to different extents, ranging from an insignificant effect for 1.6F, to modest decreases in potency (8- to 15-fold increases in IC50) for four of the antibodies (2.3E, 2.2G, LW26, and LV23), to larger increases in IC50 (80- and 170-fold) for 3.10E and 2.8F, and a >7,000-fold increase for 2.5B (Table 2). These results suggested that the RhMAbs recognize a diversity of V3 structures and that this class of QNE is generally tolerant of multiple conformations of the V3 domain.
The T8I mutation resulted in the loss of the internal V3 N-linked glycosylation site at position 306 in Env. Removal of this glycosylation site from HXB2 Env has been shown to affect coreceptor usage, tropism, and neutralization sensitivity (1, 31, 36). Introduction of this mutation into the SF162-V3ConB chimeric Env resulted in a uniform reduction in sensitivity to 2909 and all of the RhMAbs, with IC50 increases ranging from ∼100-fold (1.6F) to >900-fold (2.8F) for the MAbs with measurable IC50s. This suggested that the glycan at this position is a component of the epitope recognized by these MAbs or is masking adjacent targets required for binding or that this mutation leads to conformational changes that inhibit the formation or stability of the structure required for expression of the quaternary epitopes.
Mapping V2 determinants of the quaternary epitopes recognized by the RhMAbs.
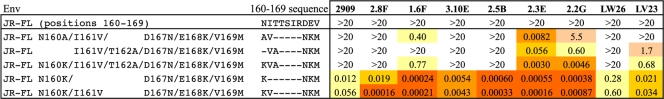
The recognition of SF162 Env by 2909 and the RhMAbs described in this study is dependent on the presence of the V2 and V3 domains but does not require the V1 domain (10, 34). All of these MAbs were specific for the SF162 sequence and did not neutralize JR-FL Env. The V2 regions of SF162 and JR-FL share considerable homology, differing in only nine sites between residues 151 and 197 (numbers correspond to the HXB2 sequence) (Fig. 2). We have previously shown that modification at two positions in V2 (Asn160Lys and Glu168Lys) was sufficient to express an enhanced form of the 2909 epitope in JR-FL Env (12). However, these substitutions were not sufficient for recognition by any of the RhMAbs (Table 3). In order to more fully map the V2 determinants of the RhMAbs, additional substitutions in the V2 region of JR-FL Env were made, and their effect on sensitivity to neutralization by these MAbs was determined.
Fig. 2.
Homology in V2 domain between SF162 and JR-FL. PNLG sites are underlined. SF162 is missing the glycosylation site at position 160 due to an N-to-K substitution and contains a glycosylation site at position 197 that is absent from JR-FL due to an N-to-D substitution. The numbering system is based on the HXB2 sequence.
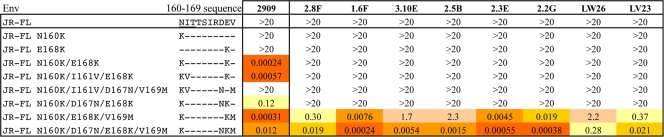
Table 3.
Effect of mutation at positions 160 and 167 to 169 on expression of QNEs in JR-FL Enva
IC50s (μg/ml) for each of the MAbs are shown. Color code is as in Table 1.
In addition to the critical substitutions at positions 160 and 168, JR-FL differs in this region from SF162 at positions 161, 167, and 169, with JR-FL containing the clade B consensus residues Ile161, Asp167, and Val169 instead of the less common Val161, Asn167, and Met169 residues present in the SF162 sequence. Adding either the Ile161Val or the Asp167Asn mutation to the Lys160/Lys168 double mutant did not lead to recognition by the RhMAbs, but adding Met169 resulted in varying levels of neutralization by all of the MAbs (Table 3). The neutralizing activity of the RhMAbs for the Lys160/Lys168/169Met triple mutant was further enhanced by the addition of Asn167, in a number of cases leading to IC50 enhancements of >4 orders of magnitude over the parental JR-FL Env. The preference of the RhMAbs for the autologous SF162 residues at positions 167 and 169 contrasted with that of 2909, which strongly preferred the clade B consensus residues at these positions (11).
Additional differences were seen in the sequence requirements of 2909 and some of the RhMAbs at positions 160 and 162 (Table 4). Whereas 2909 and four of the RhMAbs (2.8F, 3.10E, 2.5B, and LW26) were absolutely dependent on the presence of Lys160, the other four RhMAbs were to varying degrees tolerant of other residues at the 160 position, including Ala and the unmodified Asn in conjunction with the Thr162Ala substitution. Both of the latter substitutions resulted in disruption of the glycosylation signal at position 160, consistent with the absence of a glycan at this position being a key requirement for expression of these epitopes. These results suggested that the V2 determinant for the latter subset of RhMAbs was centered on residues at positions 167 to 169 and that access to this region was masked by the 160 glycan.
Table 4.
Role of the 160 glycosylation site (residues 160 to 162) on neutralization sensitivity of QNE-specific MAbsa
IC50s (μg/ml) for each of the MAbs are shown. Color code is as in Table 1.
In many cases, neutralization curves of QNE-specific antibodies are characterized by a shallow slope, with incomplete neutralization even at relatively high concentrations (12, 40). We have previously shown that for neutralization of SF162 by 2909, this resulted from a reduced avidity due to the presence of suboptimal residues in the V2 component of the epitope (12). In contrast to this, most of the rhesus QNE-specific MAbs showed classical sigmoidal curves against SF162 pseudotypes, with prominent plateaus at 100% neutralization at low antibody concentrations (see Fig. 1). However, binding patterns characteristic of reduced affinity were observed for the JR-FL Envs in which the V2 component of the epitope has been introduced (Fig. 3). A similar pattern was seen for the SF162 variant in which the V3 sequence was replaced by the consensus B sequence, suggesting that the reduced avidity of these antibodies for the JR-FL mutants was due to the presence of a suboptimal V3 sequence. This result indicated that potent neutralization by the QNE-specific MAbs required the presence of high-affinity interactions with both the V2 and V3 components of the epitope. The explanation of the ability of these MAbs to achieve 50% neutralization at low concentrations even for Envs with suboptimal substitutions is not clear, and this phenomenon may reflect heterogeneity in presentation of the quaternary epitopes in different viral subpopulations.
Fig. 3.
Representative neutralization curves for 2909 and three RhMAbs against HIV-1 pseudotypes with selected Envs.
Effect of V2 mutations on unmasking of sites in the V3 and CD4-binding domains of JR-FL.
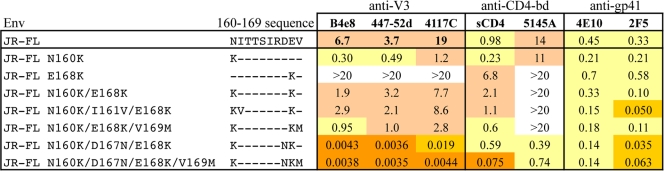
The SF162 strain is easily neutralized by common monoclonal and polyclonal antibodies, while JR-FL is relatively neutralization resistant. We have previously demonstrated that exchanging the V1/V2 domains of SF162 and JR-FL Envs resulted in a switch in their neutralization sensitivities, indicating the critical role of epitope masking by the V1/V2 domain in determining the overall neutralization phenotypes of these Envs (28). Although it is generally believed that the extent of epitope masking by the V1/V2 domain is related to the overall length and number of N-linked glycans present in these domains (19, 42), there are data showing that single amino acid substitutions that do not affect glycosylation can dramatically impact masking activity (24, 37). In order to examine the contributions of the V2 sites identified as determinants of the quaternary epitopes to conformational masking, the effect of mutations at these V2 positions on the sensitivity of JR-FL Env to neutralization by MAbs to several other domains was examined (Table 5).
Table 5.
Role of mutations at QNE-dependent V2 positions (160 to 169) on masking activity of JR-FL Enva
IC50s (μg/ml) for each of the MAbs are shown. Color code is as in Table 1.
Removal of the potential N-linked glycosylation (PNLG) site at position 160 by the Asn160Lys mutation induced a 10- to 20-fold increase in sensitivity to anti-V3 MAbs but had little effect on the CD4-bd ligands. Converting the uncommon Glu168 substitution present in JR-FL to the consensus Lys residue had the opposite effect, decreasing sensitivity to anti-V3 MAbs by >5-fold. The additive effects of these mutations resulted in a small residual increase in sensitivity to the anti-V3 MAbs for the double Asn160Lys/Glu168Lys mutant over the wild-type (wt) JR-FL sequence, and adding the Val169Met change further increased sensitivity to both V3 MAbs and sCD4 by an additional 2- to 3-fold. On the other hand, adding the Asp167Asn mutation to the Asn160Lys/Glu168Lys double mutant had a much more dramatic effect, leading to a 400- to 900-fold increase in sensitivity to the three anti-V3 MAbs and a >50-fold increase in sensitivity to the anti-CD4-bd MAb 5145A. The same mutations resulted in only small (<10-fold) increases in sensitivity of the two anti–membrane-proximal external region (anti-MPER) control MAbs, 4E10 and 2F5. These results showed that positions in the region of V2 targeted by QNE-specific antibodies are major factors in the extent of masking of the V3 and CD4-binding domains by the V1/V2 region and that the presence of individual charged residues, such as Asp167, may be stronger determinants of the masked phenotype than single glycan substitutions.
Analysis of QNE structure by complementation studies with Envs possessing either the V2 or the V3 component of the epitope.
A key question in understanding the structure of the quaternary epitopes characterized in this study as well as related structures recognized by more broadly reactive MAbs is how the V2 and V3 components of these epitopes are arranged to form the epitope. One possibility suggested by the dependence on native trimeric structures is that these epitopes are formed across two neighboring protomers in a trimer, with the V2 component of the epitope present on one subunit and the V3 component on a neighboring subunit. This would account for the loss of these epitopes in monomeric forms of Env. An alternative model is that both the V2 and V3 components are present on a single gp120 subunit, which must however be present in a conformation that is highly dependent on the trimeric state. These two models were tested by examining whether these epitopes could be detected on the surface of cells expressing combinations of Envs lacking different determinants of the epitope.
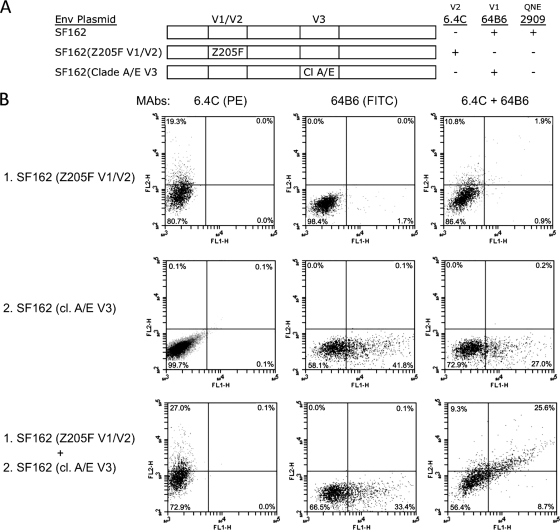
Mixed Env trimers were formed by transfection of 293 cells with combinations of fully functional chimeric or mutant SF162 Envs lacking either the V2 or the V3 component of the QNEs and analyzed by flow cytometry (Fig. 4). In each case, cell surface expression of the individual Envs was controlled by staining with MAb 2G12. In an initial experiment, the efficient coexpression of Env proteins in single cells was demonstrated by mixing antigenically distinct SF162 Env chimeras. The V1/V2 domain of the first Env was replaced with the corresponding sequence of the Z205F Env (Env 1), resulting in both the loss of reactivity with the SF162-specific QNE MAbs and the introduction of a Z205F-specific epitope recognized by MAb 6.4C (20). The quaternary epitopes were removed from the second Env by replacement of the native V3 sequence with the nonreactive clade A/E consensus sequence (Env 2). These Env chimeras contained the same backbone and possessed wild-type levels of function, so the assembly of the mixed trimers was expected to proceed efficiently. Expression of the two Envs was analyzed by flow cytometry using MAbs specific for sites in the respective V1/V2 domains: 6.4C (PE labeled) for Env 1 and 64B6 (FITC labeled), specific for a linear epitope in the SF162 V1 region (11), for Env 2. Cells transfected with the single plasmids were efficiently stained by the specific antibodies (Fig. 4B, rows 1 and 2) and the cotransfected cells were efficiently labeled by both antibodies (Fig. 4B, bottom row). Double labeling of the cotransfected cells with a mixture of the two MAbs showed that ∼25% of these cells expressed both Envs on their surfaces. Despite this efficient mixing of the two Envs in single cells, induction of these epitopes was not observed for 2909 or for any of the QNE-specific RhMAbs (Table 6).
Fig. 4.
(A) Diagram of chimeric Envs and specificity of MAbs used in this experiment. 6.4C is specific for a discontinuous epitope in the V1/V2 domain of the Z205F sequence present in Env 1 (20), while 64B6 recognizes a linear epitope in the V1 region of SF162 (wt and Env 2) (11). Substitution of either the Z205F V1/V2 region (Env 1) or the clade A/E V3 sequence (Env 2) into SF162 Env resulted in the loss of QNE expression. (B) Flow cytometry histograms showing binding profiles of MAbs to cells transfected with either single or mixed pairs of Env plasmids. Cells were pregated in the forward- and side-scatter plot to exclude dead cells and measured for fluorescence: y axis, PE-labeled 6.4C MAb, specific for Z205 V1/V2 domain (Env 1); x axis, FITC-labeled 64B6 MAb, specific for SF162 V1 epitope (Env 2). The top row contains cells transfected with SF162 (Z205F V1/V2) (Env 1), the middle row contains SF162 (cl. A/E V3) (Env 2), and the bottom row contains cells transfected with an equimolar mixture of the two Env chimeras. The first panel in each row was stained with the 6.4C (PE) MAb, the second panel with the 64B6 (FITC) MAb, and the third panel with the mixture of the two MAbs.
Table 6.
Analysis of QNE expression in mixed Env trimers expressing partial forms of the epitopesa
| Trimer(s) | Strain | % cells stained with indicated RhMAb |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2G12 | 2909 | 1.6F | 2.8F | 3.10E | 2.2G | 2.5B | 2.3E | LV23 | LW26 | ||
| wt | SF162 | 53.1 | 26.7 | 31.6 | 31.4 | 28.7 | 26.7 | 31.4 | 25.0 | 30.0 | 29.5 |
| Env 1 | SF162 (Z205F V1V2) | 38.2 | 0.0 | 0.1 | 0.1 | 0.1 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 |
| Env 2 | SF162 (CI A/E V3) | 51.9 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.1 |
| Env 1 + Env 2 | 56.8 | 0.0 | 0.2 | 0.1 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.2 | |
| Env 3 | SF162 (K160N/V161I) | 39.4 | 0.1 | 0.1 | 0.1 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.1 |
| Env 4 | SF162 (R315Q) | 38.0 | 9.8 | 9.0 | 9.1 | 4.7 | 0.1 | 0.0 | 1.6 | 0.2 | 3.8 |
| Env 3 + Env 4 | 37.7 | 2.0 | 5.5 | 2.9 | 0.4 | 0.7 | 1.0 | 2.0 | 0.0 | 0.2 | |
All MAbs were used at 5 μg/ml. Env plasmids were transfected at 1:1 ratio.
A similar lack of complementation was also observed for 2909 and the QNE-specific RhMAbs in mixing experiments with several additional combinations of Envs, including SF162 Envs in which the quaternary epitopes were eliminated by point mutations introduced into the variable domains (Envs 3 and 4) (Table 6). The lack of formation of the QNEs in these mixing experiments argues against the formation of these epitopes across neighboring protomers and suggests instead an alternate model in which the epitopes are located in a single Env molecule, which apparently must be present in a restricted conformation that is stabilized by trimer formation.
DISCUSSION
Diversity in the V3 determinants recognized by 2909 and RhMAbs.
This study provides new information about the diversity of both the V2 and V3 determinants of quaternary epitopes present in SF162 Env that are targeted by MAbs with highly potent neutralizing activities. The results suggest that while a limited number of residues in the V2 and V3 domains determine the specificity of these classes of epitope, additional sites proximal to the critical residues affect the relative affinity of the antibodies and effectiveness of neutralization. The RhMAbs varied considerably in their relative breadth for different V3 sequences. While they all preferred the autologous SF162 V3 sequence, three of the RhMAbs resembled 2909 in their recognition of multiple V3 clade consensus sequences, while the other five RhMAbs were restricted to clade B V3 sequences (Table 1). Clade B isolates contain a characteristic GPGR motif at positions 15 to 18 of the tip of the V3 loop (residues 312 to 315 in gp120), in contrast to the GPGQ motif typically found at this position in other clades. The reactivities of the RhMAbs for the clade B and clade C V3 chimeric Envs were reversed upon switching the residue at position 18, showing that the specificity of the RhMAbs for the consensus B V3 sequence was due to their dependence on a basic residue at the tip of the V3 loop (Table 2). MAb 2.5B was the most restricted and absolutely dependent on Arg18, while the other RhMAbs actually preferred Lys to Arg at this position. The RhMAbs also differed in their requirement for Pro in the GPGR motif, with 2.5B being the most dependent on Pro at position 16 (residue 313 in gp120), while the other RhMAbs tolerated the Ala substitution to varying extents. In contrast to these results, residues at the tip of the V3 loop were not critical determinants for 2909 or for the human MAbs directed against conserved quaternary epitopes PG9 and PG16 (40). These results indicated that critical recognition sites for this class of quaternary epitope can exist both at the tip of the loop and elsewhere in the V3 domain.
A feature shared by 2909 and all of the RhMAbs was their sensitivity to the Thr-to-Ile mutation at position 8 in the V3 loop (Table 2), which disrupted the N-linked glycosylation site at position 6 in the V3 domain (Asn306 in Env). This glycosylation site is highly conserved, and modifications at this site have been shown to decrease the neutralization sensitivity of HIV-1 and SHIV Envs (1, 21, 23, 32, 35) and to modify viral tropism (18, 23, 30). A similar reduction in sensitivity toward the broadly reactive quaternary-dependent MAb PG16 was reported upon the mutation of this glycosylation site in the SF162 Env bearing the K160N mutation (6). The uniform reduction in reactivity of the QNE MAbs caused by the Thr8Ile mutation could imply an interaction between these antibodies and the Thr residue at position 8 or components of the glycan at position 6 or could reflect an effect of the loss of the glycan at this position on the conformation of the V3 loop or a neighboring region that indirectly results in a reduced binding affinity. An analysis of additional mutations at these positions may help resolve these possibilities.
Variation in V2 determinants of 2909 and RhMAbs.
Key determinants in the V2 region of the quaternary epitopes recognized by the RhMAbs were characterized by examining the minimal requirements for expression of these epitopes in the heterologous JR-FL Env sequence. As was the case for the V3 determinants, the RhMAbs varied from 2909 and among each other in their specific requirements at multiple positions in V2. An important determinant for all of these MAbs, as previously described, was the residue at position 160 (34). Whereas 2909 and four of the RhMAbs required Lys at position 160, the other four RhMAbs also recognized with varying affinities the Ala160 and/or Asn160/Ala162 substitutions (Table 4). These mutations disrupted the N-linked glycosylation site at position 160, suggesting that the key requirement for reactivity of the latter group of MAbs was the absence of a glycan at this position. Whereas Lys is rare at position 160, present in only ∼3% of clade B sequences, the glycosylation signal at position 160 is present in 87% of sequences, with its absence due either to other substitutions at position 160 (9.5%) or to the presence of residues other than Ser or Thr at position 162 (3.5%) (15). In addition, there is evidence that even when the potential N-linked glycosylation site at position 160 is present, it is not always utilized (7, 44). This suggests that antibodies that tolerate the unmodified Asn and other residues at position 160, such as 2.3E, may have utility against a broader range of viruses than do antibodies that are dependent on Lys160, such as 2909.
The breadth of the RhMAbs was further restricted by their dependence on specific residues at positions 167 to 169 (Table 3). JR-FL has a rare substitution of Glu in place of the consensus Lys at position 168, which has previously been shown to be incompatible with recognition by PG9, PG16, and 2909 (11, 37). Although insertion of the double Lys160/Lys168 mutation into JR-FL Env was sufficient to induce potent neutralization by 2909, this did not result in recognition by any of the RhMAbs (Table 3). Neutralization by those MAbs required the additional substitution of Met at position 169 in place of Val and was further enhanced by the replacement of Asp at position 167 with Asn. Met169 is present in SF162 Env and in ∼22% of clade B isolates, second only to Val, which is found in approximately 50% of clade B sequences. This position contains predominantly basic residues in clade A and C sequences (15), and Lys or Arg at position 169 is an important determinant for a related quaternary-dependent neutralizing activity found in a subtype C-infected plasma that preferentially neutralized subtype A and C isolates (21). Asn167 is also relatively rare, present in only ∼10% of clade B isolates, while Asp167 is present in ∼78% of clade B sequences and in >97% of clade A and C sequences. This dependence on uncommon residues at these positions, together with the requirement of most of the RhMAbs for R315 in the V3 loop, accounts for the greatly reduced breadth of the RhMAbs, compared to 2909, against a panel of primary Envs in which Asn160 was mutated to Lys (43).
A previous study indicated that mutation of the N-linked glycosylation site at position 197 in SF162 Env resulted in the abrogation of 2909 reactivity and significantly reduced the potency of the QNE-specific RhMAbs that were tested (34). JR-FL does not possess this glycosylation site and yet expressed these epitopes when the appropriate determinants were introduced into the 160-to-169 region. Thus, it appears that a glycan at position 197 is not an invariable requisite for expression of this class of quaternary epitopes but rather that its requirement is dependent on the context of the surrounding V1/V2 sequence.
Relationship of QNE V2 determinants to conformational masking.
Previous studies have shown that the JR-FL V1/V2 domain masks sensitive neutralization targets in the V3, CD4-binding, and CD4-induced domains of this Env to antibodies against these sites that are commonly induced after infection (28). The loss of the 160 glycan due to the Asn160Lys substitution modestly increased the sensitivity of JR-FL to neutralization by these reagents (Table 5), consistent with a contribution of the 160 glycan to the masking activity of the V1/V2 domain. Converting the JR-FL-specific Glu168 residue to the consensus Lys residue at this position had the opposite effect, further reducing the sensitivity of JR-FL to neutralization, indicating that this change in charge increased V1/V2-dependent masking. Replacement of the consensus Val at position 169 by Met, the SF162-specific residue, did not significantly affect neutralization endpoints. However, adding the Asn167 mutation to the Lys160/Lys168 JR-FL mutant significantly increased sensitivity to neutralization by V3-specific MAbs, and to a lesser extent, by CD4-bd ligands, essentially converting this to a neutralization-sensitive Env. This indicated that the combination of the Lys160 and Asn167 substitutions was the major determinant of the weak masking activity of the SF162 V1/V2 domain and the corresponding high neutralization sensitivity of this isolate to common antibodies. A potent unmasking effect of the Asp167Asn substitution for epitopes in the V3 domain was previously reported in the MOKW Env (37), suggesting that this is a general effect applicable to multiple Env backbones.
The fact that specific sites in V2 (residues 160, 167, and 168) that were critical for expression of quaternary epitopes also significantly affected exposure and neutralization sensitivity of targets in the V3 and CD4-binding domains suggests a possible correlation between expression of quaternary epitopes and V1/V2-dependent masking efficiency and suggests that the QNE-specific antibodies may recognize a critical structure involved in the masking activity of the V1/V2 domain on sensitive targets elsewhere in Env. This further argues that the efficacy of conformational masking of sensitive neutralization targets may be limited by the consequent introduction of sensitive quaternary targets in the masked Env and that the interplay between these effects may be important factors in the evolution and regulation of Env structure.
Factors regulating the immunogenicity of this class of quaternary neutralization epitopes.
The SF162 type-specific quaternary epitopes characterized in this study appear to be allelic variants of those defined by two broadly reactive QNE-specific MAbs, PG9 and PG16 (41). Properties shared by these antibodies include preference for quaternary structure, dependence on sequences in both the V2 and V3 domains, unusually high neutralization potencies, and atypical neutralization curves for suboptimal forms of the epitope. The main determinant for the difference in breadth of these antibodies is the specific residue required at position 160. While PG9 and PG16 are dependent on the presence of the highly conserved 160 glycosylation site, the SF162-specific MAbs require its absence, in some cases (e.g., 2909 and 2.5B) because of an essential dependence on Lys at position 160, while in other cases (e.g., 2.3E) due to an apparent masking effect of the 160 glycan on the epitope. As discussed above, the breadth of the RhMAbs is further limited by their additional requirements for Met at position 169 and, in most cases, Arg at position 315 at the tip of the V3 loop.
The quaternary nature of these epitopes was originally defined by the inability of 2909 to bind to oligomeric forms of SF162 gp140 (10) and PG9 and PG16 to bind to several artificially trimerized gp140 proteins (40). The inability to induce these epitopes in mixed trimers that lacked either the V2 or V3 component of the epitope (Table 6) was consistent with previous analyses of the binding of PG9 and PG16 to mixtures of wild-type and mutant trimers (40) and strongly suggests that these epitopes are not expressed across two trimeric subunits but on single gp120-gp41 protomers and are restricted to specific conformations that are stabilized by trimerization. This model is consistent with a recent report indicating that PG9 and PG16 do bind efficiently to some soluble monomeric gp140s, including the SF162 K160N mutant (5). The factors that allow the recognition of these epitopes on some soluble monomeric gp140s but not others, and the generality of this result for related quaternary epitopes, are not known and merit further investigation.
The efficient isolation of multiple antibodies that recognize distinct versions of quaternary epitopes from SHIVSF162P4-infected animals suggests that these targets are highly immunogenic, and other evidence suggests that neutralizing antibodies with broad neutralizing activities specific for related QNEs are produced with reasonable frequencies by infected subjects (41), and in some cases, may represent the dominant neutralizing response generated upon infection (22). The divergence in epitope specificities of the SF162-specific MAbs and the broadly neutralizing QNE-specific MAbs shows the critical influence of the sequence of the infecting virus on the specificity and consequent breadth of such antibodies. SF162 is a heterologous strain for 2909 but is the autologous strain for the RhMAbs, and it is therefore not surprising that the latter antibodies recognized the SF162 sequence with a potency and specificity greater than that of 2909. The reactivity of 2909 for SF162 can be accounted for by the presence of Lys at position 160 in the autologous virus, while it can be presumed that the virus that induced the production of PG9 and PG16 contained the consensus N-linked glycosylation site at this position. The extent of the match between the target and immunizing viruses affects the affinity of the antibodies, which is reflected in the potency of neutralization, as measured both by the IC50s and slope and plateau levels of the neutralization curves (Fig. 3).
The superior neutralization potencies of the human and macaque antibodies directed against these epitopes argue that this general class of QNEs represents an extremely sensitive neutralization site and that the conserved forms of these epitopes may be important targets for HIV-1 vaccines. The modulating effect of many of the noncritical substitutions on the specificity and neutralization potencies of these antibodies indicates the critical contribution of sites proximal to the critical residues toward the relative affinity of the antibodies and extent of neutralization. This argues that selecting the appropriate variable-region sequences for inclusion in an immunogen could have a major impact on the ability to induce such antibodies and on the effectiveness of the resulting neutralizing responses.
ACKNOWLEDGMENTS
This work was funded by the following grants from the NIH: AI046283, AI078410, and AI088610 to A.P.; AI77451 to M.K.G.; and HL59725 to S.Z.-P.
Footnotes
Published ahead of print on 10 August 2011.
REFERENCES
- 1. Back N. K., et al. 1994. An N-glycan within the human immunodeficiency virus type 1 gp120 V3 loop affects virus neutralization. Virology 199:431–438 [DOI] [PubMed] [Google Scholar]
- 2. Cavacini L., et al. 2003. Conformational changes in env oligomer induced by an antibody dependent on the V3 loop base. AIDS 17:685–689 [DOI] [PubMed] [Google Scholar]
- 3. Changela A., et al. 2011. Crystal structure of human antibody 2909 reveals conserved features of quaternary structure-specific antibodies that potently neutralize HIV-1. J. Virol. 85:2524–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Connor R., Chen B. K., Choe S., Landau N. R. 1995. Vpr is required for efficient replication of human immunodeficiency virus type 1 in mononuclear phagocytes. Virology 206 [DOI] [PubMed] [Google Scholar]
- 5. Davenport T. M., et al. 2011. Binding interactions between soluble HIV envelope glycoproteins and quaternary-structure-specific MAbs PG9 and PG16. J. Virol. 85:7095–7107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Doores K. J., Burton D. R. 2010. Variable loop glycan dependency of the broad and potent HIV-1-neutralizing antibodies PG9 and PG16. J. Virol. 84:10510–10521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Go E. P., et al. 2008. Glycosylation site-specific analysis of HIV envelope proteins (JR-FL and CON-S) reveals major differences in glycosylation site occupancy, glycoform profiles, and antigenic epitopes' accessibility. J. Proteome Res. 7:1660–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gorny M., et al. 2006. Cross-clade neutralizing activity of human anti-V3 monoclonal antibodies derived from cells of individuals infected with non-B clades of human immunodeficiency virus type 1. J. Virol. 80:6865–6872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gorny M. K., et al. 1992. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J. Virol. 66:7538–7542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gorny M. K., et al. 2005. Identification of a new quaternary neutralizing epitope on human immunodeficiency virus type 1 virus particles. J. Virol. 79:5232–5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. He Y., et al. 2002. Efficient isolation of novel human monoclonal antibodies with neutralizing activity against HIV-1 from transgenic mice expressing human Ig loci. J. Immunol. 169:595–605 [DOI] [PubMed] [Google Scholar]
- 12. Honnen W. J., et al. 2007. Type-specific epitopes targeted by monoclonal antibodies with exceptionally potent neutralizing activities for selected strains of human immunodeficiency virus type 1 map to a common region of the V2 domain of gp120 and differ only at single positions from the clade B consensus sequence. J. Virol. 81:1424–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hu Q., et al. 2000. Identification of ENV determinants in V3 that influence the molecular anatomy of CCR5 utilization. J. Mol. Biol. 302:359–375 [DOI] [PubMed] [Google Scholar]
- 14. Kimura T., Wang X. H., Williams C., Zolla-Pazner S., Gorny M. K. 2009. Human monoclonal antibody 2909 binds to pseudovirions expressing trimers but not monomeric HIV-1 envelope proteins. Hum. Antibodies 18:35–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Korber B. T. M. 2011. HIV sequence database compendia. Los Alamos National Laboratory, Theoretical Biology and Biophysics, Los Alamos, NM: http://www.hiv.lanl.gov/content/sequence/HIV/COMPENDIUM/compendium.html [Google Scholar]
- 16. Krachmarov C. P., et al. 2006. Factors determining the breadth and potency of neutralization by V3-specific human monoclonal antibodies derived from subjects infected with clade A or clade B strains of human immunodeficiency virus type 1. J. Virol. 80:7127–7135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krachmarov C. P., Kayman S. C., Honnen W. J., Trochev O., Pinter A. 2001. V3-specific polyclonal antibodies affinity purified from sera of infected humans effectively neutralize primary isolates of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 17:1737–1748 [DOI] [PubMed] [Google Scholar]
- 18. Li Y., Rey-Cuille M. A., Hu S. L. 2001. N-linked glycosylation in the V3 region of HIV type 1 surface antigen modulates coreceptor usage in viral infection. AIDS Res. Hum. Retroviruses 17:1473–1479 [DOI] [PubMed] [Google Scholar]
- 19. Ly A., Stamatatos L. 2000. V2 loop glycosylation of the human immunodeficiency virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J. Virol. 74:6769–6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lynch R. M., et al. 2011. The B cell response is redundant and highly focused on V1V2 during early subtype C infection in a Zambian seroconverter. J. Virol. 85:905–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McCaffrey R. A., Saunders C., Hensel M., Stamatatos L. 2004. N-linked glycosylation of the V3 loop and the immunologically silent face of gp120 protects human immunodeficiency virus type 1 SF162 from neutralization by anti-gp120 and anti-gp41 antibodies. J. Virol. 78:3279–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moore P. L., et al. 2011. Potent and broad neutralization of HIV-1 subtype C viruses by plasma antibodies targeting a quaternary epitope including residues in the V2 loop. J. Virol. doi:10.1128/JVI.02658-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ogert R. A., et al. 2001. N-linked glycosylation sites adjacent to and within the V1/V2 and the V3 loops of dualtropic human immunodeficiency virus type 1 isolate DH12 gp120 affect coreceptor usage and cellular tropism. J. Virol. 75:5998–6006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O'Rourke S. M., et al. 2010. Mutation at a single position in the V2 domain of the HIV-1 envelope protein confers neutralization sensitivity to a highly neutralization-resistant virus. J. Virol. 84:11200–11209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pancera M., et al. 2010. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J. Virol. 84:8098–8110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pejchal R., et al. 2010. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc. Natl. Acad. Sci. U. S. A. 107:11483–11488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pinter A. 2007. Roles of HIV-1 Env variable regions in viral neutralization and vaccine development. Curr. HIV Res. 5:542–553 [DOI] [PubMed] [Google Scholar]
- 28. Pinter A., et al. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 78:5205–5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pinter A., Honnen W. J., Racho M. E., Tilley S. A. 1993. A potent, neutralizing human monoclonal antibody against a unique epitope overlapping the CD4-binding site of HIV-1 gp120 that is broadly conserved across North American and African virus isolates. AIDS Res. Hum. Retroviruses 9:985–996 [DOI] [PubMed] [Google Scholar]
- 30. Pollakis G., et al. 2001. N-linked glycosylation of the HIV type-1 gp120 envelope glycoprotein as a major determinant of CCR5 and CXCR4 coreceptor utilization. J. Biol. Chem. 276:13433–13441 [DOI] [PubMed] [Google Scholar]
- 31. Polzer S., Dittmar M. T., Schmitz H., Schreiber M. 2002. The N-linked glycan g15 within the V3 loop of the HIV-1 external glycoprotein gp120 affects coreceptor usage, cellular tropism, and neutralization. Virology 304:70–80 [DOI] [PubMed] [Google Scholar]
- 32. Polzer S., Muller H., Schreiber M. 2009. Effects of mutations on HIV-1 infectivity and neutralization involving the conserved NNNT amino acid sequence in the gp120 V3 loop. FEBS Lett. 583:1201–1206 [DOI] [PubMed] [Google Scholar]
- 33. Purtscher M., et al. 1994. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 10:1651–1658 [DOI] [PubMed] [Google Scholar]
- 34. Robinson J. E., et al. 2010. Quaternary epitope specificities of anti-HIV-1 neutralizing antibodies generated in rhesus macaques infected by the simian/human immunodeficiency virus SHIVSF162P4. J. Virol. 84:3443–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schonning K., Jansson B., Olofsson S., Hansen J. E. 1996. Rapid selection for an N-linked oligosaccharide by monoclonal antibodies directed against the V3 loop of human immunodeficiency virus type 1. J. Gen. Virol. 77:753–758 [DOI] [PubMed] [Google Scholar]
- 36. Schonning K., Jansson B., Olofsson S., Nielsen J. O., Hansen J. S. 1996. Resistance to V3-directed neutralization caused by an N-linked oligosaccharide depends on the quaternary structure of the HIV-1 envelope oligomer. Virology 218:134–140 [DOI] [PubMed] [Google Scholar]
- 37. Shibata J., et al. 2007. Impact of V2 mutations on escape from a potent neutralizing anti-V3 monoclonal antibody during in vitro selection of a primary human immunodeficiency virus type 1 isolate. J. Virol. 81:3757–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stiegler G., et al. 2001. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 17:1757–1765 [DOI] [PubMed] [Google Scholar]
- 39. Tilley S. A., Honnen W. J., Racho M. E., Chou T. C., Pinter A. 1992. Synergistic neutralization of HIV-1 by human monoclonal antibodies against the V3 loop and the CD4-binding site of gp120. AIDS Res. Hum. Retroviruses 8:461–467 [DOI] [PubMed] [Google Scholar]
- 40. Walker L. M., et al. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Walker L. M., et al. 2010. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 6:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wolk T., Schreiber M. 2006. N-glycans in the gp120 V1/V2 domain of the HIV-1 strain NL4-3 are indispensable for viral infectivity and resistance against antibody neutralization. Med. Microbiol. Immunol. 195:165–172 [DOI] [PubMed] [Google Scholar]
- 43. Wu X., et al. 2011. Immunotypes of a quaternary site of HIV-1 vulnerability and their recognition by antibodies. J. Virol. 85:4578–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu Z., et al. 1995. Characterization of neutralization epitopes in the V2 region of human immunodeficiency virus type 1 gp120: role of glycosylation in the correct folding of the V1/V2 domain. J. Virol. 69:2271–2278 [DOI] [PMC free article] [PubMed] [Google Scholar]