Abstract
Raltegravir (RAL) and related HIV-1 integrase (IN) strand transfer inhibitors (INSTIs) efficiently block viral replication in vitro and suppress viremia in patients. These small molecules bind to the IN active site, causing it to disengage from the deoxyadenosine at the 3′ end of viral DNA. The emergence of viral strains that are highly resistant to RAL underscores the pressing need to develop INSTIs with improved resistance profiles. Herein, we show that the candidate second-generation drug dolutegravir (DTG, S/GSK1349572) effectively inhibits a panel of HIV-1 IN variants resistant to first-generation INSTIs. To elucidate the structural basis for the increased potency of DTG against RAL-resistant INs, we determined crystal structures of wild-type and mutant prototype foamy virus intasomes bound to this compound. The overall IN binding mode of DTG is strikingly similar to that of the tricyclic hydroxypyrrole MK-2048. Both second-generation INSTIs occupy almost the same physical space within the IN active site and make contacts with the β4–α2 loop of the catalytic core domain. The extended linker region connecting the metal chelating core and the halobenzyl group of DTG allows it to enter farther into the pocket vacated by the displaced viral DNA base and to make more intimate contacts with viral DNA, compared with those made by RAL and other INSTIs. In addition, our structures suggest that DTG has the ability to subtly readjust its position and conformation in response to structural changes in the active sites of RAL-resistant INs.
Introduction
HIV-1 DNA, which is produced in the cytoplasm during reverse transcription, is transported subsequently to the nucleus and inserted into a host chromosome (Engelman, 2010). The insertion of the viral DNA is orchestrated by integrase (IN), a viral enzyme that catalyzes two distinct reactions. In a reaction termed 3′-processing, IN removes a pair of dinucleotides from both 3′ ends of the viral DNA, exposing 3′-hydroxyls attached to invariant CA dinucleotides. During strand transfer, IN joins the 3′-hydroxyls on each end of the processed viral DNA to phosphates on opposing strands of host DNA. The active sites of all retroviral INs contain triads of invariant carboxylates comprising the canonical D,DX35E active site motif (Engelman and Craigie, 1992; Kulkosky et al., 1992). The three invariant carboxylates serve to coordinate a pair of essential divalent metal cations (Mg2+ or Mn2+) (Hare at al., 2010). The intasome, comprising an IN tetramer assembled on a pair of viral DNA ends, is the smallest functional nucleoprotein complex capable of faithfully reproducing the strand transfer reaction in vitro (Li et al., 2006). Although structures of functional HIV-1 IN nucleoprotein complexes remain elusive, the corresponding orthologous assemblies from the prototype foamy virus (PFV), a retrovirus from Spumavirus genus, have been elucidated (Hare et al., 2010a; Maertens et al., 2010).
IN strand transfer inhibitors (INSTIs) represent the most recent and long-awaited addition to the arsenal of drugs to treat HIV-1 infections (Marchand et al., 2009). The first-in-class drug raltegravir (RAL) proved remarkably efficient at reducing viral loads in both treatment experienced and naive patients (Summa et al., 2008). However, viral strains with reduced susceptibility to RAL have emerged in patients receiving the drug, sometimes leading to virologic failure (Charpentier et al., 2008; Cooper et al., 2008), demonstrating the pressing need for the development of second-generation INSTIs that are active against RAL-resistant viral strains.
INSTIs only bind to the active site of IN when the enzyme is engaged with the viral DNA end (Espeseth et al., 2000). In the absence of suitable HIV-1 intasome crystals, the high degree of sequence identity within retroviral IN active sites makes it possible to use the PFV intasome as a surrogate for its HIV-1 counterpart in structural studies of INSTI binding (Hare et al., 2010a,b). The common INSTI pharmacophore comprises a triad of coplanar heteroatoms (oxygen or nitrogen), positioned to chelate a pair of divalent metal ions (Grobler et al., 2002), and a halogenated phenyl ring that invades a pocket natively occupied by the 3′-terminal base of viral DNA (Hare et al., 2010a). This mode of inhibitor binding is dependent on displacement of the viral 3′-adenosine from the active site and explains the notably slow kinetics of INSTI association and dissociation (Langley et al., 2008). The presence of a bound INSTI is not compatible with host DNA binding (Maertens et al., 2010), explaining the competition between target DNA and INSTIs for the intasome (Espeseth et al., 2000).
Clinical HIV-1 resistance to RAL involves one of three primary genetic pathways: Y143H/R/C, Q148H/R/K, or N155H (Charpentier et al., 2008; Cooper et al., 2008; Malet et al., 2008; Métifiot et al., 2010b). Of the mutated amino acids, only Tyr143 is predicted to form a direct binding contact with RAL, interacting with its oxadiazole ring via π-π stacking (Hare et al., 2010a; Krishnan et al., 2010). Other INSTIs, such as elvitegravir (Sato et al., 2006) and MK-2048, a potent INSTI with an improved resistance profile (Vacca et al., 2007), lack the oxadiazole moiety and are unaffected by Tyr143 mutations (Métifiot et al., 2011; Van Wesenbeeck et al., 2011). Crystal structures of PFV intasomes with the S217H (mimicking HIV-1 Q148H) and N224H (mimicking HIV-1 N155H) amino acid substitutions indicated that the mutant active sites must undergo significant structural rearrangements before INSTI binding (Hare et al., 2010b). Mutations at HIV-1 IN position 148 are often accompanied by a G140S/A change augmenting the level of RAL resistance (Goethals et al., 2010; Métifiot et al., 2010a). The PFV structures predict an interaction between His148 and Ser/Ala140 in mutant HIV-1 INs, explaining the coevolution of Q148H and G140A/S mutations in HIV-1 IN (Hare et al., 2010b).
We herein present functional and structural data for an advanced second-generation drug candidate dolutegravir (DTG, S/GSK1349572) (Johns et al., 2010) bound to wild-type (WT) and RAL-resistant PFV intasomes and discuss the structural basis for the compound's enhanced activity against RAL-resistant viral strains.
Materials and Methods
X-Ray Crystallography and In Vitro PFV IN Strand Transfer Assays.
DTG was synthesized as described previously (Johns et al., 2006) (see Supplemental Table S1 for the chemical structure). WT and mutant PFV intasomes, assembled and crystallized by following established protocols (Hare et al., 2010b), were soaked in 1.0 M ammonium sulfate, 25% (v/v) glycerol, 4.8% (w/v) 1,6-hexandiol, 50 mM MES-NaOH, pH 6.5, 25 mM MgCl2, and 1 mM DTG for 3 days before freezing in liquid nitrogen. X-ray diffraction data were collected at Diamond Light Source (Oxfordshire, UK) beam lines I02, I03, and I04-1. The starting points for structure refinement were Protein Data Bank IDs 3OYB, 3OYL, and 3OYN for DTG-bound WT, S217H, and N224H PFV intasomes, respectively. The topology file for DTG was generated using PRODRG2 server (Schüttelkopf and van Aalten, 2004). Iterations of manual building in Coot (Emsley and Cowtan, 2004) and restrained refinement in Refmac (Murshudov et al., 1997) were used to produce the final structures with good geometry (Table 1). Coordinates and structure factors for DTG-bound WT, S217H, and N224H intasomes were deposited with the Protein Data Bank under accession codes 3S3M, 3S3N, and 3S3O, respectively. Conditions for the PFV strand transfer assay and the quantification using real-time PCR were as described previously (Hare et al., 2010b).
TABLE 1.
Data collection and refinement statistics
Values in parentheses are for the highest resolution shell. The overall positional errors were estimated from the Cruickshank's dispersion precision indicator (eq. 31 in Cruickshank, 1999). Positional errors for the inhibitor molecule were estimated based on dispersion precision indicators and B factors (eq. 7 in Schneider, 2000).
| WT/DTG | S217H/DTG | N224H/DTG | |
|---|---|---|---|
| Data Collection | |||
| Space group | P41212 | P41212 | P41212 |
| Cell dimensions a, b, c; Å | 160.1, 160.1, 123.4 | 160.1, 160.1, 123.6 | 160.4, 160.4, 123.0 |
| Resolution range, Å | 40–2.49 (2.62–2.49) | 40–2.49 (2.62–2.49) | 40–2.55 (2.69–2.55) |
| Rmerge | 0.067 (0.691) | 0.097 (0.822) | 0.086 (0.955) |
| I/σ(I) | 13.9 (2.0) | 13.2 (2.1) | 16.7 (2.0) |
| Completeness, % | 98.7 (99.5) | 94.9 (96.9) | 99.2 (95.5) |
| Redundancy | 5.3 (5.5) | 9.3 (9.1) | 8.6 (6.7) |
| Refinement | |||
| Resolution | 39.15–2.49 | 39.16–2.49 | 39.14–2.55 |
| Reflections, total/free | 52,797/2832 | 50,479/2720 | 49,631/2639 |
| Rwork/Rfree | 0.206/0.232 | 0.209/0.232 | 0.208/0.230 |
| Positional error | |||
| Overall, Å | 0.33 | 0.34 | 0.34 |
| Inhibitor, Å | 0.23 | 0.26 | 0.21 |
| No. of Atoms | |||
| Protein | 4381 | 4357 | 4384 |
| Ligand/ion | 808 | 843 | 813 |
| Water | 235 | 210 | 199 |
| Average B-factors | |||
| Protein, Å2 | 60.4 | 61.4 | 60.4 |
| Ligand/ion, Å2 | 56.9 | 57.6 | 55.3 |
| Water, Å2 | 53.6 | 53.7 | 52.2 |
| R.M.S. Deviations | |||
| Bond lengths, Å2 | 0.012 | 0.012 | 0.011 |
| Bond angles, ° | 1.48 | 1.6 | 1.5 |
| Ramachandran | |||
| Favored, % | 96.9 | 96.3 | 96.9 |
| Outliers, % | 0 | 0 | 0.2 |
Single-Round HIV-1 Infectivity and In Vitro HIV-1 IN Strand Transfer Assays.
The human embryonic kidney cell line HEK293 was acquired from the American Type Culture Collection (Manassas, VA). The human osteosarcoma cell line was from Dr. Richard Schwartz (Michigan State University, East Lansing, MI). Cells were grown in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) supplemented with 5% (v/v) fetal bovine serum, 5% newborn calf serum, penicillin (50 U/ml), and streptomycin (50 μg/ml) at 37°C in 5% CO2 humidified atmosphere.
The HIV-1 vector pNLNgoMIVR−ΔEnv.LUC was described previously (Zhao et al., 2008). The IN coding sequence subcloned between the KpnI and SalI sites of pBluescript II KS(+) was mutagenized using the QuikChange procedure (Stratagene, La Jolla, CA). The resulting mutant IN coding sequences were cloned between KpnI and SalI sites of pNLNgoMIVR−ΔEnv.LUC to produce mutant HIV-1 vector constructs. VSV G-pseudotyped HIV-1 vector particles were produced by transfections of HEK293 cells. On the day before transfection, HEK293 cells were plated on 10-cm dishes at a density of 1.5·106 cells/plate. The cells were transfected with 16 μg of WT or mutant pNLNgoMIVR-ΔENV.LUC and 4 μg of pHCMV-G (obtained from Dr. Jane Burns, University of California, San Diego, CA) using calcium phosphate. Six hours after transfection, HEK293 cells were washed twice with phosphate-buffered saline and incubated with fresh media for 48 h. The virus-containing supernatants were clarified by low-speed centrifugation, filtered, and diluted for use in the infection assays.
On the day before infection, human osteosarcoma cells were seeded in a 96-well luminescence cell culture plate at a density of 4000 cells in 100 μl per well. Cells, preincubated with compounds (10 μM to 0.5 nM) for 3 h, were infected with 100 μl virus stocks diluted to achieve a maximum luciferase signal between 0.2 and 1.5 relative light units for 48 h. Virus infectivities were measured by using the steadylite plus luminescence reporter gene assay system (PerkinElmer Life and Analytical Sciences, Waltham, MA) and normalized to those in the absence of compounds. KaleidaGraph (Synergy Software, Reading, PA) was used to perform regression analysis on the data. In vitro HIV-1 IN strand transfer assays were performed for 2 h at 37°C using 400 nM HIV-1 IN (WT or mutant) and 20 nM 32P-labeled preprocessed HIV-1 U5 DNA and quantified as described previously (Métifiot et al., 2010a).
Results
Crystal Structure of the WT PFV Intasome Bound to DTG.
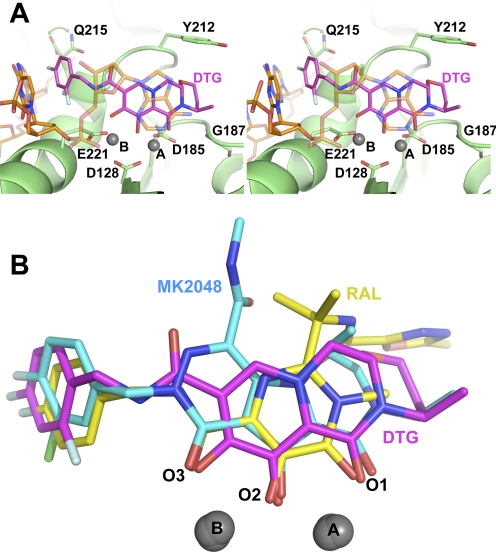
To elucidate the binding mode of DTG, we soaked crystals of the PFV intasome in the presence of the compound and Mg2+ and refined the structure to a resolution of 2.49 Å (Table 1; Supplemental Fig. S1). The small molecule binds to the intasome active site similarly to other INSTIs, with the three coplanar oxygen atoms (herein referred to as O1, O2, and O3) coordinated to Mg2+ cations (with metal ion A coordinated between O1, O2, Asp128, and Asp185 and ion B between O2, O3, Asp128, and Glu221). The difluorophenyl group of the bound inhibitor occupies a pocket vacated by the adenine base of the 3′-terminal viral DNA nucleotide (Fig. 1A). Compared with apo or Mn2+-bound intasomes (Protein Data Bank IDs 3L2Q and 3OY9), the most significant rearrangement of the intasome structure upon DTG binding is the displacement of this terminal nucleotide.
Fig. 1.
A, wall-eye stereo view of DTG bound to the active site of the WT PFV intasome. Carbon atoms of the INSTI are colored magenta, the protein green, and the DNA orange, whereas other atoms follow standard coloration: blue for nitrogen, red for oxygen, and pale blue for fluorine. Magnesium atoms are represented as gray spheres. Viral DNA, DTG, and selected protein residues are shown as sticks; for clarity, the 3′-nucleotide is displayed in semitransparent mode. Pale green arrowheads indicate alternative conformations for Gln215. B, comparison of the structures of RAL (yellow), MK-2048 (cyan), and DTG (magenta) when bound to the WT PFV intasome.
There are several subtle differences in the binding modes of DTG and other INSTIs to the PFV intasome. One intriguing feature of DTG is the length and flexibility of the linker bridging its tricyclic metal-chelating core and the difluorophenyl ring. Six bonds connect the halogenated phenyl ring of DTG to O3 compared with four in RAL and most other INSTIs. In this respect, DTG may seem similar to elvitegravir, in which the phenyl group is separated from O3 by six bonds. However, flexibility of elvitegravir is limited to only two exocyclic bonds in the linker compared with four in DTG. We note, however, that the torsional flexibilities of the two bonds closest to the metal chelating core of DTG are restricted because of π-conjugation of the amide and aromatic systems. The longer linker, coupled with reduced bulk in this region of the inhibitor, allows the difluorophenyl ring of DTG to enter deeper into the pocket vacated by the adenine group, permitting a better fit and potentially a more favorable stacking with the cytidine base of the preceding viral nucleotide (Fig. 1B; Supplemental Movie S1). The 4-fluoro atom of DTG approaches C6 of guanine from the C/G base pair as close as 2.9 Å, which is 0.3 and 0.6 Å closer than the analogous distances in RAL and MK-2048 structures, respectively.
The 2-fluoro atom of the difluorophenyl group of the bound DTG makes Van der Waals contacts with the Cγ and Cδ atoms of the Glu221 side chain (Fig. 1), a residue corresponding to Glu152 in HIV-1 IN. The deeper position of the halogenated phenyl ring also affects the structure of the underlying protein. Specifically, the orientation of the side chain of Gln215 (which corresponds to Gln146 in HIV-1) seems to be less stable than in other INSTI-bound and apo intasomes and exists in equilibrium between two rotamers, one of which angles away from DTG (Fig. 1A). The extended linker between the metal chelating core and the halogenated phenyl group of DTG includes an amide moiety. Although RAL also contains an amide at the same position relative to its fluorophenyl group, it uses its amide oxygen atom as part of the metal chelating core (Fig. 1B). Tyr212 is the PFV equivalent of Tyr143 of HIV-1 IN, and although this residue does contact bound DTG, as it does with most INSTIs (Hare et al., 2010b), it forms a Van der Waals interaction rather than the π-π stacking interaction uniquely observed with the oxadiazole ring of RAL.
The binding mode of DTG is strikingly similar to that of the tricyclic hydroxypyrrole MK-2048 (Vacca et al., 2007; Hare et al., 2010b). With the exception of the methylamide of MK-2048 (which is not present in DTG) and despite containing different tricyclic ring systems and engaging the Mg2+ ion pair at different angles, the second-generation INSTIs occupy almost the same physical space in the active site of the intasome, and both extend toward Gly187 (Fig. 1B; Supplemental Movie S1), a residue that corresponds to Gly118 in HIV-1 IN. In addition, the displaced 3′-terminal base of viral DNA packs against the ring systems of these INSTIs in a similar way. However, the 3′-nucleotide displays variable degrees of disorder in the INSTI cocrystal structures, and the relevance of its positions relative to the bound INSTIs remains unclear.
Susceptibility of Different HIV-1 IN Variants to DTG.
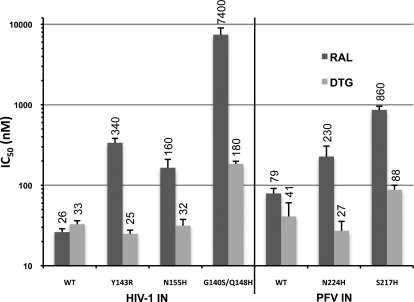
We tested the activity of DTG against purified recombinant WT HIV-1 IN and the main variants associated with clinical RAL resistance, Y143R, N155H, and G140S/Q148H. The IC50 values of RAL and DTG against these INs were determined using an in vitro strand transfer assay with radiolabeled oligonucleotides mimicking the processed end of HIV-1 U5 DNA end (Fig. 2). RAL inhibited WT HIV-1 IN with an IC50 of 26 nM. The Y143R, N155H, and G140S/Q148H IN mutants displayed approximately 13-, 6-, and 280-fold decreased susceptibility, respectively. DTG inhibited WT HIV-1 IN with an IC50 of 33 nM (Fig. 2). Neither Y143R nor N155H imparted significant resistance to DTG, whereas G140S/Q148H, the mutant with the highest level of RAL resistance, displayed only a 5.6-fold decrease in susceptibility (Fig. 2).
Fig. 2.
Activities of RAL and DTG against HIV-1 and PFV INs in in vitro strand transfer assays. IC50 values of RAL and DTG for the different INs are shown as dark and light gray bars, respectively. Error bars represent the S.D. of five (HIV-1) and three (PFV) independent measurements. Mean IC50 values are given atop the bars.
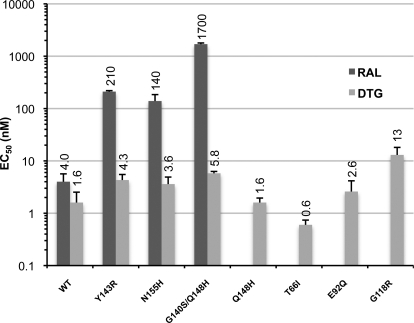
We also investigated the effect of these IN mutations on the susceptibility of HIV-1 to DTG in a single-cycle infectivity assay. The EC50 of RAL against WT HIV-1 was 4 nM, and viruses carrying Y143R, N155H, and G140S/Q148H mutations caused, respectively, a 50-, 30-, and 400-fold decrease in susceptibility to the drug (Fig. 3). However, DTG, which inhibited WT HIV-1 with an EC50 of 1.6 nM, retained a high potency against the Y143R, N155H, and G140S/Q148H mutants, with less than 4-fold decrease in activity in all cases (Fig. 3). Overall, there was a good correlation between the results of the infectivity assays and the in vitro strand transfer assays, indicating that the amino acid substitutions within IN are responsible for the observed phenotypic resistance.
Fig. 3.
Single-cycle infectivity assays. EC50 values of RAL are shown as dark gray bars and those of DTG as light gray bars. S.D. values are calculated from four independent measurements are indicated. Mean EC50 values are given atop the bars.
We also used the single-cycle virus assay to measure the effects of the Q148H mutation alone, which causes a modest loss of HIV-1 susceptibility to RAL (Métifiot et al., 2010a), the T66I and E92Q mutations known to cause a substantial loss of susceptibility to elvitegravir (Goethals et al., 2008), and the G118R mutation, which has been reported to affect susceptibility to MK-2048 (Bar-Magen et al., 2010). The elvitegravir resistance mutations and single Q148H mutation did not change the susceptibility to DTG, whereas G118R caused an 8-fold reduction in susceptibility, the largest fold change in activity of the mutations tested here (Fig. 3).
Susceptibility of the PFV IN Mutants N224H and S217H to DTG.
To further validate the DTG-bound PFV intasome crystal structure as a model of drug binding to HIV-1 IN, we tested activity of the compound in PFV IN in vitro strand transfer assays. DTG inhibited WT PFV IN with an IC50 of 40 nM. The PFV IN mutant N224H, which is equivalent to HIV-1 IN N155H, retained full susceptibility to DTG (Fig. 2); we showed previously that the N224H mutant is ∼4-fold less susceptible to RAL than WT PFV IN (Hare et al., 2010b). The HIV-1 IN double mutant G140S/Q148H is mimicked by the PFV IN single mutant S217H, because Gly140 in HIV-1 IN is equivalent to Ser209 in PFV. Similar to the HIV-1 Q148H/G140S mutant, S217H PFV IN showed only a modest decrease in susceptibility to DTG (approximately 2-fold) but was ∼10-fold less susceptible to RAL (Fig. 2) (Hare et al., 2010b). Thus, N224H and S217H PFV IN mutants seem to mimic the relative susceptibilities of N155H and Q148H/G140S HIV-1 INs to RAL and the second-generation INSTIs MK-2048 (Hare et al., 2010b) and DTG (Fig. 2).
Crystal Structures of DTG-Bound N224H and S217H PFV Intasomes.
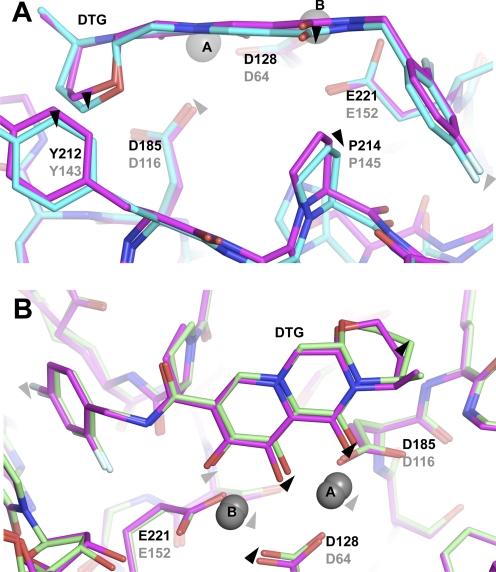
To elucidate the effects of the mutations associated with RAL resistance on DTG binding, crystals of S217H and N224H PFV intasomes were soaked in the presence of the compound and Mg2+. The two drug-bound structures were refined to resolutions of 2.49 and 2.55 Å, respectively (Table 1). Our previous S217H and N224H intasome structures obtained in the presence and absence of MK-2048 showed that these amino acid substitutions cause subtle shifts in active site geometry and that binding of the INSTI causes remodeling of the mutant active sites (Hare et al., 2010b). In the structure of S217H intasome bound to MK-2048, the 310 helix in the active site is shifted by more than 1 Å compared with the WT/MK-2048 complex, whereas INSTI binding to the N224H intasome perturbs a salt bridge between the side chain of His224 and the phosphate group of the 3′-nucleotide (Hare et al., 2010b). Similar changes in active site geometry are also observed when DTG binds to the mutant intasomes, supporting our previous observations.
Intriguingly, in both mutant structures, DTG exists in slightly different positions and conformations compared with those in the WT intasome (Fig. 4, A and B). When the asymmetric units of the refined models are superposed by the Cα atoms of the inner chain catalytic core domains (residues 117–303 of chain A), the positional root mean square deviations of the 30 nonhydrogen atoms of DTG in WT/DTG versus S217H/DTG and N224H/DTG structures are 0.37 and 0.30 Å, respectively. Some of the individual positional shifts are quite considerable (Supplemental Table S1). For instance, the oxygen atom of the exocyclic amido group of DTG is displaced by as much as 0.52 Å between N224H and S217H cocrystal structures. The estimated positional errors for the bound inhibitor in the refined structures, based on Cruickshank's diffraction-component precision index and B factors (Cruickshank, 1999; Schneider, 2000), are in the range of 0.21 to 0.26 Å (Table 1). Thus, we can consider positional shifts significant if they exceed 0.35 Å for WT/DTG structure versus S217H/DTG, 0.31 Å for WT/DTG versus N224H/DTG, and 0.33 Å for S217H/DTG versus N224H/DTG.
Fig. 4.
Comparisons of DTG binding to WT and mutant PFV intasomes. A, superposition of WT (magenta) and S217H (cyan) structures. Ribbon representations are used for the protein backbones, residues mentioned in the text, and DTG shown as sticks, and Mg2+ ions as gray spheres. Black labels indicate the PFV residue numbers and mutations; with the corresponding HIV-1 residues/mutations in gray. B, superposition of WT (magenta) and N224H (green) structures. Black arrowheads indicate significant positional shifts (more than 0.35 and 0.31 Å for WT/DTG versus S217H/DTG and N224H/DTG, respectively); gray arrowheads indicate atomic displacements that may be important yet fall short of reaching significance based on the diffraction-component precision indices of the refined models.
In the S217H/DTG structure, Pro214 (corresponding to Pro145 in HIV-1 IN) located on the 310 helix is displaced by 0.49 Å compared with its position in the WT/DTG structure (Fig. 4A; Supplemental Table S1). This change in the protein structure is accompanied by a significant movement of the DTG linker atoms (0.40 Å at the exocyclic amide oxygen atom; Fig. 4A; Supplemental Table S1), which allows the inhibitor to preserve close Van der Waals interactions with Pro214, keeping two of its linker atoms within 0.38 Å from the Cγ atom of this residue. In the N224H/DTG complex, the metal chelating oxygen atoms of the drug move by up to 0.35 Å (the O1 atom) in response to shifts of the metal cofactors brought about by small displacements of the catalytic side chains (Fig. 4B; Supplemental Table S1); this movement makes room for the mutant histidine residue, which replaces the less bulky asparagine. The shifts within the linker and the metal chelating group are accompanied by more subtle movements of the difluorophenyl group of DTG. The aromatic group seems to readjust its position in response to mutations, with some of its atoms shifting by up to 0.43 Å, comparing S217H and N224H cocrystal structures (Supplemental Table S1). We also note that there seems to be a difference in the pucker of the saturated six-membered ring of DTG when the small molecule is bound to the WT and mutant intasomes (Fig. 4), leading to individual DTG atom displacements in this region of up to 0.66 Å (Fig. 4; Supplemental Table S1). The conformation of this ring is likely influenced by Van der Waals interactions with the side chain of Tyr212 (Fig. 4A).
Discussion
The use of surrogates either as native orthologous proteins or as optimized chimeras, which can be readily crystallized and soaked with small molecules, has been validated by its success in other fields, particularly in guiding kinase inhibitor development (Ikuta et al., 2001; Breitenlechner et al., 2004). One should bear in mind, however, that the surrogate approach has its limitations, and local structural differences may have considerable effect on ligand binding (Davies et al., 2007). HIV-1 and PFV INs are fully orthologous, with identical canonical domain folds and stoichiometry. Fortuitously, all intasome atoms (protein, DNA, metal ions) that are in contact with the soaked INSTI molecules are invariant between HIV-1 and PFV (Hare et al., 2010a). Thus, we expect that structural differences between HIV-1 and PFV intasomes that are directly relevant to INSTI binding will be small. An unbiased test of this idea can be made by comparing the active sites in isolated catalytic core domains from different INs. Although the IN active site is not fully structured in the absence of viral DNA, because of disorder of the flexible loop and the active site glutamic acid, the remainder of the local structure superposes remarkably well across retroviral genera (Valkov et al., 2009). These arguments present a very strong case for using PFV as a reliable surrogate system to study HIV-1 IN inhibitors. We note, however, that functionally important structural variations between Lentivirus and Spumavirus intasomes outside of their active sites are to be expected. The variations would account for the different sizes of the host DNA duplications generated during integration of these retroviral genera (5 and 4 bp, respectively) and the fact that Spumavirus INs do not bind LEDGF (Cherepanov, 2007; Valkov et al., 2009).
The results of our in vitro enzymatic and virus infectivity assays confirm that the canonical HIV-1 IN mutations associated with RAL resistance have much more modest effects on the potency of DTG. Likewise, infection assays show that the elvitegravir resistance mutations T66I and E92Q do not pose a challenge to DTG. These observations are in accord with preliminary reports from other groups and collectively suggest that DTG could be used to treat patients who suffer virological failure during treatment with RAL or elvitegravir (Johns et al., 2010; Seki et al., 2010; Kobayashi et al., 2011). Although the overall binding modes of all of the INSTIs are conserved, the exact positioning of DTG in the active site is most similar to that of another second-generation drug candidate, the tricyclic hydroxypyrrole MK-2048. Both compounds extend toward the β4–α2 loop in IN (Fig. 1B; Supplemental Movie S1). This explains why the G118R mutation in HIV-1 IN, which has been selected in vitro with MK-2048 (Bar-Magen et al., 2010), also affects the activity of DTG (Fig. 3). Bound DTG approaches to within 3.9 Å of the Cα atom of PFV IN residue Gly187 (the structural equivalent to Gly118 in HIV-1 IN) (Fig. 1A). The substitution of a more bulky residue for this glycine would result in steric hindrance to the drug binding. However, even this mutation has a relatively modest effect on the activity of DTG, which maintains an impressive EC50 of 13 nM against the G118R mutant virus (Fig. 3).
The intasome structures clearly show why mutations of Tyr143 affect the susceptibility to RAL to a much greater extent than the other INSTIs: only RAL possesses a heterocyclic substituent, which makes a π-π stacking interaction with this residue (Hare et al., 2010b; Métifiot et al., 2011). The major RAL resistance mutations N155H and G140S/Q148H seem to affect drug potency indirectly by causing subtle changes in the geometry of the active site (Hare et al., 2010b). Binding of an INSTI molecule forces the mutant active site to adopt a conformation similar to that seen for the WT intasome. A highly optimized ligand that makes the most favorable contacts within the active site would be able to offset the energetic penalty associated with this remodeling. Our crystal structures of the mutant intasomes bound to DTG suggest an additional, complementary mechanism, in which the inhibitor may be able to adapt its position and/or conformation to the altered geometry of the mutant active sites. The positions of some DTG atoms differ by more than 0.5 Å when the inhibitor is bound to intasomes assembled with different PFV IN variants (Supplemental Table S1); such structural adjustments of the INSTI may help to optimize its binding to the different underlying surfaces of the intasomes. The ability of the HIV-1 reverse transcriptase inhibitor rilpivirine (TMC278) to wiggle (shift) and jiggle (flex) within the non-nucleoside inhibitor-binding pocket allows the molecule to adapt to structural changes caused by mutations and effectively inhibit viral strains resistant to other inhibitors of the same class (Das et al., 2008). Likewise, DTG may be able to adopt positions and conformations that better match active sites of WT and RAL-resistant intasomes. The extended linker of the DTG allows its difluorophenyl group to enter farther into the pocket vacated by the base of the viral 3′-deoxyadenosine (Fig. 1B). This leads to more intimate Van der Waals contacts within the pocket, compared with those formed by RAL and most other INSTIs, and may allow better stacking with the underlying cytidine base. It also seems important that this planar group is allowed to wiggle within the pocket (Fig. 4; Supplemental Table S1), adjusting to local structural changes brought about by the mutations. Although the resolution of the structures is not sufficient to reliably estimate the contributions of wiggling and jiggling of DTG to the observed atomic positional shifts of its atoms, the torsional flexibility within the linker region in the small molecule is likely to be critical. Relatively minor torsion angle adjustments (≤15°) in the four bonds of the linker are sufficient for the inhibitor to adopt the conformations observed in the refined models. The conservation of the HIV-1 and PFV IN active sites argue that structural changes in the HIV-1 intasome brought about by RAL resistance mutations will be similar to those observed in the corresponding PFV IN structures (Hare et al., 2010b). Although the magnitudes and contributions of individual atomic shifts may differ between the two systems, it seems likely that the ability of DTG to adjust its structure and position within the intasome active site will be relevant to its ability to inhibit HIV-1 mutants.
One of the prominent features of the INSTI binding site is the viral 3′-deoxyadenosine, which must be displaced to allow the drugs to bind. The absence of base pairing to the complementary deoxythymidine (Hare et al., 2010a) probably contributes to the mobility of the 3′-nucleotide, which is the Achilles' heel of the retroviral integration machinery exploited by the INSTIs. Measurements using in vitro assembled HIV-1 IN-DNA complexes showed that the 3′-deoxyadenosine controls INSTI binding, and that removing the nucleotide drastically increases the kinetics of INSTI binding and dissociation (Langley et al., 2008). In our PFV intasome/INSTI cocrystal structures, the viral 3′-deoxyadenosine displays significant disorder and its conformation is not conserved. It seems likely that upon INSTI binding, the nucleotide exists in a range of conformations, most of which would interfere with the dissociation of the small molecule. We note that in MK-2048- and DTG-bound intasome structures, the predominant pose of the 3′-deoxyadenosine seems to be similar, with the base packed against the metal chelating core of the small molecules. It remains to be seen whether this property will be conserved in future generations of INSTIs. Based on our observations, we suggest that it will probably be advantageous for future generations of INSTIs to incorporate the strategic flexibility of the linker region, retain contacts with the β4-α2 IN loop (Gly118 in HIV-1 IN), and, possibly, to strengthen interactions with the displaced deoxyadenosine at the 3′ end of the viral DNA.
Supplementary Material
Acknowledgments
We thank Alan Engelman and Maxwell D. Cummings for critical reading of the manuscript and the staff of I02, I03, and I04-1 beam lines (Diamond Light Source, Oxfordshire, UK) for assistance with X-ray data collection.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This work was supported by the UK Medical Research Council [Grant G1000917]; the National Institutes of Health Intramural Program, Center for Cancer Research, National Cancer Institute; the National Institutes of Health Intramural AIDS Targeted Program; and the Wellcome Trust Value in People Award.
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.111.073189.
- IN
- integrase
- DTG
- dolutegravir
- INSTI
- IN strand transfer inhibitor
- MES
- 2-(N-morpholino) ethanesulfonic acid
- PFV
- prototype foamy virus
- RAL
- raltegravir
- WT
- wild type
- HEK
- human embryonic kidney
- TMC278
- rilpivirine
- MK-2048
- (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide.
Authorship Contributions
Participated in research design: Hare, Smith, Métifiot, Jaxa-Chamiec, Pommier, Hughes, and Cherepanov.
Conducted experiments: Hare, Smith, and Métifiot.
Contributed new reagents or analytic tools: Jaxa-Chamiec.
Performed data analysis: Hare, Smith, Métifiot, Jaxa-Chamiec, Pommier, Hughes, and Cherepanov.
Wrote or contributed to the writing of the manuscript: Hare, Smith, Métifiot, Pommier, Hughes, and Cherepanov.
References
- Bar-Magen T, Sloan RD, Donahue DA, Kuhl BD, Zabeida A, Xu H, Oliveira M, Hazuda DJ, Wainberg MA. (2010) Identification of novel mutations responsible for resistance to MK-2048, a second-generation HIV-1 integrase inhibitor. J Virol 84:9210–9216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitenlechner CB, Wegge T, Berillon L, Graul K, Marzenell K, Friebe WG, Thomas U, Schumacher R, Huber R, Engh RA, et al. (2004) Structure-based optimization of novel azepane derivatives as PKB inhibitors. J Med Chem 47:1375–1390 [DOI] [PubMed] [Google Scholar]
- Charpentier C, Karmochkine M, Laureillard D, Tisserand P, Bélec L, Weiss L, Si-Mohamed A, Piketty C. (2008) Drug resistance profiles for the HIV integrase gene in patients failing raltegravir salvage therapy. HIV Med 9:765–770 [DOI] [PubMed] [Google Scholar]
- Cherepanov P. (2007) LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res 35:113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DA, Steigbigel RT, Gatell JM, Rockstroh JK, Katlama C, Yeni P, Lazzarin A, Clotet B, Kumar PN, Eron JE, et al. (2008) Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N Engl J Med 359:355–365 [DOI] [PubMed] [Google Scholar]
- Cruickshank DW. (1999) Remarks about protein structure precision. Acta Crystallogr D Biol Crystallogr 55:583–601 [DOI] [PubMed] [Google Scholar]
- Das K, Bauman JD, Clark AD, Jr, Frenkel YV, Lewi PJ, Shatkin AJ, Hughes SH, Arnold E. (2008) High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: strategic flexibility explains potency against resistance mutations. Proc Natl Acad Sci USA 105:1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies TG, Verdonk ML, Graham B, Saalau-Bethell S, Hamlett CC, McHardy T, Collins I, Garrett MD, Workman P, Woodhead SJ, et al. (2007) A structural comparison of inhibitor binding to PKB, PKA and PKA-PKB chimera. J Mol Biol 367:882–894 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- Engelman A. (2010) Reverse transcription and integration, in Retroviruses: Molecular Biology, Genomics and Pathogenesis (Kurth R, Bannert N. eds) pp 129–159, Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- Engelman A, Craigie R. (1992) Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol 66:6361–6369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeseth AS, Felock P, Wolfe A, Witmer M, Grobler J, Anthony N, Egbertson M, Melamed JY, Young S, Hamill T, et al. (2000) HIV-1 integrase inhibitors that compete with the target DNA substrate define a unique strand transfer conformation for integrase. Proc Natl Acad Sci USA 97:11244–11249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goethals O, Clayton R, Van Ginderen M, Vereycken I, Wagemans E, Geluykens P, Dockx K, Strijbos R, Smits V, Vos A, et al. (2008) Resistance mutations in human immunodeficiency virus type 1 integrase selected with elvitegravir confer reduced susceptibility to a wide range of integrase inhibitors. J Virol 82:10366–10374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goethals O, Vos A, Van Ginderen M, Geluykens P, Smits V, Schols D, Hertogs K, Clayton R. (2010) Primary mutations selected in vitro with raltegravir confer large fold changes in susceptibility to first-generation integrase inhibitors, but minor fold changes to inhibitors with second-generation resistance profiles. Virology 402:338–346 [DOI] [PubMed] [Google Scholar]
- Grobler JA, Stillmock K, Hu B, Witmer M, Felock P, Espeseth AS, Wolfe A, Egbertson M, Bourgeois M, Melamed J, et al. (2002) Diketo acid inhibitor mechanism and HIV-1 integrase: implications for metal binding in the active site of phosphotransferase enzymes. Proc Natl Acad Sci USA 99:6661–6666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. (2010a) Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464:232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare S, Vos AM, Clayton RF, Thuring JW, Cummings MD, Cherepanov P. (2010b) Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc Natl Acad Sci USA 107:20057–20062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikuta M, Kamata K, Fukasawa K, Honma T, Machida T, Hirai H, Suzuki-Takahashi I, Hayama T, Nishimura S. (2001) Crystallographic approach to identification of cyclin-dependent kinase 4 (CDK4)-specific inhibitors by using CDK4 mimic CDK2 protein. J Biol Chem 276:27548–27554 [DOI] [PubMed] [Google Scholar]
- Johns B, Kawasuiji T, Taishi T, Yoshida H, Garvey E, Spreen W, Underwood M, Sato A, Yoshinaga T, Fujiwara T. (2010) The discovery of S/GSK1349572: a once-daily next generation integrase inhibitor with a superior resistance profile, in 17th Conference on Retroviruses and Opportunistic Infections, 2010 Feb 16–19, San Francisco, CA Conference on Retroviruses and Opportunistic Infections, Alexandria, VA [Google Scholar]
- Johns BA, Kawasuiji T, Taishi T, Taoda Y. (2006), inventors; Shionogi & Co., assignee. Polycyclic carbamoylpyridone derivative having HIV integrase inhibitory activity. World patent WO2006116764 2006 Nov 2
- Kobayashi M, Yoshinaga T, Seki T, Wakasa-Morimoto C, Brown KW, Ferris R, Foster SA, Hazen RJ, Miki S, Suyama-Kagitani A, et al. (2011) In Vitro antiretroviral properties of S/GSK1349572, a next-generation HIV integrase inhibitor. Antimicrob Agents Chemother 55:813–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan L, Li X, Naraharisetty HL, Hare S, Cherepanov P, Engelman A. (2010) Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc Natl Acad Sci USA 107:15910–15915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkosky J, Jones KS, Katz RA, Mack JP, Skalka AM. (1992) Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol 12:2331–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley DR, Samanta HK, Lin Z, Walker MA, Krystal MR, Dicker IB. (2008) The terminal (catalytic) adenosine of the HIV LTR controls the kinetics of binding and dissociation of HIV integrase strand transfer inhibitors. Biochemistry 47:13481–13488 [DOI] [PubMed] [Google Scholar]
- Li M, Mizuuchi M, Burke TR, Jr, Craigie R. (2006) Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J 25:1295–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens GN, Hare S, Cherepanov P. (2010) The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature 468:326–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malet I, Delelis O, Valantin MA, Montes B, Soulie C, Wirden M, Tchertanov L, Peytavin G, Reynes J, Mouscadet JF, et al. (2008) Mutations associated with failure of raltegravir treatment affect integrase sensitivity to the inhibitor in vitro. Antimicrob Agents Chemother 52:1351–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand C, Maddali K, Métifiot M, Pommier Y. (2009) HIV-1 IN inhibitors: 2010 update and perspectives. Curr Top Med Chem 9:1016–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métifiot M, Maddali K, Naumova A, Zhang X, Marchand C, Pommier Y. (2010a) Biochemical and pharmacological analyses of HIV-1 integrase flexible loop mutants resistant to raltegravir. Biochemistry 49:3715–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métifiot M, Marchand C, Maddali K, Pommier Y. (2010b) Resistance to Integrase Inhibitors. Viruses 2:1347–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métifiot M, Vandegraaff N, Maddali K, Naumova A, Zhang X, Rhodes D, Marchand C, Pommier Y. (2011) Elvitegravir overcomes resistance to raltegravir induced by integrase mutation Y143. AIDS 25:1175–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53:240–255 [DOI] [PubMed] [Google Scholar]
- Sato M, Motomura T, Aramaki H, Matsuda T, Yamashita M, Ito Y, Kawakami H, Matsuzaki Y, Watanabe W, Yamataka K, et al. (2006) Novel HIV-1 integrase inhibitors derived from quinolone antibiotics. J Med Chem 49:1506–1508 [DOI] [PubMed] [Google Scholar]
- Schneider TR. (2000) Objective comparison of protein structures: error-scaled difference distance matrices. Acta Crystallogr D Biol Crystallogr 56:714–721 [DOI] [PubMed] [Google Scholar]
- Schüttelkopf AW, van Aalten DM. (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 60:1355–1363 [DOI] [PubMed] [Google Scholar]
- Seki T, Kobayashi M, Wakasa-Morimoto C, Yoshinaga T, Sato A, Fujiwara T, Underwood M, Garvey E, Johns B. (2010) S/GSK1349572 is a potent next generation HIV integrase inhibitor and demonstrates a superior resistance profile substantiated with 60 integrase mutant molecular clones, in 17th Conference on Retroviruses and Opportunistic Infections, 2010 Feb 16–19, San Francisco, CA Conference on Retroviruses and Opportunistic Infections, Alexandria, VA [Google Scholar]
- Summa V, Petrocchi A, Bonelli F, Crescenzi B, Donghi M, Ferrara M, Fiore F, Gardelli C, Gonzalez Paz O, Hazuda DJ, Jones P, Kinzel O, Laufer R, Monteagudo E, Muraglia E, Nizi E, Orvieto F, Pace P, Pescatore G, Scarpelli R, Stillmock K, Witmer MV, Rowley M. (2008) Discovery of raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. J Med Chem 51:5843–5855 [DOI] [PubMed] [Google Scholar]
- Vacca J, Wai J, Fisher T, Embrey M, Hazuda D, Miller M, Felock P, Witmer M, Gabryelski L, Lyle T. (2007) Discovery of MK-2048—subtle changes confer unique resistance properties to a series of tricyclic hydroxypyrrole integrase strand transfer inhibitors, in 4th International AIDS Society Conference, 2007 July 22–25, Sydney, Australia International AIDS Society, Geneva, Switzerland [Google Scholar]
- Valkov E, Gupta SS, Hare S, Helander A, Roversi P, McClure M, Cherepanov P. (2009) Functional and structural characterization of the integrase from the prototype foamy virus. Nucleic Acids Res 37:243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wesenbeeck L, Rondelez E, Feyaerts M, Verheyen A, Van der Borght K, Smits V, Cleybergh C, De Wolf H, Van Baelen K, Stuyver LJ. (2011) Cross-resistance profile determination of two second-generation HIV-1 integrase inhibitors using a panel of recombinant viruses derived from raltegravir-treated clinical isolates. Antimicrob Agents Chemother 55:321–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XZ, Semenova EA, Vu BC, Maddali K, Marchand C, Hughes SH, Pommier Y, Burke TR., Jr (2008) 2,3-dihydro-6,7-dihydroxy-1H-isoindol-1-one-based HIV-1 integrase inhibitors. J Med Chem 51:251–259 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.