Abstract
Warfarin is a widely used anticoagulant with a narrow therapeutic index and large interpatient variability in the dose required to achieve target anticoagulation. Common genetic variants in the cytochrome P450-2C9 (CYP2C9) and vitamin K–epoxide reductase complex (VKORC1) enzymes, in addition to known nongenetic factors, account for ~50% of warfarin dose variability. The purpose of this article is to assist in the interpretation and use of CYP2C9 and VKORC1 geno-type data for estimating therapeutic warfarin dose to achieve an INR of 2–3, should genotype results be available to the clinician. The Clinical Pharmacogenetics Implementation Consortium (CPIC) of the National Institutes of Health Pharmacogenomics Research Network develops peer-reviewed gene–drug guidelines that are published and updated periodically on http://www.pharmgkb.org based on new developments in the field.1
Focused Literature review
The Supplementary Notes online include a systematic literature review of CYP2C9 and VKORC1 genotype and warfarin dosing, which forms the basis for this guideline.
DRUG: WARFARIN
Warfarin (Coumadin and others) is the most commonly used oral anticoagulant worldwide, with annual prescriptions typically equaling 0.5–1.5% of the population. It is prescribed for treatment and prevention of thrombotic disorders.2 Although highly efficacious, warfarin's narrow therapeutic index and wide interindividual variability make its dosing notoriously challenging.3–5 Complications from inappropriate warfarin dosing are among the adverse events most frequently reported to the US Food and Drug Administration (FDA) and one of the most common reasons for emergency room visits.6
Warfarin is often dosed empirically: an initial dose is prescribed, typically followed by at least weekly measurement of the INR and subsequent dose adjustment. The initial dose is often based on population averages (e.g., 3–5 mg/day), but stable doses to achieve an INR of 2–3 can range from 1–20 mg/ day. The iterative process to define the appropriate dose can take weeks to months, and during this period patients are at increased risk of over- or under-anticoagulation and thus at risk of thromboembolism or bleeding.
Warfarin pharmacology and pharmacokinetics
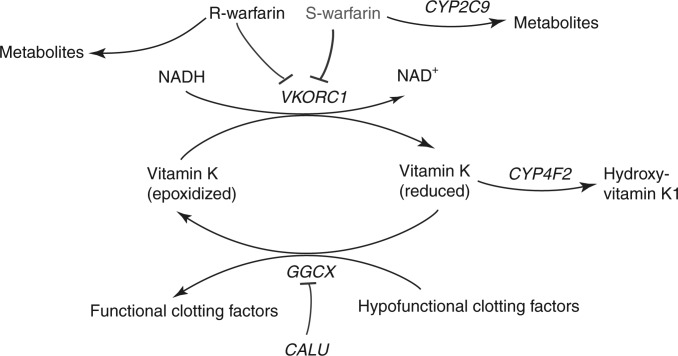
Figure 1 highlights key elements of warfarin pharmacology and pharmacokinetics. Warfarin inhibits vitamin K–epoxide reductase complex7 and is administered as a racemic mixture, with S-warfarin being more potent than R-warfarin.2
Figure 1.
Schematic representation of warfarin metabolism and its mechanism of action. Warfarin is administered via a racemic mixture of the R- and S- stereoisomers. S-warfarin is three to five times more potent than R-warfarin and is metabolized predominantly to 7- and 6-hydroxyl metabolites via CYP2C9. Warfarin exerts its anticoagulant effect through inhibition of its molecular target, VKORC1, which in turn limits availability of reduced vitamin K, leading to decreased formation of functionally active clotting factors. these clotting factors are glycoproteins that are postranslationally carboxylated by γ-glutamyl carboxylase (GGCX) to Gla-containing proteins. the endoplasmic reticulum chaperone protein calumenin (CALU) can bind to and inhibit GGCX activity. the metabolism of reduced vitamin K to hydroxyvitamin K1 is catalyzed by CYP4F2, which removes vitamin K from the vitamin K cycle. Adapted from the warfarin pharmacokinetics (PK) and pharmacodynamics (PD) pathways provided at PharmGKB, <http://www.pharmgkb.org/do/serve?objId=PA451906&objcls=Drug#tabview=tab4>.
GENES: CYP2C9 AND WARFARIN
CYP2C9 is a hepatic drug-metabolizing enzyme in the CYP450 superfamily8 and is the primary metabolizing enzyme of S-warfarin (Figure 1). The CYP2C9 gene has more than 30 known variant alleles (http://www.cypalleles.ki.se/cyp2c9.htm; Supplementary Tables S1 and S2 online). Individuals homozygous for the reference CYP2C9 allele (CYP2C9*1) have the “normal metabolizer” phenotype. Each named CYP2C9 star (*) allele is defined by a genotype at one or more specific single-nucleotide polymorphisms (SNPs) and is associated with enzyme activity (Supplementary Table S1). The two most common variants with reduced enzyme activity among individuals of European ancestry are CYP2C9*2 (rs1799853) and CYP2C9*3 (rs1057910).8 The frequencies of the CYP2C9 variant alleles differ between racial/ethnic groups.8,9
In vitro and ex vivo studies suggest that CYP2C9*2 and *3 impair metabolism of S-warfarin by ~30–40% and ~80–90%, respectively.8 As compared with patients who are homozygous for CYP2C9*1, individuals who inherit one or two copies of CYP2C9*2 or *3 are at greater risk of bleeding during warfarin therapy,5,10,11 require lower doses to achieve similar levels of anticoagulation, and require more time to achieve a stable INR.10,12 Additional CYP2C9 variant alleles with reduced activity (CYP2C9*5, *6, *8, and *11) contribute to dose variability among African Americans (Supplementary Table S3). Including these additional CYP2C9 variants in dosing algorithms for warfarin may improve predictability for African Americans.
VKORC1 AND WARFARIN
VKORC1 encodes the vitamin K–epoxide reductase protein, the target enzyme of warfarin.7,13 VKORC1 catalyzes the conversion of vitamin K–epoxide to vitamin K, which is the rate-limiting step in vitamin K recycling.14
A common noncoding variant (−1639G>A, rs9923231) is significantly associated with warfarin sensitivity and reduced dose requirements, as −1639A carriers require lower initial warfarin doses than −1639G carriers.4,9,15–18 The −1639G>A polymorphism alters a VKORC1 transcription factor binding site, leading to lower protein expression.4,16
Including other common VKORC1 SNPs or haplotypes in dosing algorithms does not further improve warfarin dose prediction.9,17
The −1639G>A allele frequency varies among different ethnic groups (Supplementary Table S2 online) and largely explains the differences in average dose requirements among whites, blacks, and Asians.18 Several rare nonsynonymous VKORC1 variants confer warfarin resistance (high-dose requirements); these are detailed in Supplementary Table S3.19
LINKING GENETIC VARIABILITY TO VARIABILITY IN DRUG-RELATED PHENOTYPES
CYP2C9 and VKORC1 polymorphisms account for up to 18 and 30%, respectively, of the variance in stable warfarin dose among patients of European ancestry,9,17,18,20 but these variants explain less of the dose variability in patients of Asian or African ancestry. Other genes of potential importance are discussed in Supplementary Note S1 online.
In 2007, the FDA modified the warfarin label, stating that CYP2C9 and VKORC1 genotypes may be useful in determining the optimal initial dose of warfarin.21 The label was further updated in 2010 to include a table (Table 1) describing recommendations for initial dosing ranges for patients with different combinations of CYP2C9 and VKORC1 genotypes.
Table 1.
Recommended daily warfarin doses (mg/day) to achieve a therapeutic INR based on CYP2C9 and VKORC1 genotype using the warfarin product insert approved by the US Food and Drug Administration
| VKORC1:–1639G>A | CYP2C9*1/*1 | CYP2C9*1/*2 | CYP2C9*1/*3 | CYP2C9*2/*2 | CYP2C9*2/*3 | CYP2C9*3/*3 |
|---|---|---|---|---|---|---|
| GG | 5–7 | 5–7 | 3–4 | 3–4 | 3–4 | 0.5–2 |
| GA | 5–7 | 3–4 | 3–4 | 3–4 | 0.5–2 | 0.5–2 |
| AA | 3–4 | 3–4 | 0.5–2 | 0.5–2 | 0.5–2 | 0.5–2 |
Reproduced from updated warfarin (Coumadin) product label.
Genetic test interpretation
Most clinical laboratories report CYP2C9 genotype using the star (*) allele nomenclature and may provide interpretation of the patient's predicted metabolizer phenotype. Alleles other than *2 and *3 might not be tested, influencing the accuracy of the genotype-based dose prediction, which is particularly relevant in those of African ancestry who more commonly carry other CYP2C9 variant alleles. VKORC1 is typically reported by −1639G>A (or the linked 1173C>T) genotype, with accompanying interpretation of warfarin sensitivity. Of note, most commercial genotyping platforms do not detect rare CYP2C9 and VKORC1 variants that may influence warfarin dosing (Supplementary Note S2 and Supplementary Table S3).
Genetic test options
Commercially available genetic testing options change frequently, but several platforms are available for CYP2C9/VKORC1 genotyping, some of which have been approved by the FDA (Supplementary Note S2).
Incidental findings
No diseases have been linked to CYP2C9 variants independent of drug metabolism and response. Similarly, no diseases have been consistently linked to common VKORC1 variants routinely interrogated in warfarin pharmacogenetics tests.
RECOMMENDATIONS FOR WARFARIN MAINTENANCE (CHRONIC) DOSAGE BASED ON GENETIC INFORMATION
We use the three-tiered rating system described previously (and in Supplementary Note S3 online)1 in which ratings of A, strong; B, moderate; and C, optional are applied based on the evidence reviewed. The recommendations for dosing based on genotype contained herein are rated as level A, or strong, and are derived from numerous observational studies and some prospective studies that suggest the ability to more accurately identify stable therapeutic warfarin dose requirements through use of both genetic and clinical information. However, there are limited prospective data from randomized trials on the use of genetic information to guide warfarin dosing (summarized in Supplementary Note S4), and the impact on clinical outcomes is unknown, although several such studies are currently ongoing, the largest of which are described in Supplementary Note S5.
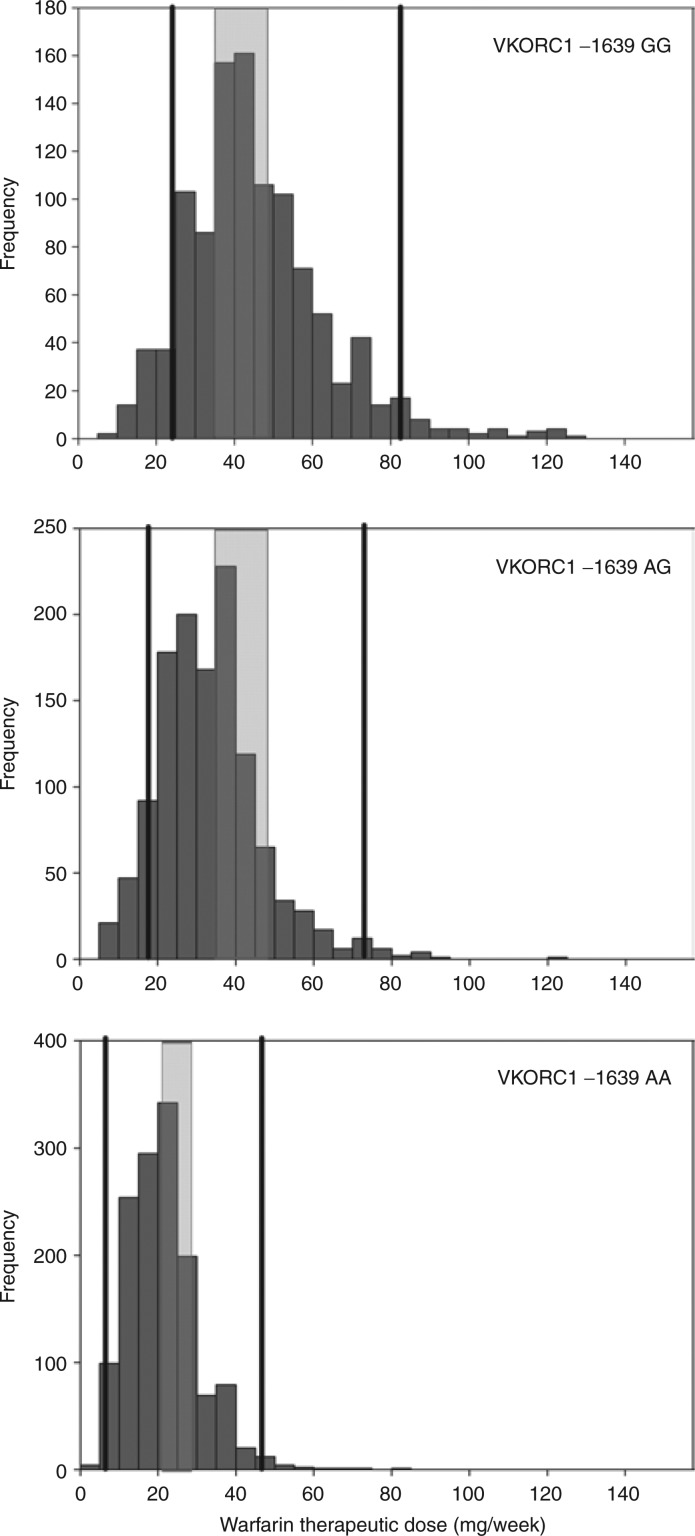
Numerous studies have derived warfarin-dosing algorithms that use both genetic and nongenetic factors to predict warfarin dose.17,18,20 Two algorithms perform well in estimating stable warfarin dose across different ethnic populations;17,18 these were created using more than 5,000 subjects. Dosing algorithms using genetics outperform nongenetic clinical algorithms and fixed-dose approaches in dose prediction.17,18 The greatest benefit of genetics in warfarin dosing is likely to be in patients requiring less than 21 mg/week or more than 49 mg/week (constituting >40% of all patients). Genetics-based algorithms also better predict warfarin dose than the FDA-approved warfarin label table.22 Therefore, the use of pharmacogenetic algorithm-based dosing is recommended when possible, although if electronic means for such dosing are not available, the table-based dosing approaches (Table 1) are suggested. The range of doses by VKORC1 geno-type and the range of dose recommendations/predictions by the FDA table and algorithm are shown in Figure 2.
Figure 2.
Frequency histograms of stable therapeutic warfarin doses in mg/week, stratified by VKORC1 −1639G>A genotype in 3,616 patients recruited by the International Warfarin Pharmacogenetics consortium (IWPC) who did not carry the CYP2C9*2 or *3 allele (i.e., coded as *1*1 for US Food and Drug Administration (FDA) table and algorithm dosing). the range of doses within each genotype group recommended on the FDA table is shown via the shaded rectangle. the range of doses predicted using the IWPc dosing algorithm in these 3,616 patients is shown by the solid lines.
Pharmacogenetic algorithm-based warfarin dosing
The best way to estimate the expected stable dose of warfarin is to use the algorithms available on http://www.warfarindosing.org (offering both high-performing algorithms17,18). The dosing algorithm published by the International Warfarin Pharmacogenetics Consortium is also online, at <http://www.pharmgkb.org/do/serve?objId=PA162372936&objCls=Dataset#tabview=tab2>. The two algorithms provide very similar dose recommendations. The clinical and genetic information used in one or both algorithms is shown in Box 1. These algorithms compute the anticipated stable daily warfarin dose to one decimal, and the clinician must then prescribe a regimen (e.g., an estimate of 4.3 mg/day might be given as 4 mg daily except 5 mg two days per week).
Approach to pharmacogenetics-based warfarin dosing without access to dosing algorithms
Table 1 summarizes the average dose ranges based on CYP2C9*2 and *3 and VKORC1 genotypes, as recommended in the FDA-approved warfarin (Coumadin) product label. The specific dose selected within that range should take into account other important variables, such as patient age, body size, and interacting drugs. The published evidence strongly supports our level A (strong) recommendation that, if genetic information is available, warfarin dosing should be estimated using a pharmacogenetic dosing algorithm, but in the absence of access to such an algorithm, a genotype dosing table is superior to other approaches that ignore genetic information in predicting stable warfarin dose.22
Other considerations
Supplementary Note S6 summarizes other considerations in the dosing of warfarin, including clinical factors and interacting drugs, some of which are included in the pharmacogenetic dosing algorithms (see Box 1). Other genes of potential importance are detailed in Supplementary Note S1 and Tables S3 and S4, including a nonsynonymous SNP in CYP4F2. Most clinical genotyping platforms do not yet include this SNP, and neither do the dosing tables or algorithms. Supplementary Note S6 also discusses incorporation of genetic information into the initial dose, as well as alternatives to warfarin.
Potential benefits and risks for the patient
Several ongoing, randomized controlled clinical trials are evaluating the risks and benefits of using genetics to dose warfarin (Supplementary Note S5). Incorporation of genetic information has the potential to shorten the time to attain stable INR, increase the time within the therapeutic INR range, and reduce underdosing or overdosing during the initial treatment period.23 If these benefits are realized, they may combine to reduce the risk of bleeding and thromboembolic events.24 There are also potential risks. For example, the use of genetic information to guide dosing may lead to false security and inadequate INR monitoring. Genetics-guided dosing may increase the risk for overdosing or underdosing, although current evidence suggests that this is not likely.18 Genetic information may affect patient decisions about warfarin—those requiring lower than average warfarin doses may believe they can take warfarin less frequently. The cost–benefit of genetics-guided therapy depends on the cost of genotyping and the reduction in adverse events,25 and most insurance plans currently do not pay for warfarin pharmacogenetic testing. Although there is substantial evidence associating CYP2C9 and VKORC1 variants with warfarin dosing, there is sparse randomized clinical trial evidence showing better outcomes. Although CYP2C9/VKORC1 genotyping is reliable when performed in qualified laboratories, an additional risk to the patient is an error in genotyping or reporting of genotype. Genotypes are lifelong test results, so such an error could have adverse health implications for the life of the patient.
Caveats: appropriate use and/or potential misuse of genetic tests
Many pharmacogenetic dosing algorithms are focused on a target INR of 2–3,18 so their utility outside this range is limited; however, some algorithms accommodate the target INR explicitly.17 Pharmacogenetics-guided warfarin dosing should not alter the requirements for regular INR monitoring. There are very few data on warfarin pharmacogenetics and the performance of dosing algorithms in children,26 so no recommendations are made in this regard. There are patients for whom genetic testing is likely to be of little or no benefit, including those who have already had long-term treatment with stable warfarin doses and those who are unable to achieve stable dosing owing to variable adherence or dietary vitamin K intake. The greatest potential benefit is early in the course of therapy (before therapy initiation or in the first few days);27 however, there may also be benefit several weeks into therapy.28 It is likely that patients on therapy for many weeks to months, with careful INR monitoring, will derive little benefit from subsequent warfarin pharmacogenetics testing.29
Disclaimer
CPIC guidelines reflect expert consensus based on clinical evidence and peer-reviewed literature available at the time they are written and are intended only to assist clinicians in decision making and to identify questions for further research. New evidence may have emerged since the time a guideline was submitted for publication. The guidelines are limited in scope and are not applicable to interventions or diseases not specifically identified. They do not account for all individual variations among patients and cannot be considered inclusive of all proper methods of care or exclusive of other treatments. It remains the responsibility of the health-care provider to determine the best course of treatment for a patient. Adherence to any guideline is voluntary, with the ultimate determination regarding its application to be made solely by the clinician and the patient. CPIC assumes no responsibility for any injury to persons or damage to persons or property arising out of or related to any use of CPIC's guidelines, or for any errors or omissions.
Supplementary Material
Box 1 Patient characteristics utilized in the Gage algorithm,17 the International Warfarin Pharmacogenetics Consortium18 algorithm, or both.
Age
Sex
Race
Weight
Height
Smoking status
Warfarin indication
Target international normalized ratio
-
Interacting drugs
Inhibitors: amiodarone, statins, sulfamethoxazole, azole antifungals
Inducers: rifampin, phenytoin, carbamazepine
-
Genetic variables
CYP2C9 genotype
VKORC1 genotype
The Gage algorithm can also incorporate CYP4F2 and GGCX genotypes
ACKNOWLEDGMENTS
We acknowledge and thank Sherry Xie and Brian Gawronski for helping to create Figure 2 and our colleagues in the Clinical Pharmacogenetics Implementation Consortium for their helpful comments, particularly Issam Zineh, Nita Limdi, and John Callaghan. the effort of the authors was supported in part by grants U01 GM074492 (J.A.J.), U01 GM061374 (C.M.S.); R01 HL097036 (B.F.G.); R01 HL066176 (S.E.K.); R24 GM061374 (R.B.A. and T.E.K.); CLZ RR029885 (S.A.S.); R01 HL097036; and NSC99-3112-B-001-023 (M.T.M.L.). M.P. is an NIHR Senior Investigator in the UK and acknowledges the support of the UK Department of Health, the Wolfson Foundation, and the MRC; M.W. is supported by the Swedish Heart and Lung Foundation, the Swedish Research Council (Medicine 523-2008-5568), and Clinical Research Support (ALF) at Uppsala University. Both M.P. and M.W. are supported by EU-FP7 (HEALTH-F2-2009-223062).
Footnotes
SUPPLEMENTARY MATERIAL is linked to the online version of the paper at http://www.nature.com/cpt
CONFLICT OF INTEREST
All authors are members of the International Warfarin Pharmacogenetics consortium. S.E.K. is the principal investigator for the US-based COAG trial; J.L.A., B.F.G., and J.A.J. are members of the COAG executive committee; and S.A.S. and c.M.S. are investigators in COAG. B.F.G. is principal investigator of the GIFT trial, and J.L.A. is an investigator. M.P. is the UK chief investigator for the EU-PACT trial, and M.W. is the principal investigator in Sweden; the views presented here are their own and are not presented on behalf of the EU-PACT investigators collectively. the EU-PACT trial is funded by the EU-FP7 framework. M.T.M.L. is the principal investigator for the Taiwan-based pharmacogenetic dosing of warfarin randomized trial. S.E.K. receives additional funding from the National Institutes of Health for research on warfarin pharmacogenetics, has received funding from the Aetna Foundation for warfarin-related research, has received an honorarium for a talk on warfarin from Ortho McNeill, and has received grants from and served as a consultant to several pharmaceutical companies, all unrelated to warfarin. J.A.J. is on an advisory committee for Medco. R.B.A. is a consultant to 23andme.com.
References
- 1.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin. Pharmacol. Ther. 2011;89:464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansell J, Hirsh J, Hylek E, Jacobson A, Crowther M, Palareti G. American College of Chest Physicians. Pharmacology and management of the vitamin K antagonists: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines. Chest. (8th Edition) 2008;133:160S–198S. doi: 10.1378/chest.08-0670. [DOI] [PubMed] [Google Scholar]
- 3.Jacobs LG. Warfarin pharmacology, clinical management, and evaluation of hemorrhagic risk for the elderly. Clin. Geriatr. Med. 2006;22:17–32. vii. doi: 10.1016/j.cger.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Rieder MJ, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N. Engl. J. Med. 2005;352:2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 5.Higashi MK, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA. 2002;287:1690–1698. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- 6.Shehab N, Sperling LS, Kegler SR, Budnitz DS. National estimates of emergency department visits for hemorrhage-related adverse events from clopidogrel plus aspirin and from warfarin. Arch. Intern. Med. 2010;170:1926–1933. doi: 10.1001/archinternmed.2010.407. [DOI] [PubMed] [Google Scholar]
- 7.Rost S, et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004;427:537–541. doi: 10.1038/nature02214. [DOI] [PubMed] [Google Scholar]
- 8.Lee CR, Goldstein JA, Pieper JA. Cytochrome P450 2C9 polymorphisms: a comprehensive review of the in-vitro and human data. Pharmacogenetics. 2002;12:251–263. doi: 10.1097/00008571-200204000-00010. [DOI] [PubMed] [Google Scholar]
- 9.Limdi NA, et al. International Warfarin Pharmacogenetics Consortium Warfarin pharmacogenetics: a single VKORC1 polymorphism is predictive of dose across 3 racial groups. Blood. 2010;115:3827–3834. doi: 10.1182/blood-2009-12-255992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aithal GP, Day CP, Kesteven PJ, Daly AK. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet. 1999;353:717–719. doi: 10.1016/S0140-6736(98)04474-2. [DOI] [PubMed] [Google Scholar]
- 11.Limdi NA, et al. Influence of CYP2C9 and VKORC1 1173C/T genotype on the risk of hemorrhagic complications in African-American and European-American patients on warfarin. Clin. Pharmacol. Ther. 2008;83:312–321. doi: 10.1038/sj.clpt.6100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindh JD, Holm L, Andersson ML, Rane A. Influence of CYP2C9 genotype on warfarin dose requirements–a systematic review and meta-analysis. Eur. J. Clin. Pharmacol. 2009;65:365–375. doi: 10.1007/s00228-008-0584-5. [DOI] [PubMed] [Google Scholar]
- 13.Li T, Chang CY, Jin DY, Lin PJ, Khvorova A, Stafford DW. Identification of the gene for vitamin K epoxide reductase. Nature. 2004;427:541–544. doi: 10.1038/nature02254. [DOI] [PubMed] [Google Scholar]
- 14.Wajih N, Hutson SM, Owen J, Wallin R. Increased production of functional recombinant human clotting factor IX by baby hamster kidney cells engineered to overexpress VKORC1, the vitamin K 2,3-epoxide-reducing enzyme of the vitamin K cycle. J. Biol. Chem. 2005;280:31603–31607. doi: 10.1074/jbc.M505373200. [DOI] [PubMed] [Google Scholar]
- 15.Wadelius M, et al. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics J. 2005;5:262–270. doi: 10.1038/sj.tpj.6500313. [DOI] [PubMed] [Google Scholar]
- 16.Yuan HY, et al. A novel functional VKORC1 promoter polymorphism is associated with inter-individual and inter-ethnic differences in warfarin sensitivity. Hum. Mol. Genet. 2005;14:1745–1751. doi: 10.1093/hmg/ddi180. [DOI] [PubMed] [Google Scholar]
- 17.Gage BF, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin. Pharmacol. Ther. 2008;84:326–331. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klein TE, et al. International Warfarin Pharmacogenetics Consortium. Estimation of the warfarin dose with clinical and pharmacogenetic data. N. Engl. J. Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott SA, Edelmann L, Kornreich R, Desnick RJ. Warfarin pharmacogenetics: CYP2C9 and VKORC1 genotypes predict different sensitivity and resistance frequencies in the Ashkenazi and Sephardi Jewish populations. Am. J. Hum. Genet. 2008;82:495–500. doi: 10.1016/j.ajhg.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wadelius M, et al. the largest prospective warfarin-treated cohort supports genetic forecasting. Blood. 2009;113:784–792. doi: 10.1182/blood-2008-04-149070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gage BF, Lesko LJ. Pharmacogenetics of warfarin: regulatory, scientific, and clinical issues. J. Thromb. Thrombolysis. 2008;25:45–51. doi: 10.1007/s11239-007-0104-y. [DOI] [PubMed] [Google Scholar]
- 22.Finkelman BS, Gage BF, Johnson JA, Brensinger CM, Kimmel SE. Genetic warfarin dosing: tables versus algorithms. J. Am. Coll. Cardiol. 2011;57:612–618. doi: 10.1016/j.jacc.2010.08.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caraco Y, Blotnick S, Muszkat M. CYP2C9 genotype-guided warfarin prescribing enhances the efficacy and safety of anticoagulation: a prospective randomized controlled study. Clin. Pharmacol. Ther. 2008;83:460–470. doi: 10.1038/sj.clpt.6100316. [DOI] [PubMed] [Google Scholar]
- 24.Odén A, Fahlén M. Oral anticoagulation and risk of death: a medical record linkage study. BMJ. 2002;325:1073–1075. doi: 10.1136/bmj.325.7372.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eckman MH, Rosand J, Greenberg SM, Gage BF. cost-effectiveness of using pharmacogenetic information in warfarin dosing for patients with nonvalvular atrial fibrillation. Ann. Intern. Med. 2009;150:73–83. doi: 10.7326/0003-4819-150-2-200901200-00005. [DOI] [PubMed] [Google Scholar]
- 26.Nowak-Göttl U, et al. In pediatric patients, age has more impact on dosing of vitamin K antagonists than VKORC1 or CYP2C9 genotypes. Blood. 2010;116:6101–6105. doi: 10.1182/blood-2010-05-283861. [DOI] [PubMed] [Google Scholar]
- 27.Lenzini P, et al. Integration of genetic, clinical, and INR data to refine warfarin dosing. Clin. Pharmacol. Ther. 2010;87:572–578. doi: 10.1038/clpt.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Epstein RS, et al. Warfarin genotyping reduces hospitalization rates results from the MM-WES (Medco-Mayo Warfarin Effectiveness study). J. Am. Coll. Cardiol. 2010;55:2804–2812. doi: 10.1016/j.jacc.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 29.Ferder NS, et al. Ability of VKORC1 and CYP2C9 to predict therapeutic warfarin dose during the initial weeks of therapy. J. Thromb. Haemost. 2010;8:95–100. doi: 10.1111/j.1538-7836.2009.03677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.