Abstract
Timely intervention for cancer requires knowledge of its earliest genetic aberrations. Sequencing of tumors and their metastases reveals numerous abnormalities occurring late in progression. A means to temporally order aberrations in a single cancer, rather than inferring them from serially acquired samples, would define changes preceding even clinically evident disease. We integrate DNA sequence and copy number information to reconstruct the order of abnormalities as individual tumors evolve for two separate cancer types. We detect vast, unreported expansion of simple mutation sharply demarcated by recombinative loss of the second copy of TP53 in cutaneous squamous cell carcinomas (cSCCs) and serous ovarian adenocarcinomas, in the former surpassing 50 mutations per megabase. In cSCCs, we also report diverse secondary mutations in known and novel oncogenic pathways, illustrating how such expanded mutagenesis directly promotes malignant progression. These results reframe paradigms in which TP53 mutation is required later, to bypass senescence induced by driver oncogenes.
Keywords: mutation, p53, cancer genetics, genomic, Notch
Introduction
Molecular characterization of human cancers usually profiles a single point in time, yielding a catalogue of genomic and epigenetic abnormalities reflecting years of somatic change. Although recent efforts reveal some mutations associated with metastasis and recurrence(1), timing of events early in tumorigenesis remains difficult. Precursor dysplastic lesions may be sampled and compared against invasive malignancies, but in many cancer types, early lesions are not clinically identifiable, nor is it obvious which lesions will actually progress. Additionally, the increasingly apparent heterogeneity of human cancers suggests such comparisons will require very large sample sizes to reconstruct progression(2).
Primary cSCCs rank among the most common human malignancies, with an annual incidence in Caucasians of more than 150 in 100,000 individuals(3). These tumors arise in anatomic site and demographic proportionate to sunlight exposure and acquire a mutational spectrum reflecting significant ultraviolet radiation damage(4). Although many are excised without complication, cSCCs sometimes behave aggressively with recurrence and regional spread, especially in immunosuppressed and repair-deficient genetic backgrounds (3). We hypothesized that the long-term mutational stress on these tumors might offer unique insight into the progressive events determining a cancer’s individuality.
Results
Exome-level sequencing of eight primary cSCCs and matched normal tissue revealed a very large mutation burden of approximately 1,300 somatic single-nucleotide variants per cSCC exome, (1 per ~30,000 base pairs of coding sequence, Supplementary Figure 1, Supplementary Methods and Appendix) making cSCCs the most highly mutated human malignancy. Of mutations assessed by capillary sequencing across our series, 75/75 (100%) confirmed the originally detected mutations, including four instances of dinucleotide substitution. C > T transition base substitutions at dipyrimidine sites were by far the most common change (>85%), consistent with UV damage. Past analysis of selected TP53 exons suggests that the gene is mutated in 50–90% of cSCCs(5). Our study identified TP53 mutations in 7/8 skin cancers, all coinciding with previously reported changes in the COSMIC database(6). Known changes were also found in CDKN2A, encoding the p16/p14ARF bifunctional tumor suppressor, the HRAS small GTPase, and instances of numerous COSMIC mutations not previously described in cSCCs (Table 1; full list of base substitutions in Supplementary Table 1). We also detected 42 discrete chromosomal abnormalities, about five per sample (Supplementary Table 2).
Table 1.
Description of sample cases and selected mutations of interest.
| Sex | Age | Site | Immune status |
TP53 | CDKN2A | NOTCH1 | NOTCH2 | NOTCH3 | NOTCH4 | Other known COSMIC mutations |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. | M | 76 | scalp | + | R248W | P135L | Q610* | W330*, R1838* |
EP300, WT1 | ||
| 2. | M | 87 | scalp | + | E285K | P1770S, R1594Q |
P226S | WTI | |||
| 3. | M | 84 | left dorsal hand |
+ |
E224 (splice site) |
C478F | R1333C | ||||
| 4. | F | 61 | left cheek | + | Y220N | W309* | PIK3CG | ||||
| 5. | M | 83 | left cheek | + | H179Y, P278S |
W1769* | Q1634*, T2278I |
S1602F | |||
| 6. | M | 85 | right temple | + | P142N, H179Y |
P133L | (splice site) | S1836F, E297K |
EZH2 | ||
| 7. | M | 58 | left helix | − | E286K, T329I, E349* |
Q1924* | Q1616*, G488D |
HRAS | |||
| 8. | M | 63 | lower lip | + | none§ | HSPB2 |
Bold-face notes loss of wild-type allele and *notes nonsense mutations, shaded mutations are found in the COSMIC database. TP53 mutations in samples 1–4 are duplicated through CN-LOH on chromosome 17p.
In sample 8, no mutation in TP53 was seen, but early, duplicated mutation was detected in in ATM at G2024R, in the PI-kinase related domain. Given their sheer number, some TP53 mutations may act in true dominant negative fashion, but were not detected in our series. COSMIC Notch family mutations are activating in hematologic malignancies; the truncations and disabling mutations reported here have not been previously described.
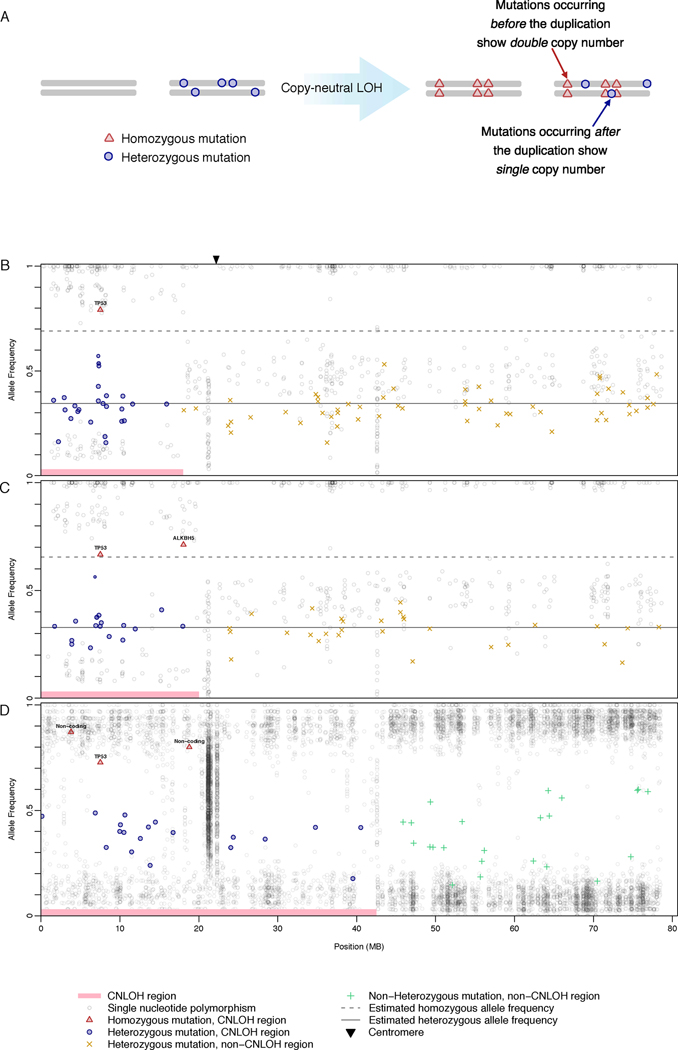
The ability to temporally order successive molecular changes within an individual tumor, beginning in the initial stages of tumorigenesis, would allow discrimination between mutations forming precancerous lesions from those producing invasive carcinomas. The high prevalence of both simple mutations and copy number abnormalities abnormalities in cSCCs and ovarian cancers enabled us to reconstruct the evolutionary order of some somatic changes based on the following idea: if a mutation precedes a regional duplication, its copy number is doubled. Mutations following a duplication event appear in haploid copy number(7). Therefore 1) simple mutations preceding a chromosomal duplication event show discretely higher copy number compared to those occurring after duplication (Figure 1) and 2) the ratio of heterozygous to homozygous mutations ρ, in a region of CN-LOH, directly measures the age of the duplication (in evolutionary time) (Figure 2, Supplementary Figure 2).
Figure 1.
Loss of second TP53 wild-type allele precedes simple mutations in cutaneous squamous cell carcinomas and ovarian cancers. (A) Schematic illustrating acquisition of discretely higher copy number for mutations preceding a regional duplication, enabling temporal ordering of mutations during tumorigenesis. For representative samples, mutant allele frequency (y-axis) shown plotted against physical position on chromosome 17 (x-axis) for cutaneous squamous cell (B,C) and ovarian cancers (D). More than 50 independent reads were required for inclusion of a mutation. Estimated allele frequencies are shown as lines for heterozygous (solid) and homozygous (dotted) mutations (See Supplementary Methods and Appendix for details). Regions without CN-LOH in ovarian cancers often demonstrate complex ploidy, generating greater variance in mutation frequency; these simple frequency estimates were therefore not applied. Mutation frequencies are centered below 1.0 for homozygotes and 0.5 for heterozygotes because of the effects of partial dilution with non-tumorous cells (see Supplementary Methods and Appendix for more details).
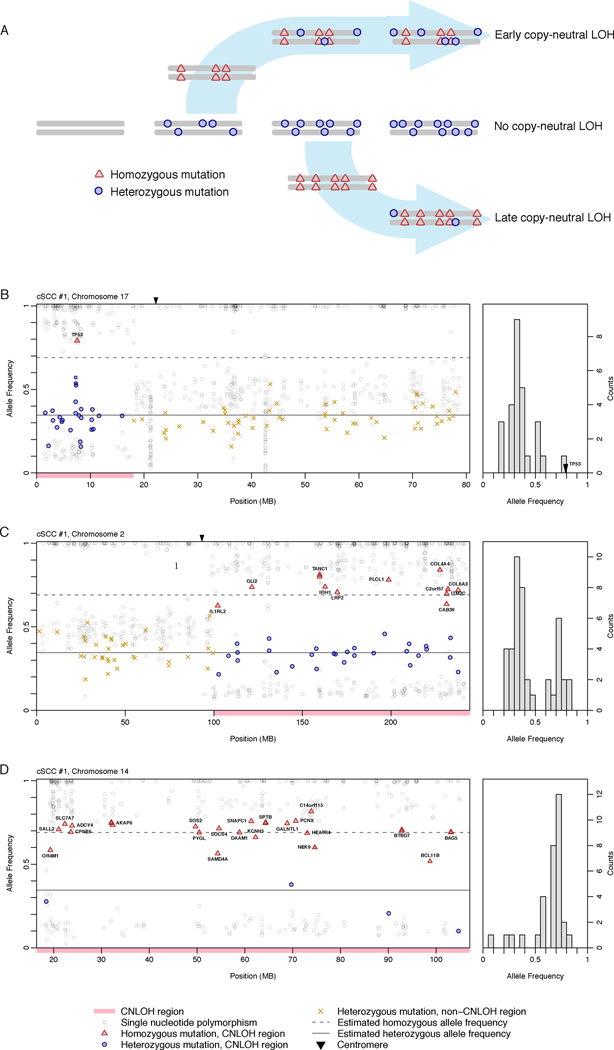
Figure 2.
Conceptual framework for temporal ordering of chromosomal aberrations. (A) Schematic for differential mutant allele frequencies in areas of copy-neutral loss-of-heterozygosity, based on relative age of chromosomal event during tumorigenesis. Earlier events allow greater time for accumulation of heterozygous mutations, whereas later events are predominated by homozygous mutations. Three events are ordered in a single sample, earliest on chromosome 17 (B), later on chromosome 2 (C), and last on chromosome 14 (D). Relative mutant allele frequency (y-axis) is plotted against chromosomal physical position (x-axis) for cutaneous squamous cell cancers. To the right of each panel, a histogram of mutant allele frequencies for each CN-LOH event shows density (y-axis) plotted against allele frequency (x-axis). A modified binomial distribution function (Supplementary Methods and Appendix) distinguishes the proportion of heterozygote and homozygote mutant frequencies ρ, ordering duplications in chromosome 17 (0.00–0.12), chromosome 2 (0.17–0.45), and 14 (0.65–0.94).
We first utilized this principle to investigate the specific temporal order of mutations in areas of copy-neutral loss-of-heterozygosity (CN-LOH), in which a regional chromosomal duplication replaces the matching portion of the paired chromosome(8; 9). Of the cSCCs in our series, 4/8 showed CN-LOH at chromosome 17p, all harboring TP53 mutations reported in COSMIC. Remarkably, all four TP53 mutations were present at high allelic abundance in the CN-LOH region compared to other somatic mutations, indicating that TP53 mutations occurred and were duplicated before other mutations arose (Figure 1B,C). In aggregate, 59/63 mutations in 17p appear after loss of the second TP53 wild-type allele, 15-fold greater than those preceding loss. CN-LOH events at 17p represent 2% of coding sequence and show normalized mutation frequencies reflective of the remainder of the exome (Supplementary Figure 3). While studies establish some p53 mutations as gain-of-function with respect to cancer type(10), or biochemically dominant negative, ours is the first report that the vast majority of simple mutation – tens of thousands genome-wide in the case of cSCCs – appear sharply gated by elimination of the second copy of TP53. We further detect at least partial persistence of active DNA repair, suggesting a profound loss of damage surveillance contributes to the high number of observed mutations (Supplementary Figure 1, Supplementary Methods and Appendix) (11).
Three samples without CN-LOH at 17p demonstrate at least two distinct TP53 mutations, presumably causing biallelic mutation. In the sample in which TP53 mutations were not detected, a regionally duplicated mutation in the ATM kinase domain was observed, suggesting an alternate means of escaping damage surveillance mechanisms during telomere crisis(12).
We sought to validate our observations in an additional cancer type. Recently, full genomic sequence and copy number changes were determined for 10 ovarian serous adenocarcinomas by The Cancer Genome Atlas Project. Ovarian cancers generally show more complex karyotypic abnormalities than cSCCs(13). In the three samples with a clear, informative CN-LOH event at 17p, we again found clear evidence for complete loss of TP53 as the earliest event (Figure 1D). These initial events in ovarian tumorigenesis could not have been determined through sequencing of precursor lesions and invasive cancers(1; 14), as the asymptomatic nature of early disease precludes tissue collection.
Integrative analyses of copy number and exome sequence also reveal information about the temporal order of chromosomal abnormalities within an individual cancer(7). As described above, the ratio of heterozygous to homozygous mutations ρ, in a given region of CN-LOH, provides a direct measure of the relative age of the duplication (Figure 2A). In other words, duplications with higher ρ occur earlier than those with lower ρ. We found that ρ varied widely among regions of CN-LOH (Figure 2B–D, Figure 3) and could statistically distinguish the temporal order of aberrations within a sample (Figure 2). Overall, seven informative duplications co-occurring with 17p CN-LOH all showed a substantially lower relative ρ (Supplementary Table 2) and thus likely occurred after 17p duplication. Therefore loss of the second TP53 allele appears to precede not only a vast expansion of simple mutations, but also the development of chromosomal aberrations. As a general principle, any regional copy gain acquires a heterozygote mutation frequency uniquely reflective of the time of gain. For selected instances in our series, extension of this principle enabled temporal dissection of more complex copy gains (Figure 3), revealing that these alterations also follow complete TP53 loss.
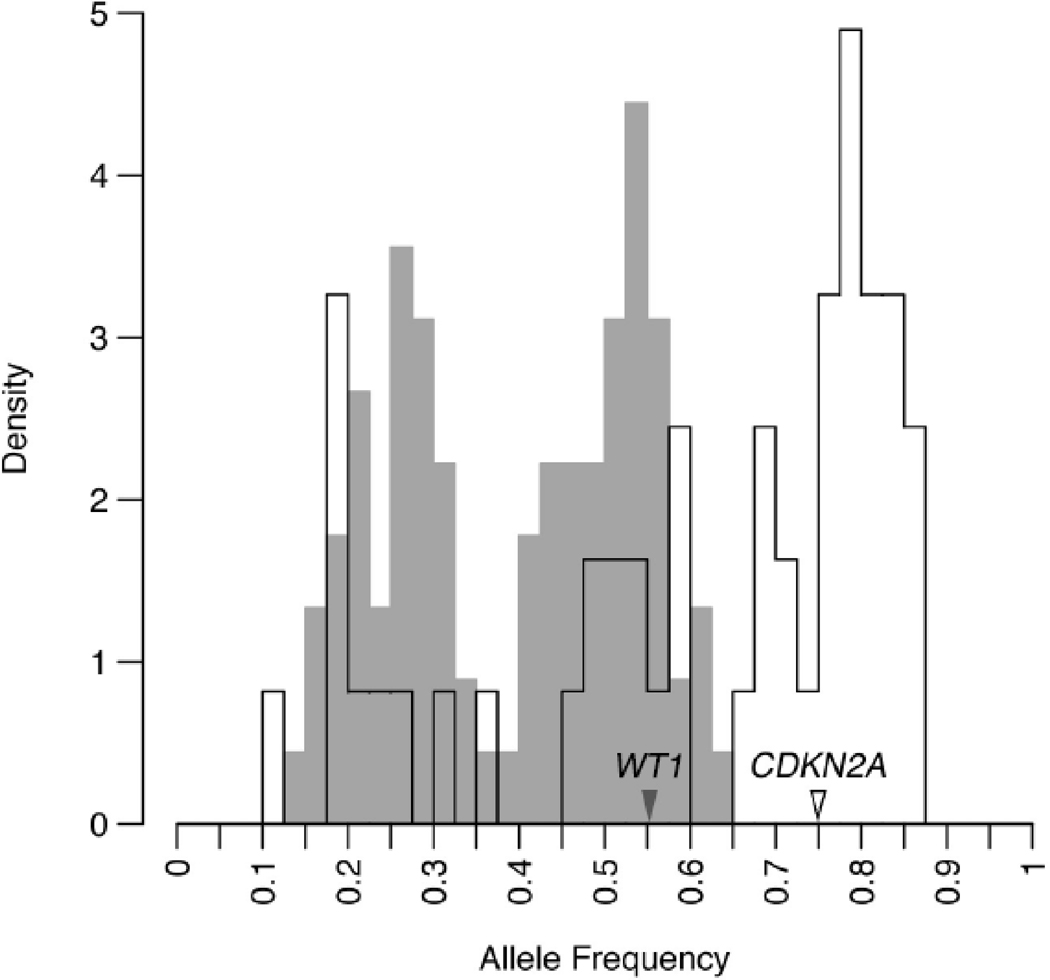
Figure 3.
For two regional chromosomal aberrations, mutant allele frequencies reveal distinct evolutionary histories. On chromosome 11, two peaks in mutation frequency are seen, one for heterozygous and one for homozygous mutations, reflect a simple regional gain, including for a COSMIC mutation in WT1. In contrast, three separate frequencies are detected on chromosome 9, culminating in a triploid state for mutant CDKN2A and loss of all wild-type copies. Three discrete allele frequencies can only be achieved by two gains and one loss, with the highest frequency (triploid) peak representing the first gain, and the diploid peak containing mutations from the interval preceding the second gain. The disproportionate size of the triploid peak (p < 0.03) indicates that the first gain occurred more than halfway through in tumor evolution, as measured by the evolutionary clock, and the second gain even later (see Supplementary Methods and Appendix for more detail).
In cSCCs, we found 486 non-synonymous mutations that were sequenced deeply enough to determine copy number (> 50 independent reads) and that fell at least once in a region of CNLOH. These included known mutations in CDKN2A, WT1, and HRAS (Table 1) each of which showed multiple instances of either wild-type allele loss or biallelic mutation, as seen for TP53. Interestingly, this pattern of recurrent biallelic inactivation was also detected at high prevalence for the suspected epithelial tumor suppressors NOTCH1 and NOTCH2 and the polycystic kidney disease gene PKHD1 (Supplementary Methods and Appendix). NOTCH1 demonstrates three instances of early truncation, two of which show wild-type loss, one case of multiple mutation, and two other mutations, one of which occurs in a splice site (Table 1). NOTCH2 shows multiple mutations in 4/8 samples, and three of these contain at least one truncating mutation.
Discussion
We trace the mutational evolution of individual tumors using a novel, sequence-based assessment strategy and in doing so, provide an individualizable, patient-centric complement to more traditional “mutation-by-stage” approaches(2; 14). Our results illuminate key aspects of timing in cancer evolution without requiring large sample series, for which precursor lesions are often inaccessible. TP53 is often mutated in precursor lesions, but paradigms of oncogenesis propose p53 loss as a late requirement, overcoming senescence programs activated by prior activation of driver oncogenes(15; 16) and surviving telomere crisis(17). Furthermore, biallelic TP53 loss occurs frequently, despite evidence that p53 mutants behave dominantly structurally and also functionally with respect to phenotypes such as tumor formation(18; 10).
Our data show that decades of UV damage and inactivation of a single TP53 allele only result in about one hundred mutations in the epithelial exome. This tenacious genetic stability explains the benign behavior of clonal keratinocyte proliferations, harboring heterozygous TP53 mutation, that commonly form in sun-exposed skin(19; 20). Subsequent elimination of the second TP53 allele, often through recombination, sharply demarcates a vast expansion in simple mutation, in cSCCs reaching 50 per megabase (150,000 per genome) and making them the most mutagenized human cancers known. Because DNA repair remains at least partially active, this vast mutation burden might result from the collaborative effects of ongoing DNA damage (from intrinsic and exogenous insults) coupled with disabled DNA damage-induced apoptosis. The reproduction of this phenomenon in ovarian adenocarcinoma suggests that the vast majority of mutations follow second TP53 allele loss, irrespective of mode of DNA damage or tissue of origin.
Classic studies report that precursor lesions and invasive cancers both carry mutated driver oncogenes, but find TP53 inactivation more frequent in invasive disease (21; 22), suggesting p53 inactivation to be a late event. Activation of a key oncogene prior to biallelic TP53 loss in our series is formally possible, but few coding mutations precede 17p duplication and none recur in established oncogenes. While apparently contradictory, these findings could be reconciled by a temporal requirement that TP53 mutation precede driver oncogene mutation in precursor lesions destined to progress to invasive cancer. In this model, precursors lesions that activate oncogenes first (before TP53 inactivation) fail to progress, but would nonetheless be detected in “mutation frequency-by-stage” surveys. Alternatively, different cancer types might demonstrate distinct temporal ordering of key mutations. Application of our approach to sequence data from other cancer types, such as colon adenocarcinoma, should help distinguish these possibilities.
In selected instances we are able to show mutant TP53 duplication occurring before dosage changes in mutant alleles, such as for CDKN2A and WT1 (Figure 3). The consequences of such expanded mutagenesis and chromosomal instability emerge dramatically in the Notch signaling pathway, in which multiple family members develop mutations and wild-type alleles are lost frequently. Constitutive Notch protein activation drives subsets of acute leukemias(23), but a clear tumor suppressor phenotype has also been established in keratinocytes(24), with attentuated expression producing proliferation and invasive morphologies. Further study should clarify the breadth of epithelial cancers harboring these somatic changes, as well as their specific functional effects. Our data also confirms low-prevalence activation of known oncogenes such as Ras in human cSCCs, raising the possibility that numerous mutations in other pathways may serve as the functional oncogene in this setting(25).
Taken together, these insights imply that targeting activated oncogenes (e.g. with small molecule inhibitors) fails to address a fundamental, detectable abnormality in cancer genomes that accelerates evolution toward clinical resistance. Such temporal dissection of tumorigenesis provides both early, assayable diagnostic markers and also illuminates the specific biological consequences of these aberrations. We demonstrate the utility of this method for the CN-LOH and copy gains that comprise most chromosomal aberrations. Because many cancer types carry rearrangement of substantial proportions of the genome, especially those spanning key oncogenes, extension of this method should rapidly reveal additional ordered events. The described reconstruction of genomic aberration history can be applied immediately to any cancer for which sequence data and copy number is available.
Materials and Methods
We obtained eight matched cSCC and normal tissue samples as part of a skin cancer study protocol, with all subjects providing informed consent according to procedures approved by the University of California, San Francisco Committee on Human Research. Diagnosis of cutaneous squamous cell carcinoma was confirmed for all tumors via histological examination of a standard biopsy specimen by a board-certified dermatopathologist. DNA was extracted from tumor and control samples, and allele-specific copy number analysis was performed using Affymetrix Genome-Wide Human SNP Array 6.0 chips. About 40 megabases of coding region were isolated from each sample using oligonucleotide-based hybrid capture and sequenced using the Illumina sequencing-by-synthesis platform (Supplementary Figure 4). Mutation detection was performed as previously described (see Supplementary Methods and Appendix for detail). Eighty mutations were independently validated using Sanger sequencing; 100% confirmed the originally identified somatic chance. Sequence for all non-synonymous mutations will be deposited in dbGAP. Patient information and genomic profiling for ovarian serous adenocarcinomas analyzed here have been described previously(26).
Chromosomal regions with aberrant copy number, including copy neutral loss-of-heterozygosity (CN-LOH) and simple copy gains and losses, were identified based on discrete shifts in single nucleotide polymorphism copy number from both SNP array and exome sequencing data. The type of abnormality was further confirmed by assessing raw copy number depth from SNP array data. Mutations were called after alignment of sequence reads to a reference genome (NCBI36). The fraction of chromosomal copies carrying a mutation was estimated as the fraction of all independent sequence reads containing that mutation. (A detailed description of patient consent, methods and reagents used for tissue acquisition, genomic profiling, and statistical analysis of mutational evolution is provided in the Supplementary Methods and Appendix.)
Significance.
Our approach reveals sequential ordering of oncogenic events in individual cancers, based on chromosomal rearrangements. Identifying the earliest abnormalities in cancer represents a critical step in timely diagnosis and deployment of targeted therapeutics.
Supplementary Material
Acknowledgements
We thank Allan Balmain, Boris Bastian, Jeffrey Cheng, Douglas Brash, and Dennis Oh for early support and helpful discussions. Henrik Bengtsson, Pierre Neuvial, Hubert Stoppler, Matthew Akana, Connie Ha, Lauren Lee, Annie Poon, and Eric Dybbro provided technical assistance. W.L. is supported by a Dermatology Foundation Psoriasis Career Development Award, J.C. by the University of California Cancer Coordinating Committee, S.T.A. by NIH/NCRR/OD UCSF-CTSI Grant Number KL2 RR024130, a Canary Foundation/American Cancer Society Postdoctoral Fellowship for the Early Detection of Cancer, and a Dermatology Foundation Career Development Award, R.J.C. by a Dermatology Foundation Career Development Award and as a Samsung Biotechnology Scholar-in-Residence.
This research was supported under Joe W Gray (by the Director, Office of Science, Office of Biological & Environmental Research, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231, by the National Institutes of Health, National Cancer Institute grants P50 CA 58207, the U54 CA 112970, the NHGRI U24 CA 126551, by the Department of the Army, award: W81XWH-07-1-0663 [The U.S. Army Medical Research Acquisition Activity, 820 Chandler Street, Fort Detrick, MD 21702-5014 is the awarding and administering acquisition office], and by the Stand Up To Cancer-American Association for Cancer Research Dream Team Translational Cancer Research Grant SU2C-AACR-DT0409. The content of this information does not necessarily reflect the position or the policy of the Government, and no official endorsement should be inferred.); Eric A. Collisson (NIH/NCI K08 CA137153); Paul T. Spellman (NIH/NCI U24 CA1437991); Raymond J. Cho (by an unrestricted gift grant from the Samsung Advanced Institute of Technology); Theodoro M. Mauro (by NIH grants AR051930 and R01AG028492, and the Medical Research Service, Department of Veterans Affairs); and Allan Balmain, (by NIH/NIAMS Program Project Grant 5-P01-AR050440-05).
Abbreviations
- CN-LOH
copy-neutral loss-of-heterozygosity
- COSMIC
Catalogue of Somatic Mutations In Cancer
- dbGAP
database of Genotypes and Phenotypes
- DNA
deoxyribonucleic acid
- cSCC
cutaneous squamous cell carcinoma
- SNP
single nucleotide polymorphism
- UV
ultraviolet
Footnotes
Financial disclosure: None reported
References
- 1.Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bozic I, Antal T, Ohtsuki H, Carter H, Kim D, Chen S, et al. Accumulation of driver and passenger mutations during tumor progression. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18545–18550. doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Madan V, Lear JT, Szeimies R. Non-melanoma skin cancer. Lancet. 2010;375:673–685. doi: 10.1016/S0140-6736(09)61196-X. [DOI] [PubMed] [Google Scholar]
- 4.Ziegler A, Jonason AS, Leffell DJ, Simon JA, Sharma HW, Kimmelman J, et al. Sunburn and p53 in the onset of skin cancer. Nature. 1994;372:773–776. doi: 10.1038/372773a0. [DOI] [PubMed] [Google Scholar]
- 5.Leffell DJ. The scientific basis of skin cancer. J. Am. Acad. Dermatol. 2000;42:18–22. doi: 10.1067/mjd.2000.103340. [DOI] [PubMed] [Google Scholar]
- 6.Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, Cole C, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–D657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Keefe C, McDevitt MA, Maciejewski JP. Copy neutral loss of heterozygosity: a novel chromosomal lesion in myeloid malignancies. Blood. 2010;115:2731–2739. doi: 10.1182/blood-2009-10-201848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuna M, Knuutila S, Mills GB. Uniparental disomy in cancer. Trends Mol Med. 2009;15:120–128. doi: 10.1016/j.molmed.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Wu X, Levine AJ. p53 and E2F-1 cooperate to mediate apoptosis. Proc. Natl. Acad. Sci. U.S.A. 1994;91:3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooke SL, Ng CKY, Melnyk N, Garcia MJ, Hardcastle T, Temple J, et al. Genomic analysis of genetic heterogeneity and evolution in high-grade serous ovarian carcinoma. Oncogene. 2010;29:4905–4913. doi: 10.1038/onc.2010.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 16.Hruban RH, Goggins M, Parsons J, Kern SE. Progression model for pancreatic cancer. Clin. Cancer Res. 2000;6:2969–2972. [PubMed] [Google Scholar]
- 17.Chin L, Artandi SE, Shen Q, Tam A, Lee SL, Gottlieb GJ, et al. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- 18.Milner J, Medcalf EA. Cotranslation of activated mutant p53 with wild type drives the wild-type p53 protein into the mutant conformation. Cell. 1991;65:765–774. doi: 10.1016/0092-8674(91)90384-b. [DOI] [PubMed] [Google Scholar]
- 19.Jonason AS, Kunala S, Price GJ, Restifo RJ, Spinelli HM, Persing JA, et al. Frequent clones of p53-mutated keratinocytes in normal human skin. Proc. Natl. Acad. Sci. U.S.A. 1996;93:14025–14029. doi: 10.1073/pnas.93.24.14025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren ZP, Ahmadian A, Pontén F, Nistér M, Berg C, Lundeberg J, et al. Benign clonal keratinocyte patches with p53 mutations show no genetic link to synchronous squamous cell precancer or cancer in human skin. Am. J. Pathol. 1997;150:1791–1803. [PMC free article] [PubMed] [Google Scholar]
- 21.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 22.Baker SJ, Preisinger AC, Jessup JM, Paraskeva C, Markowitz S, Willson JK, et al. p53 gene mutations occur in combination with 17p allelic deletions as late events in colorectal tumorigenesis. Cancer Res. 1990;50:7717–7722. [PubMed] [Google Scholar]
- 23.Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 24.Lefort K, Mandinova A, Ostano P, Kolev V, Calpini V, Kolfschoten I, et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKα kinases. Genes & Development. 2007;21:562–577. doi: 10.1101/gad.1484707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon MM, Sliutz G, Luger TA. Proto-oncogenes and oncogenes in epidermal neoplasia. Exp Dermatol. 1995;4:65–73. doi: 10.1111/j.1600-0625.1995.tb00224.x. [DOI] [PubMed] [Google Scholar]
- 26.The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. doi: 10.1038/nature10166. (in press, 2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.