ABSTRACT
The genus Cyanothece comprises unicellular cyanobacteria that are morphologically diverse and ecologically versatile. Studies over the last decade have established members of this genus to be important components of the marine ecosystem, contributing significantly to the nitrogen and carbon cycle. System-level studies of Cyanothece sp. ATCC 51142, a prototypic member of this group, revealed many interesting metabolic attributes. To identify the metabolic traits that define this class of cyanobacteria, five additional Cyanothece strains were sequenced to completion. The presence of a large, contiguous nitrogenase gene cluster and the ability to carry out aerobic nitrogen fixation distinguish Cyanothece as a genus of unicellular, aerobic nitrogen-fixing cyanobacteria. Cyanothece cells can create an anoxic intracellular environment at night, allowing oxygen-sensitive processes to take place in these oxygenic organisms. Large carbohydrate reserves accumulate in the cells during the day, ensuring sufficient energy for the processes that require the anoxic phase of the cells. Our study indicates that this genus maintains a plastic genome, incorporating new metabolic capabilities while simultaneously retaining archaic metabolic traits, a unique combination which provides the flexibility to adapt to various ecological and environmental conditions. Rearrangement of the nitrogenase cluster in Cyanothece sp. strain 7425 and the concomitant loss of its aerobic nitrogen-fixing ability suggest that a similar mechanism might have been at play in cyanobacterial strains that eventually lost their nitrogen-fixing ability.
IMPORTANCE
The unicellular cyanobacterial genus Cyanothece has significant roles in the nitrogen cycle in aquatic and terrestrial environments. Cyanothece sp. ATCC 51142 was extensively studied over the last decade and has emerged as an important model photosynthetic microbe for bioenergy production. To expand our understanding of the distinctive metabolic capabilities of this cyanobacterial group, we analyzed the genome sequences of five additional Cyanothece strains from different geographical habitats, exhibiting diverse morphological and physiological attributes. These strains exhibit high rates of N2 fixation and H2 production under aerobic conditions. They can generate copious amounts of carbohydrates that are stored in large starch-like granules and facilitate energy-intensive processes during the dark, anoxic phase of the cells. The genomes of some Cyanothece strains are quite unique in that there are linear elements in addition to a large circular chromosome. Our study provides novel insights into the metabolism of this class of unicellular nitrogen-fixing cyanobacteria.
Introduction
Cyanobacteria constitute a fascinating group of photosynthetic prokaryotes that have inhabited almost every sunlit ecosystem of the earth for ~3 billion years. The remarkable success of this group of microbes in adapting to a wide range of environmental and ecological conditions has largely been attributed to the presence of an extraordinarily flexible repertoire of metabolic pathways (1, 2). Cyanobacteria possessed an efficient cellular machinery to function in anaerobic environments that prevailed during the mid-/late Archaean era, and their metabolic activities are credited for the transitioning of the earth into the present-day oxygen-rich environment (3). In fact, many of the archaic metabolic traits have been retained in extant cyanobacterial species, enabling them to thrive in many diverse ecological niches.
The metabolic feats of cyanobacteria are exemplified by the ability of some strains to fix molecular nitrogen, a process sensitive to oxygen (4) and not found in any other known oxygenic organism (5, 6). Cyanobacteria have adapted various strategies to meet the cellular demands of nitrogen fixation, the most critical being the protection of the oxygen-sensitive nitrogenase enzyme (7). While some filamentous strains have developed specialized cells, called heterocysts, to accommodate this process, unicellular strains make use of the diurnal cycle to separate oxygen-evolving photosynthesis from oxygen-sensitive nitrogen fixation (8, 9). Recent studies have demonstrated the importance of unicellular nitrogen-fixing cyanobacteria in the marine nitrogen and carbon cycle (10, 11). The efficiency of nitrogen fixation exhibited by these microbes during the dark period of a day/night cycle suggests that they must have the ability to harvest and store sufficient solar energy during the day, which in turn fuels the energy-intensive nitrogen-fixing process at night. The suboxic intracellular conditions that are maintained during the nitrogen fixation period also facilitate various fermentative processes that are commonly observed in many facultative and obligate anaerobes (12, 13).
Cyanothece is a genus of morphologically diverse, unicellular cyanobacteria that are known to inhabit a variety of ecological niches. This genus was created by Komárek (14) to include unicellular cyanobacteria with distinct morphological, ultrastructural, and genomic features (15–17). Cyanothece strains from various ecotypes have been studied in the past for their robust circadian rhythm, fermentative capabilities, and other biotechnological applications (12, 18, 19). Cyanothece sp. ATCC 51142 [hereinafter referred to as Cyanothece 51142], a prototypic member of this genus, was isolated from the Texas Gulf Coast and is one of the most potent diazotrophic strains yet characterized (20). System-level studies with Cyanothece 51142 revealed many novel metabolic traits of this unicellular cyanobacterium which led to the determination of its genome sequence at the Washington University sequencing center (21). The studies revealed robust diurnal and circadian cycling of central metabolic processes in this strain, as well as a strong coordination of correlated processes at the transcriptional level (22). Interestingly, genome analysis of Cyanothece 51142 uncovered the presence of a 430-kb functional linear chromosomal element, the first such element to be identified in any photosynthetic bacterium. The arrangement of genes on this chromosome suggested a specific role for it in energy metabolism, and it was hypothesized that such linear elements with regulatory functions might be a distinctive trait of the genus Cyanothece (21). Also interesting from the genomic perspective is the finding that some atypical nitrogen-fixing strains, such as the endosymbiont spheroid body of the eukaryotic diatom Rhopalodia gibba and the unicellular marine cyanobacterium UCYN-A, which lacks photosystem II, have genomes closely related to those of Cyanothece spp. (23, 24). In particular, the nitrogenase gene clusters in both of these organisms is highly similar to that in Cyanothece 51142. It has been hypothesized that these organisms may have evolved as a result of targeted gene loss (loss of genes involved in photosynthesis while maintaining an elaborate gene cluster involved in nitrogen fixation) from a Cyanothece-like ancestor (23), thus suggesting a highly plastic nature of Cyanothece genomes as well as the robustness of their nitrogen-fixing machinery.
The most striking of the unique metabolic capabilities of Cyanothece 51142 is that cells can exhibit high rates of nitrogenase-mediated H2 production under aerobic conditions, an unusual metabolic trait in oxygenic phototrophs (25). Furthermore, the metabolic versatility of this strain was demonstrated by its ability to switch between photoautotrophic and photoheterotrophic modes of metabolism depending on the availability of external carbon sources and the presence of an atypical alternative citramalate pathway for isoleucine biosynthesis (26).
In an effort to unravel the genomic basis of the observed metabolic traits of unicellular diazotrophic cyanobacteria, the genomes of five additional members of the genus Cyanothece (Cyanothece sp. strains PCC 7424, PCC 7425, PCC 7822, PCC 8801, and PCC 8802 [hereinafter referred to as Cyanothece 7424, 7425, 7822, 8801, and 8802]) were sequenced at the Joint Genome Institute, U.S. Department of Energy. The strains were collected from different geographical locations and exhibit considerable diversity with respect to cell size and pigment composition. A comparison of the genomes of the different Cyanothece strains revealed that members of this genus are metabolically versatile, each member having acquired unique metabolic capabilities. The capability of aerobic nitrogen fixation and the presence of a large, contiguous nif gene cluster distinguish this group of unicellular photosynthetic microbes. Analysis of the genes common and unique to five of the six Cyanothece strains revealed that the core Cyanothece genomes is an amalgamation of genes from strains associated with fermentative capabilities, such as Microcystis and Microcoleus strains, and from aerobic nitrogen-fixing filamentous strains. The key to the success of this group of organisms appears to lie in their ability to retain such useful metabolic traits as nitrogen fixation and anaerobic fermentation while simultaneously adapting and accommodating advanced cellular features of contemporary photosynthetic organisms.
RESULTS
General features of the Cyanothece genomes.
Table 1 summarizes the general characteristics of the six sequenced Cyanothece genomes. Cyanothece 51142 is a marine (benthic) strain, whereas Cyanothece 7424, 7425, 7822, 8801, and 8802 were collected from different tropical and subtropical rice fields in Asia and Africa. The genome sizes of the six strains show considerable variation, ranging between ~4.8 and 7.8 Mbp. Cyanothece 7822 has the largest cell size (8 to 10 µm), as well as the largest genome, with ~6,600 protein-coding genes, whereas Cyanothece 8801 and 8802 have the smallest genomes, with ~4,400 open reading frames. The genome of each strain consists of a circular chromosome and several (3 to 6) smaller plasmids. The plasmids range in size from ~10 kb (smallest plasmid in Cyanothece 51142) to ~330 kb (largest plasmid in Cyanothece 7424).
TABLE 1 .
General characteristics of Cyanothece genomesa
| Characteristic of Cyanothece strains | ATCC 51142
|
PCC 7424
|
PCC 7425
|
PCC 7822
|
PCC 8801
|
PCC 8802
|
|---|---|---|---|---|---|---|
| Cell size (µm) | 4–5 | 7–8 | 3–4 | 8–10 | 4–5 | 4–5 |
| Site of isolation | Port Aransas, TX | Rice field, Senegal | Rice field, Senegal | Rice field, India | Rice field, Taiwan | Rice field, Taiwan |
| Size (Mbp) | 5.46 | 6.55 | 5.79 | 7.84 | 4.79 | 4.80 |
| No. of coding sequences | 5,304 | 5,710 | 5,327 | 6,642 | 4,367 | 4,444 |
| % G+C | 37.1 | 38.0 | 49.9 | 39.6 | 39 | 39 |
| No. of plasmids | 4 | 6 | 3 | 3 | 3 | 4 |
| No. of linear chromosomal elements | 1 | 0 | 0 | 3 | 0 | 0 |
| No. of pseudogenes | 6 | 177 | 131 | 341 | 200 | 206 |
Size bar, 2 μm.
Like Cyanothece 51142, Cyanothece 7822 also carries linear chromosomal elements in its genomes. The single linear chromosome in Cyanothece 51142 did not exhibit significant synteny with any sequenced cyanobacterial genome except the partially sequenced genome of Cyanothece sp. strain CCY 0110. Based on this observation, it was suggested that the linear chromosome might be specific to the genus Cyanothece (21). Interestingly, our analyses showed that Cyanothece 7822 has 3 linear chromosomal elements in its genome. The largest of these is an 880-kb element with 595 coding sequences, followed by a 474-kb fragment with 422 coding sequences. The third linear element is 14 kb long with 13 coding sequences. About 50% of the coding genes in the largest linear chromosome are unique to Cyanothece 7822, and 229 of these are without any paralogs elsewhere in the genome. A large fraction of these genes encode ABC transporters, with two operons containing genes involved in phosphate and molybdenum transport (see Table S1 in the supplemental material). In addition, this linear element has a significant number of genes involved in carbohydrate metabolism, including a cluster of genes encoding carbohydrate degradation and glycosylation proteins. Several genes encoding proteins with regulatory functions, transposons, and CRISP-R-associated proteins, as well as proteins involved in aromatic compound degradation, are also present in this chromosome. A cluster of cytochrome oxidase (cox) genes involved in respiration is common to the linear chromosomes of both Cyanothece 51142 and 7822. About 65% of the genes on the second-largest linear element are unique to Cyanothece 7822, with 52% of the genes unique to this chromosomal element.
The GC content of five of the sequenced Cyanothece genomes is close to 40 percent. Cyanothece 7425 is an exception, with a GC content of 50%. The terrestrial Cyanothece strains have a high percentage of pseudogenes in their genome (Table 1). Although Cyanothece 51142 has only 6 pseudogenes, Cyanothece 7822 contains 341, whereas in Cyanothece 8801 and 8802, the strains with the smallest genomes, the ~200 pseudogenes account for ~4% of the genome.
Phylogenetic analysis.
Phylogenetic analysis of 61 cyanobacterial genomes using 226 homolog protein groups (see Materials and Methods) revealed various novel aspects of the evolutionary history of the Cyanothece strains. Based on this analysis, five of the six Cyanothece strains (Cyanothece 51142, Cyanothece 8801, Cyanothece 8802, Cyanothece 7822, and Cyanothece 7424) branch into a single clade together with three other nitrogen-fixing unicellular cyanobacteria, Cyanothece CCY 0110, Crocosphaera watsonii WH 8501, and UCYN-A (Fig. 1). These eight strains are likely to have evolved from a common ancestor, from which the two nondiazotrophic Microcystis strains also seem to have branched. Two other nondiazotrophic strains, Synechococcus sp. strain 7002 and Synechocystis sp. strain 6803, form a distant branch within this unicellular nitrogen-fixing group (Cyanothece 51142 to Synechococcus sp. strain JA-2-3Ba). Striking in this analysis is the position of Cyanothece 7425, which appears to have evolved separately and is phylogenetically close to Acaryochloris marina MBIC 11017 (a chlorophyll-d-containing strain), compared to any other Cyanothece strain. Cyanothece 7425 branched off earlier than most other nitrogen fixers except for three anaerobic nitrogen-fixing Synechococcus strains (Synechococcus sp. strain 7335, Synechococcus sp. strain JA-3-3AB [2–13], and Synechococcus JA-2-3Ba).
FIG 1 .
Phylogenetic tree of cyanobacteria. The tree was generated from 226 homologous protein groups, coorthologous in all 61 of the analyzed strains. The diazotrophic strains are colored as follows: green, Cyanothece strains; red, other diazotrophic cyanobacterial strains. S. elongatus, Synechococcus elongatus; M. vaginatus, Microcoleus vaginatus; T. erythraeum, Trichodesmium erythraeum; A. maxima, Arthrospira maxima; L. majuscula, Lyngbya majuscula; G. violaceus, Gloeobacter violaceus.
One of the two main branches in the phylogenetic tree contains only non-nitrogen-fixing cyanobacteria (consisting of Synechococcus sp. strain 6301, Synechococcus sp. strain 7942, and all alpha-cyanobacteria). The other branch (from Nodularia spumigena CCY 9414 to Synechococcus sp. JA-2-3Ba), although consisting predominantly of nitrogen fixers, is interspersed with non-nitrogen-fixing strains. As suggested by earlier phylogenetic studies of diazotrophic cyanobacteria (21, 27), it is likely that some of the cyanobacteria in the second branch lost their nitrogen-fixing capability in the course of evolution. The position of the newly sequenced Cyanothece 7425 (with a functional nitrogenase cluster), which branched off from a common ancestor with A. marina (a non-nitrogen-fixing strain), strengthens this premise.
Shared and unique genes in Cyanothece genomes.
Based on NCBI protein BLAST analysis (see Materials and Methods for details), we identified 1,705 homologous gene groups that are shared by all of the six Cyanothece strains (see Table S2 in the supplemental material). Using the classification scheme in the CyanoBase database (28), 1,003 (59%) of these genes are associated with known functional categories. Genes related to nitrogen fixation, central carbon metabolism, photosynthesis, respiration, and most common amino acid biosynthetic pathways are included in this shared group of genes. When the protein sequences of these homologous genes were BLAST-aligned against all sequenced genomes (excluding those of three closely related strains; see Materials and Methods), >99.5% of the 1,705 groups had homologs in other cyanobacterial strains. These genes were distributed among several cyanobacterial strains (Table S2 in the supplemental material), with the highest number (more than 85%) of homologues in Microcystis sp. and filamentous nitrogen-fixing strains. In contrast, few of these genes (less than 50%) had a homolog in members of the alpha-cyanobacterial group.
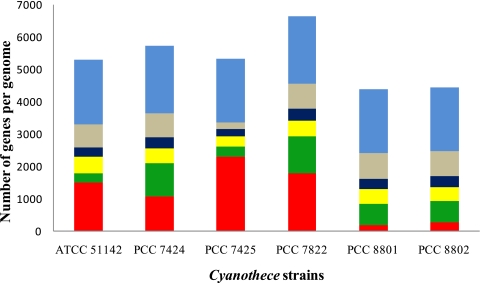
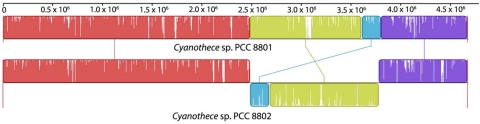
Cyanothece 7425 is the most disparate among the six strains, with more than 2,300 unique genes (Fig. 2) representing 43% of its protein coding genes. Cyanothece 8801 and 8802 have the same geographical origin and are more than 90% identical to each other at the genome level, with a single large region of inversion (Table 2; Fig. 3). Since most of the genes are shared between these two strains, they have very few unique genes (183 in Cyanothece 8801 and 263 in Cyanothece 8802). Sixty-eight percent of the genes in Cyanothece 8801 and 8802 have homologs in Cyanothece 51142, and their close phylogenetic association is also reflected in their proximity in the tree (Fig. 1). Also in accordance with their position in the tree, Cyanothece 7424 is closest to Cyanothece 7822, sharing more than 60% of the genes in their individual genomes. Although Cyanothece 7822 has the largest genome, only 27% of the genes are unique to this strain. Interestingly, even though Cyanothece 7424 and 7425 have a common ecological origin (Table 1), unlike Cyanothece 8801 and 8802, their genomes are very diverse.
FIG 2 .
Number of shared and unique genes in the genomes of the six Cyanothece strains. Each bar represents the total number of genes in a genome. Protein sequences of the individual Cyanothece strains were compared with each other in order to identify the distribution of orthologs among different strains (see Materials and Methods). The number of genes in each genome having orthologs in one or more of the remaining five strains was determined and grouped accordingly. These gene groups are colored as follows: light blue, genes shared by all six genomes; grey, shared by 5 of the 6 genomes; dark blue, shared by 4 genomes; yellow, shared by 3 genomes; green, shared by two genomes;: red, genes unique to each Cyanothece strain. Orthologous sequences for about 1,700 genes are found in all genomes. Cyanothece 7425 has the highest number of unique genes, whereas Cyanothece 8801 and 8802 have the least, with most genes being shared between the two.
TABLE 2 .
Ortholog comparison between pairs of Cyanothece genomes
| Cyanothece strain | No. (%) of orthologsa |
|||||
|---|---|---|---|---|---|---|
| ATCC 51142 | PCC 7424 | PCC 7425 | PCC 7822 | PCC 8801 | PCC 8802 | |
| ATCC 51142 | 4,864 | 2,791 (57) | 2,059 (42) | 2,739 (56) | 2,823 (58) | 2,838 (58) |
| PCC 7424 | 2,791 (54) | 5,163 | 2,273 (44) | 3,707 (60) | 2,711 (52) | 2,700 (52) |
| PCC 7425 | 2,059 (44) | 2,273 (48) | 4,729 | 2,275 (48) | 2,055 (43) | 2,064 (44) |
| PCC 7822 | 2,739 (47) | 3,707 (64) | 2,275 (39) | 5,787 | 2,743 (47) | 2,740 (47) |
| PCC 8801 | 2,823 (68) | 2,711 (66) | 2,055 (50) | 2,743 (66) | 4,132 | 3,844 (93) |
| PCC 8802 | 2,838 (68) | 2,700 (65) | 2,064 (49) | 2,740 (66) | 3,844 (92) | 4,181 |
Paralogs excluded.
FIG 3 .
Alignment of the genomes of Cyanothece 8801 and Cyanothece 8802. The two genomes were aligned using the Mauve software program (54), using its default parameters. A high level of sequence similarity is observed between the two strains, as shown by the colored portions of the aligned regions. However, the two genomes differ from each other due to a 1.3-Mbp-long inverted region spanning from 2.5 × 106 to 3.8 × 106 locations of the genomes.
A BLAST analysis of the individual protein sequences from each homologous group showed that the top hits for these sequences are spread among a number of cyanobacterial strains, indicating differences in the evolutionary pathways of the six Cyanothece strains. Interestingly, for all strains except Cyanothece 7425, the top hits for more than 70% of the group of 1,705 homologous genes were mostly from Microcystis aeruginosa, Microcoleus chthonoplastes, and Synechocystis 6803. In contrast, top BLAST hits for protein sequences of Cyanothece 7425 (67% of the homologous groups) were mostly from Acaryochloris marina, Thermosynechococcus elongatus, Microcoleus chthonoplastes, Oscillatoria sp. strain PCC 6506, and Nostoc punctiforme.
In contrast, BLAST results for the protein sequences unique to Cyanothece 51142, 7424, 7822, 8801, and 8802, against the entire sequence database, showed that more than 50% of them did not have any significant hits in any other organism. Furthermore, the top BLAST hits for the remaining unique proteins in these Cyanothece strains were spread among several organisms and could not be associated with any specific organism, as was seen with the shared genes. About 40% of the unique genes in Cyanothece 7425 did not show any significant hits with any other organism. More than 10% of the unique genes showed top hits in A. marina and N. punctiforme, with the remainder spread among several organisms. About 10% of the unique genes in the Cyanothece strains have homologs in several nonoxygenic bacteria, many of which are diazotrophic strains.
Due to the genomic diversity observed in Cyanothece 7425, its distant location in the phylogenetic tree compared to the other Cyanothece strains, and its proximity to the three anaerobic nitrogen-fixing Synechococcus strains, we assessed its relationship to all sequenced cyanobacterial strains. BLAST analysis of all protein sequences of Cyanothece 7425 against those of all other sequenced cyanobacteria revealed that 1,885 of its 5,327 genes had homologs in the other five Cyanothece strains. In contrast, only 1,335 genes are shared with the three Synechococcus strains. Considering all sequenced cyanobacterial strains, Cyanothece 7425 shares the highest number of genes with Nostoc sp. strain PCC 7120 (49%), N. punctiforme PCC 73102 (49%), Cyanothece 7822 (49%), Anabaena variabilis ATCC 29413 (48%), Cyanothece 7424, and A. marina MBIC 11017 (46%).
Metabolic traits of Cyanothece. (i) Nitrogen fixation and hydrogen production.
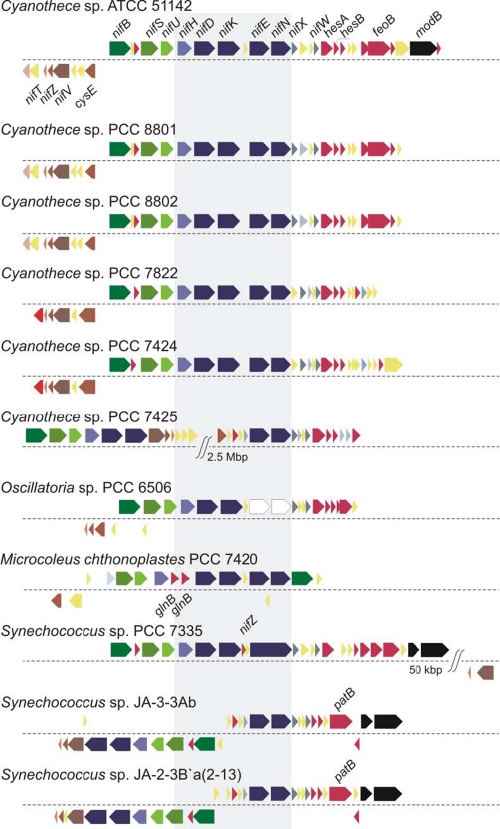
Our analysis revealed the presence of a large nitrogenase (nif) gene cluster in each of the sequenced Cyanothece genomes, thereby establishing this genus as a group of unicellular diazotrophic cyanobacteria. Comparison of the nif gene clusters in all sequenced diazotrophic cyanobacterial strains showed that the largest contiguous cluster is present in Cyanothece 51142, consisting of 35 genes arranged in two adjacent regulons (Fig. 4). The nif clusters in Cyanothece 8801 and 8802 are identical to each other and closely resemble the cluster in Cyanothece 51142, with only three missing genes: the molybdate ABC transporter permease protein gene modB, the hypothetical gene between nifK and nifE, and the hypothetical gene between the ferrous iron transport protein gene feoA2 and modB. The synteny of this nif cluster is also largely maintained in Cyanothece 7424 and 7822, although it is somewhat shortened by gene losses in these two strains. In contrast, the nitrogenase cluster in Cyanothece 7425 is interrupted by a 2.5-Mbp insertion in the middle of the cluster, separating nifHDK from nifE. Also, in contrast to the other five Cyanothece strains, which possess the hup genes, encoding an uptake hydrogenase, an enzyme associated with nitrogenase activity and nitrogenase-mediated hydrogen production, the Cyanothece 7425 genome does not have genes for this enzyme.
FIG 4 .
Alignment of clusters of nitrogen fixation-related genes in the six Cyanothece strains and in five other sequenced anaerobic nitrogen-fixing strains. The cluster in Cyanothece 7425 is significantly different from the clusters in the other Cyanothece strains, with a 2.5-Mbp insertion between nifK and nifN. The nifVZT regulon also shows an inversion similar to sequences in two other Synechococcus strains (Synechococcus JA-3-3Ab and Synechococcus JA-2-3B). In Oscillatoria sp. 6506, the nifE and nifN genes are pseudogenes, shown in white. Microcoleus has a shorter cluster with nifB translocated next to nifN. Synechococcus 7335 has a 50-kbp insertion between nifV and nifB and has nifZ translocated between nifK and nifE.
We assessed the abilities of the six Cyanothece strains to fix nitrogen and produce hydrogen. Cyanothece 51142 showed the highest nitrogenase activity, as well as the highest rates of hydrogen production, followed by Cyanothece 8802 and 8801 (Table 3). All Cyanothece strains except Cyanothece 7425 exhibited nitrogenase activity and hydrogen production capacity under aerobic incubation conditions (incubation with air in the headspace). In contrast, Cyanothece 7425 exhibited nitrogenase activity and hydrogen production ability only when an anaerobic environment was provided (incubation with argon in the headspace).
TABLE 3 .
Nitrogenase activity and hydrogen production in the six Cyanothece strainsa
| Strain | Nitrogenase activity (C2H4 production [µmol/mg Chl ⋅ h]) |
Hydrogen production (µmol/mg Chl ⋅ h) |
||
|---|---|---|---|---|
| Aerobic | Anaerobic | Aerobic | Anaerobic | |
| Cyanothece ATCC 51142 | 148.03 ± 19.06 | 202.33 ± 23.41 | 132.04 ± 33.03 | 308.62 ± 42.01 |
| Cyanothece PCC 7424 | 40.63 ± 9.53 | 160.53 ± 28.6 | 59.64 ± 17.32 | 201.77 ± 35.6 |
| Cyanothece PCC 7425 | 0 | 40.35 ± 10.76 | 0 | 54.20 ± 12.04 |
| Cyanothece PCC 7822 | 52.49 ± 11.07 | 112.32 ± 18.28 | 47.83 ± 9.8 | 133.6 ± 30.13 |
| Cyanothece PCC 8801 | 101.22 ± 11.65 | 187.65 ± 43.12 | 51.64 ± 9.34 | 186.2 ± 40.23 |
| Cyanothece PCC 8802 | 98.5 ± 18.12 | 192.12 ± 36.78 | 43.87 ± 6.24 | 176.17 ± 38.78 |
Chl, chlorophyll.
A comparison of the nitrogenase cluster of all the sequenced nitrogen-fixing cyanobacterial strains revealed an unusual arrangement of the nifVZT regulon in Cyanothece 7425, closely resembling the cluster in two other anaerobic nitrogen-fixing strains, Synechococcus JA-2-3Ba and Synechococcus JA-3-3Ab, which exhibit an inversion between nifV and nifE. While the organization of this cluster is known to be largely conserved among all nonheterocystous nitrogen-fixing strains (21), an alteration in the arrangement of the regulons or an inversion/ insertion in the region was also observed in other anaerobic nitrogen-fixing cyanobacterial strains investigated in this study (Synechococcus 7335, Oscillatoria PCC 6506, and M. chthonoplastes) (Fig. 4). Interestingly, as with Cyanothece 7425, the genomes of these other anaerobic nitrogen-fixing strains also do not possess any gene for an uptake hydrogenase.
(ii) Other metabolic characteristics of the genus Cyanothece. (a) Photosynthesis.
As expected from their ability for oxygenic photosynthesis, the genomes of all the sequenced Cyanothece strains contain most of the genes encoding the core cyanobacterial proteins related to photosynthesis (29). However, a BLAST analysis of the Cyanothece genomes with all the annotated genes in the KEGG database showed that certain low-molecular-mass proteins associated with PSI and PSII are missing in some of the Cyanothece strains. While all six genomes encode genes for biosynthesis of the light-harvesting pigment phycocyanin, Cyanothece 7424, 7822, and 8801 also have genes encoding phycoerythrin, a pigment which imparts a brownish-green color to these strains. The core cyanobacterial genes encoding chlorophyll biosynthesis enzymes, Calvin cycle enzymes, and regulatory proteins (29) are present in all six Cyanothece genomes. Interestingly, Cyanothece 7424, Cyanothece 7425, and Cyanothece 7822 have two very similar copies of the psaB gene, a trait shared by Synechococcus 7335 and Nostoc azollae 0708 among the 61 sequenced cyanobacteria. Cyanothece 7822 was the only strain to have the second psaB gene contiguous to the psaAB operon.
(b) Carbon metabolism.
Members of the genus Cyanothece have been documented to synthesize and store large amounts of carbohydrates in the form of glycogen granules (30) when grown under light/dark cycles. BLAST analysis of the Cyanothece proteins against the KEGG database showed that several genes involved in glycogen synthesis, degradation, and metabolism are present in the core group of Cyanothece genes (see Table S2 in the supplemental material). A cluster of four genes involved in the metabolism of polyhydroxyalkanoic acid is present in Cyanothece 7424, 7822, and 7425 (Cyan7424_0494-0497, Cyan7822_1326-1330, and Cyan7425_4054-4057) and in M. aeruginosa. In addition, the Cyanothece genomes encode several genes involved in the synthesis and utilization of diverse sugar molecules. All the Cyanothece strains have genes for cellulose synthesis and metabolism. Cyanothece 7424 and 7425 encode genes for sucrose synthase, whereas Cyanothece 51142, 7424, 7822, 8801, and 8802 have genes for trehalose metabolism.
All of the sequenced Cyanothece strains have genes encoding enzymes for complete glycolytic and pentose phosphate pathways and an incomplete tricarboxylic acid (TCA) cycle. Noteworthy is the presence of a gene encoding phosphoenolpyruvate carboxykinase in five of the six sequenced Cyanothece strains. This enzyme, involved in the gluconeogenic conversion of oxaloacetate to phosphoenolpyruvate, is not very common among other cyanobacteria, occurring only in Arthrospira, Microcystis, and Microcoleus. Our analysis revealed two unusual genes in Cyanothece 7424 and 7822, encoding isocitrate lyase (PCC7424_4054 and Cyan7822_2461) and malate synthase (PCC7424_4055 and Cyan7822_2460), enzymes that are involved in the glyoxylate shunt of the TCA cycle. No other sequenced cyanobacterial genome has genes for these two enzymes.
(c) Nitrogen metabolism.
The six Cyanothece strains exhibit diversity in various nitrogen metabolism pathways. Cyanophycin, a nitrogen reserve molecule in cyanobacteria, is a polymer of arginine and asparagine. Catabolism of l-arginine can serve as a source of nitrogen, carbon, and energy for the cells (31). Differences are observed in this catabolic pathway, suggesting that the pathway fulfills diverse roles in these Cyanothece strains. Although all six strains have an arginine decarboxylase that catalyzes the conversion of arginine to agmatine, the fate of agmatine appears to differ. Like most cyanobacterial strains, Cyanothece 7822 and 7424 possess an agmatinase enzyme that converts agmatine into putrescine and urea. The genomes of these two strains have an operon of seven genes (the largest among all sequenced cyanobacteria) encoding urease and its accessory proteins (Cyan7424_4411-4417 and Cyan7822_2693-2699), as well as genes involved in the conversion of putrescine into spermine and spermidine. Cyanothece 7425, 8801, and 8802, in contrast, convert agmatine to putrescine via an agmatine deaminase and N-carbamoylputrescine amidase. Cyanothece 8801 and 8802 can further process putrescine into spermidine and spermine. The Cyanothece 51142 genome does not have genes that can convert agmatine to putrescine, suggesting that agmatine is the preferred polyamine for this strain. It also does not contain any gene for urea metabolism. Carbamate kinase, an unusual cyanobacterial enzyme involved in the production of ATP from ADP and carbamoyl phosphate in the final step of the fermentative degradation of arginine (32), is present in Cyanothece 7822, 8801, and 8802 and in Synechocystis 6803.
Other interesting differences in amino acid metabolism pathways include the presence of the kynurenine pathway of tryptophan degradation in Cyanothece 7822, 7425, 8801, and 8802 and the methionine salvage pathway in Cyanothece 7424, 7822, 8801, and 8802.
(d) Anaerobic metabolic capabilities.
Our earlier studies have shown that Cyanothece 51142 exhibits high levels of anaerobic metabolism capacity (22, 25). In fact, all of the Cyanothece strains show several biochemical pathways associated with anaerobic metabolism. Cyanothece 51142, 7424, and 7822 have genes for fermentative lactate production, and both Cyanothece 7424 and 7822 perform mixed acid fermentation with formate as the end product (13). Cyanothece 7822 has been shown to have a capacity for mixed acid fermentation (12), a pathway also observed in the genus Microcystis. Pathways for ethanol, acetate, and hydrogen production are found in most of the Cyanothece strains. Also, an anaerobic chlorophyll biosynthesis pathway involving protoporphyrin IX cyclase (BchE) is present in Cyanothece 7425 and 7822. This gene has a homolog in the filamentous cyanobacterial strain Cylindrospermopsis raciborskii and in noncyanobacterial strains like Heliobacillus mobilis and Rhodopseudomonas palustris. While a gene for BchE has been identified in Synechocystis 6803 (29), this gene has little sequence similarity with the Cyanothece gene. Many of the genes in the five Cyanothece strains (except Cyanothece 7425) had top hits to M. aeruginosa (>700 genes) and M. chthonoplastes (>260 genes), both of which are associated with anaerobic environments and have been extensively studied for fermentative processes. Furthermore, a significant number of unique genes found in each of the Cyanothece strains have homologs in either facultative or obligate anaerobic bacteria.
(e) Other novel aspects of Cyanothece metabolism.
Cyanothece strains have acquired or retained diverse metabolic traits that make them interesting model organisms for studying various biological processes. For example, Cyanothece 8801 and 8802 differ from the other Cyanothece strains and most other cyanobacterial strains in possessing genes that encode a V-type ATPase (a six-gene operon, Cyan8801_3221-3226 and Cyan8802_2894-2899). This operon is also present in Cyanobium species and Synechococcus sp. strain WH 5701. It is also important to note that Cyanothece 8801 and 8802 have a number of genes encoding proteins involved in phosphonate metabolism. These include a three-gene operon encoding phosphonate transporters. Part of this operon is an amidohydrolase gene that is involved in phosphonate metabolism. In addition, the C–P lyase system involved in phosphonate metabolism is also present in these Cyanothece strains. In some ecosystems, phosphonates comprise a significant proportion of the available phosphorous (33), and consequently some strains might have evolved the capability to metabolize them.
Cytochrome P450s in cyanobacteria have been implicated in several metabolic processes involved in natural product synthesis, and members of the Cyanothece genus have been shown to be particularly enriched for some of these heme oxygenases (34). Our analysis revealed several unique cytochrome P450s in the Cyanothece strains, some with homologs in A. marina. In particular, the Cyanothece 8801 and 8802 genomes have several genes (Cyan8801_2436, Cyan8801_1896, Cyan8802_3674, and Cyan8802_1920) encoding these proteins, and interestingly, these strains also have large operons encoding nonribosomal peptide synthetase modules and related proteins (Cyan8801_3021-3032 and Cyan8802_3090-3101).
Most of the Cyanothece strains (except Cyanothece 7424 and 7822) also possess an alkane biosynthetic pathway involving aldehyde decarbonylase and an acyl-ACP reductase (cce_0778 and cce_1430, Cyan7425_0398 and Cyan7425_0399, PCC8801_0455, and PCC8801_0872, and Cyan8802_0468 and Cyan8802_0898) (35). Pathways involved in the nonfermentative synthesis of higher alcohols have also been identified in all the Cyanothece strains.
(iii) Characteristics of unicellular nitrogen-fixing strains.
In order to identify the genes that might influence the metabolism of unicellular nitrogen-fixing cyanobacteria, we BLAST-aligned the group of 1,705 homologous genes common to all Cyanothece strains against the entire sequenced cyanobacterial genome database. Among these, 59 genes were identified that are common to all unicellular and filamentous N2-fixing strains but are lacking in most (<6 out of 39 strains) non-N2-fixing strains. These included most of the core N2 fixation-related genes from the nif cluster, several genes encoding regulatory proteins, and genes involved in energy metabolism. In addition, several hypothetical and conserved hypothetical proteins (see Table S2 in the supplemental material) are found to be restricted to the N2-fixing group. Among this group of 1,705 homologous genes, 51 genes were found to be present exclusively in the unicellular N2-fixing strains. Most of these gene products are hypothetical or conserved hypothetical proteins with domains implicated in regulatory functions. Some transporters and regulatory proteins are also found to be present only in the unicellular strains.
Earlier studies have shown that many of the genes are differentially regulated under diazotrophic growth conditions in Cyanothece 51142. We BLAST-aligned the genes known to be diurnally regulated in Cyanothece 51142 (22, 36, 37) against all sequenced cyanobacterial genomes. Our results show that the core nitrogen fixation genes (~15 genes) are present in both unicellular and filamentous nitrogen-fixing strains (see Table S3). Another ~50 of these diurnally regulated genes are mostly restricted to the unicellular N2-fixing strains. These include genes encoding proteins with regulatory functions (transcriptional and translational regulators and two component system proteins), transporters, signaling proteins, peroxiredoxins, and peroxidases and several hypothetical and conserved hypothetical proteins.
DISCUSSION
Our analyses of the six completely sequenced Cyanothece genomes revealed that many key metabolic features were conserved during evolution, while considerable diversity was also gained (Fig. 2). The metabolic traits common to the six Cyanothece strains are shared by many other cyanobacteria, suggesting that they must have been acquired from an ancient ancestor and retained in the extant strains. The plasticity of the Cyanothece genomes is evident from the fact that the strains have acquired many novel metabolic capabilities, which is reflected in their diverse genotypes and phenotypes (such as cell size, shape, and pigment composition). Two of the Cyanothece strains possess linear chromosomal elements, a feature not observed in any other photosynthetic bacteria studied to date. These chromosomal elements seem to accommodate specific adaptive features that might impart niche-specific advantages to the strains, as is suggested by the presence of a large number of genes encoding transposons and CRISP-R-associated proteins.
The significant difference observed in the numbers of predicted coding sequences between the Cyanothece strains suggests a substantial amount of loss or gain of genetic material over evolutionary time. Cyanothece 8801 and 8802 possess the smallest genomes and have a high percentage of pseudogenes, indicating that they might be undergoing a reductive genome evolution. A high percentage of pseudogenes is also observed in the genome of N. azollae, a strain that has undergone significant gene loss to adapt to a symbiotic lifestyle (38). Despite their small genomes, Cyanothece 8801 and 8802 possess many novel genes that are missing in the other Cyanothece strains and in most sequenced cyanobacteria, suggesting that they must have been acquired in response to some selective pressure. An outstanding example is the presence of the V-type ATPases, involved in numerous energy transduction pathways and known to be indispensable for plant growth, especially under different stress conditions (39, 40). Another plant-like feature in Cyanothece is the presence of a two-gene operon in Cyanothece 7424 and 7822 encoding enzymes involved in the glyoxylate cycle. In plants this cycle is implicated in the conversion of storage lipids into carbohydrates (41). This cycle is also known to impart metabolic versatility to some bacterial strains. However, no other cyanobacterial strain sequenced to date is known to possess the glyoxylate cycle.
Our phylogenetic analysis showed that Cyanothece 7425 separated from the other Cyanothece strains at an early stage of evolution. Cyanothece 7425 cells are smaller than those of the other Cyanothece strains and are more cylindrical. A GC content of ~40% is characteristic of the genus Cyanothece, and Cyanothece 7425 is an anomaly in this regard as well, with a GC content of ~50%. In contrast to the other five Cyanothece strains, Cyanothece 7425 fixes nitrogen only under anaerobic conditions and appears to share a common ancestor with three other anaerobic nitrogen-fixing Synechococcus strains. In contrast to the nif gene cluster in the five Cyanothece strains, the cluster in Cyanothece 7425 is disrupted by the insertion of a large fragment and exhibits inversions similar to those of the clusters in the Synechococcus strains. However, our protein BLAST analysis did not show significant homology of the Cyanothece 7425 genes with those of any Synechococcus strain. Also, unlike the other Cyanothece strains, very few of the Cyanothece 7425 genes had homologs in Microcystis and Microcoleus. These results indicate that Cyanothece sp. PCC 7425 represents a cyanobacterial strain that is losing its nitrogen-fixing ability and evolving independently of the other Cyanothece strains.
Another interesting observation in this study is the absence of an uptake hydrogenase in all the sequenced anaerobic nitrogen-fixing cyanobacteria, which suggests that this enzyme must be associated with aerobic nitrogen fixation in nonheterocystous cyanobacterial strains. Raphidiopsis brookii, a strain that has lost the ability to fix nitrogen and has eliminated most of the nitrogen fixation related genes (42), also does not have genes encoding the uptake hydrogenase. Cyanothece 7425 is phylogenetically closest to A. marina and shares a common ancestor with this strain, indicating that the latter lost its nitrogen-fixing ability in the course of evolution. Similarly, T. elongatus, a unicellular nitrogen-fixing strain, located between two anaerobic nitrogen fixers, appears to have lost its nitrogen-fixing ability. These evolutionary trends suggest that strains that have not adapted for functioning under aerobic conditions may not succeed in a predominantly oxygen-rich environment and therefore lose this ability with the simultaneous elimination of the nitrogenase cluster. Therefore, the nitrogenase cluster of Cyanothece, which appears to have evolved to function efficiently under ambient conditions, is evolutionarily selected for, as is seen in strains like UCYNA and the endosymbiont of R. gibba.
Cyanothece cells are unique in their ability to provide a platform for both aerobic and anaerobic metabolic processes at alternate phases of the diurnal cycle. While the unicellular Microcystis cells also have the capability to create an anoxic intracellular environment, they do not have genes required for nitrogen fixation. Five Cyanothece strains exhibit high rates of nitrogenase-mediated hydrogen production under aerobic conditions, indicating that an anaerobic intracellular environment is created to protect the oxygen-sensitive nitrogenase enzyme. The Cyanothece genomes also contain many genes for catalases and peroxidases, enzymes which protect oxygen-sensitive cellular constituents required for anaerobic metabolism. A large, contiguous nif gene cluster and the ability to perform aerobic nitrogen fixation distinguish the unicellular Cyanothece cells from all other cyanobacteria. The presence of versatile metabolic pathways, such as nitrogen fixation and oxygenic photosynthesis, and the ability to generate anoxic cellular environments under diazotrophic growth conditions make members of the genus Cyanothece attractive model systems for studying various sunlight-driven biofuel-yielding pathways which entail microaerobic conditions.
MATERIALS AND METHODS
Genome annotation.
The Cyanothece 51142 genome was annotated at Washington University in St. Louis, MO (21), whereas the genomes of the other five Cyanothece strains were annotated at the Joint Genome Institute, U.S. Department of Energy. In these five strains, genes were identified using the Prodigal software program (43) as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline (44). The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database and the UniProt, TIGRFam, Pfam, PRIAM, KEGG, COG, and InterPro databases. These data sources were combined to assert a product description for each predicted protein. Noncoding genes and miscellaneous features were predicted using tRNAscan-SE (45), RNAMMer (46), Rfam (47), TMHMM (48), and signalP (49).
Intergenome BLAST analysis.
Sequences of protein-coding genes of six completed Cyanothece genomes (Cyanothece 51142, 7424, 7425, 7822, 8801, and 8802) were downloaded from NCBI (http://www.ncbi.nlm.nih.gov/) (as of 14 October 2010). Homolog genes between different strains were identified using NCBI protein-protein BLAST analysis (BLASTP 2.2.22 [50]). Two genes are defined to be homologs to each other if their reciprocal BLAST hits resulted in the following: (i) an E value of <1E−4, (ii) a ratio between length of the BLAST hit region and length of the complete protein >2/3, and (iii) a ratio between the raw score for two-protein BLAST and the raw score for “self-self” BLAST >1/3.5. All BLAST runs were conducted with the additional parameters “-num_descriptions 99999 -num_alignments 999999 -comp_based_stats F -seg No” in order to ensure all relevant alignments are analyzed.
Based on these analyses, all genes in the six Cyanothece strains could be associated with 11,607 homolog groups. Among them, 1,705 homolog gene groups are shared by all six strains and are defined as the core genome (see Table S2 in the supplemental material). In addition, unique genes in individual strains were also identified.
In order to identify the evolutionary history of the Cyanothece family, these common and unique genes were BLAST-aligned against the NCBI nonredundant protein database. These BLAST runs excluded three additional cyanobacterial strains, namely, Crocosphaera 8501, Cyanothece CCY 0110, and the uncultured cyanobacterium UCYN-A. Among these, the Crocosphaera 8501 and Cyanothece CCY 0110 genomes are incomplete. Further, the draft versions of genomes of the two strains reveal that 64% (3,799/5,958) and 69% (2,009/6,475), respectively, of the probable protein-coding genes in these strains share homologs with one or more of the other Cyanothece strains.
Phylogenetic tree construction.
Orthologous sets of proteins were identified across 61 cyanobacterial strains through an all-versus-all BLASTP v2.2.23 (50, 51) analysis of their respective proteomes. Orthology was defined as reciprocal best-match hits between proteomes, matching 66% of the length of the longer of the two proteins, with scores 1/10 of the higher of the self-self scores. Any highest-scoring protein with multiple identically scoring hits was discarded. Sets of orthologs were considered to be conserved if ≥ 75% of proteins within the set were orthologous to one another, resulting in 226 sets of genes with orthologs in all 61 proteomes. Each of the 226 sets of 61 proteins was individually aligned using the MAFFT v6.811b software program (52) and then concatenated into a single alignment, removing all columns containing gaps. The PHYLIP v3.64 (5) software package was used to generate the final consensus tree using the Fitch-Margoliash method with 100 bootstraps, global rearrangement, and 1 jumble per bootstrap. Distances were then back fit to the resulting consensus tree using maximum-likelihood estimates from the original concatenated alignment. The resultant tree was rendered using the Archaeopteryx v0.957b software program (53).
Hydrogen production and nitrogenase activity measurement.
Hydrogen production and nitrogenase activity were measured following the protocol published in the work of Bandyopadhyay et al. (25).
KEGG pathway mapping.
In order to identify genes that may be involved in different metabolic reactions, individual protein-coding genes were BLAST-aligned against the KEGG pathway database (http://www.genome.jp/kegg/pathway.html). For each reaction, protein sequences of all currently annotated genes from different organisms were BLAST-aligned against the Cyanothece genomes. Following the same criteria utilized to identify the homolog genes in intergenome BLAST analysis, genes were assigned to relevant KEGG reactions if they were homolog to any of the currently annotated genes in the KEGG.
Genome alignments.
Whole-genome alignments were performed using “ProgressiveMauve” (54) with default parameter values.
SUPPLEMENTAL MATERIAL
Genes present in the two largest linear chromosomes of Cyanothece 7822. For many genes in these two chromosomes, no homologous sequences were found in the remaining five Cyanothece strains. Further, many of these genes did not have any paralog sequences in its main chromosome. Some of the functional groups overrepresented in the linear chromosome are highlighted.
Homolog gene groups (1,705) common to six Cyanothece strains. Based on NCBI protein BLAST (See Materials and Methods for more details), 1,705 homologous gene groups shared by six Cyanothece strains (Cyanothece 51142, Cyanothece 7424, Cyanothece 7425, Cyanothece 7822, Cyanothece 8801, and Cyanothece 8802) were identified. Gene identifiers and their annotations corresponding to each homologous group are provided. Representative protein sequences selected from Cyanothece 51142 were BLAST-aligned against all currently sequenced cyanobacterial strains to identify the presence/absence of homologous genes. Genes specific to N2-fixing strains (identified as those genes with homologs in at least 8 unicellular or filamentous N2-fixing strains but at most 6 non-N2-fixing strains) were further classified as those commonly found in both unicellular and filamentous strains (≥8 in unicellular strains and ≥8 in filamentous strains; blue) and those specific to unicellular strains (≥8 in unicellular strains and ≤3 in filamentous strains; green). Remaining genes specific to N2-fixing strains are in red.
Genes specific to N2-fixing organisms, selected using diurnally regulated genes in Cyanothece 51142. Protein sequences of diurnally regulated genes in Cyanothece 51142 (22, 37) were blasted against 61 cyanobacterial genomes to identify the presence of homologous genes. Genes specific to N2-fixing strains were defined as those genes, for which, homologous sequences are present in at least 8 unicellular or filamentous nitrogen-fixing strains and in at most six non-nitrogen-fixing strains. We also identified genes specific to unicellular N2-fixing strains (highlighted in light green) and genes commonly found in both unicellular and filamentous N2-fixing strains (highlighted in light blue).
ACKNOWLEDGMENTS
This work was supported by funding from DOE-BER (DE-FC02-07ER64694). The work conducted by the U.S. Department of Energy Joint Genome Institute was supported by the Office of Science of the U.S. Department of Energy under contract no. DE-AC02-05CH11231.
We acknowledge the excellent work of the following Joint Genome Institute personnel: from the Los Alamos National Lab, Olga Chertkov, David Sims, David Bruce, Chris Detter, Elizabeth Saunders (deceased), Roxanne Tapia, and Shunsheng Han; from JGI-Walnut Creek, James Han, Tanja Woyke, Sam Pitluck, Len Pennacchio, and Matt Nolan; and annotation authors Natalia Ivanova, Miriam Land (Oak Ridge National Lab), Natalia Mikhailova, Galina Ovchinikova, and Amrita Pati.
Footnotes
Citation Bandyopadhyay A, et al. 2011. Novel metabolic attributes of the genus Cyanothece, comprising a group of unicellular nitrogen-fixing cyanobacteria. mBio 2(5):e00214-11. doi:10.1128/mBio.00214-11.
REFERENCES
- 1. Vermaas WFJ. 2001. Photosynthesis and respiration in cyanobacteria, p 245–251 In Encyclopedia of Life Sciences. Nature Publishing Group, London, United Kingdom: http://dx.doi.org/10.1038/npg.els.0001670 [Google Scholar]
- 2. Stal LJ. 1995. Physiological ecology of cyanobacteria in microbial mats and other communities. New Phytol. 131:1–32 [DOI] [PubMed] [Google Scholar]
- 3. Kasting JF. 2001. Earth history. The rise of atmospheric oxygen. Science 293:819–820 [DOI] [PubMed] [Google Scholar]
- 4. Gallon JR. 1981. The oxygen sensitivity of nitrogenase—a problem for biochemists and microorganisms. Trends Biochem. Sci. 6:19–23 [Google Scholar]
- 5. Berman-Frank I, et al. 2001. Segregation of nitrogen fixation and oxygenic photosynthesis in the marine cyanobacterium Trichodesmium. Science 294:1534–1537 [DOI] [PubMed] [Google Scholar]
- 6. Berman-Frank I, Lundgren P, Falkowski P. 2003. Nitrogen fixation and photosynthetic oxygen evolution in cyanobacteria. Res. Microbiol. 154:157–164 [DOI] [PubMed] [Google Scholar]
- 7. Raymond J, Siefert JL, Staples CR, Blankenship RE. 2004. The natural history of nitrogen fixation. Mol. Biol. Evol. 3:541–554 [DOI] [PubMed] [Google Scholar]
- 8. Fay P. 1992. Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol. Rev. 56:340–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Falcón LI, Cipriano F, Chistoserdov AY, Carpenter EJ. 2002. Diversity of diazotrophic unicellular cyanobacteria in the tropical North Atlantic Ocean. Appl. Environ. Microbiol. 68:5760–5764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Montoya JP, et al. 2004. High rates of N2 fixation by unicellular diazotrophs in the oligotrophic Pacific Ocean. Nature 430:1027–1032 [DOI] [PubMed] [Google Scholar]
- 11. Moisander PH, et al. 2010. Unicellular cyanobacterial distributions broaden the oceanic N2 fixation domain. Science 327:1512–1514 [DOI] [PubMed] [Google Scholar]
- 12. van der Oost J, Bulthuis BA, Feitz S, Krab K, Kraayenhof R. 1989. Fermentation metabolism of the unicellular cyanobacterium Cyanothece PCC 7822. Arch. Microbiol. 152:415–419 [Google Scholar]
- 13. Stal LJ, Moezelaar R. 1997. Fermentation in cyanobacteria. FEMS Microbiol. Rev. 21:179–211 [Google Scholar]
- 14. Komárek J. 1976. Taxonomic review of the genera Synechocystis Sauv. 1892, Synechococcus Näg. 1849, and Cyanothece gen. nov. (Cyanophyceae). Arch. Protistenkd. 118:119–179 [Google Scholar]
- 15. Reddy KJ, Haskell JB, Sherman DM, Sherman LA. 1993. Unicellular, aerobic nitrogen-fixing cyanobacteria of the genus Cyanothece. J. Bacteriol. 175:1284–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Komárek J, Cepák V. 1998. Cytomorphological characters supporting the taxonomic validity of Cyanothece (Cyanoprokaryota). Plant Syst. Evol. 210:25–39 [Google Scholar]
- 17. Porta D, Rippka R, Hernández-Mariné M. 2000. Unusual ultrastructural features in three strains of Cyanothece (cyanobacteria). Arch. Microbiol. 173:154–163 [DOI] [PubMed] [Google Scholar]
- 18. Huang T, Tu J, Chow T, Chen T. 1989. Circadian rhythm of the prokaryote Synechococcus sp. RF-1. Plant Physiol. 92:531–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Margheri MC, Bosco M, Giovannetti L, Ventura S. 1999. Assessment of the genetic diversity of halotolerant coccoid cyanobacteria using amplified 16S rDNA restriction analysis. FEMS Microbiol. Lett. 173:9–16 [Google Scholar]
- 20. Sherman LS, Min H, Toepel J, Pakrasi HB. 2010. Better living through Cyanothece—unicellular diazotrophic cyanobacteria with highly versatile metabolic systems. Adv. Exp. Med. Biol. 675:275–290 [DOI] [PubMed] [Google Scholar]
- 21. Welsh EA, et al. 2008. The genome of Cyanothece 51142, a unicellular diazotrophic cyanobacterium important in the marine nitrogen cycle. Proc. Natl. Acad. Sci. U. S. A. 105:15094–15099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stöckel J, et al. 2008. Global transcriptomic analysis of Cyanothece 51142 reveals robust diurnal oscillation of central metabolic processes. Proc. Natl. Acad. Sci. U. S. A. 105:6156–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kneip C, Voss C, Lockhart PJ, Maier UG. 2008. The cyanobacterial endosymbiont of the unicellular algae Rhopalodia gibba shows reductive genome evolution. BMC Evol. Biol. 8:30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zehr JP, et al. 2008. Globally distributed uncultivated oceanic N2-fixing cyanobacteria lack oxygenic photosystem II. Science 322:1110–1112 [DOI] [PubMed] [Google Scholar]
- 25. Bandyopadhyay A, Stockel J, Min H, Sherman LA, Pakrasi HB. 2010. High rates of photobiological H2 production by a cyanobacterium under aerobic conditions. Nat. Commun. 1:139. [DOI] [PubMed] [Google Scholar]
- 26. Feng X, et al. 2010. Mixotrophic and photoheterotrophic metabolism in Cyanothece sp. ATCC 51142 under continuous light. Microbiology 156:2566–2574 [DOI] [PubMed] [Google Scholar]
- 27. Swingley WD, Blankenship RE, Raymond J. 2008. Integrating Markov clustering and molecular phylogenetics to reconstruct the cyanobacterial species tree from conserved protein families. Mol. Biol. Evol. 25:643–654 [DOI] [PubMed] [Google Scholar]
- 28. Nakamura Y, Kaneko T, Hirosawa M, Miyajima N, Tabata S. 1998. CyanoBase, a www database containing the complete nucleotide sequence of the genome of Synechocystis sp. strain PCC6803. Nucleic Acids Res. 26:63–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mulkidjanian AY, et al. 2006. The cyanobacterial genome core and the origin of photosynthesis. Proc. Natl. Acad. Sci. U. S. A. 103:13126–13131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schneegurt MA, Sherman DM, Nayar S, Sherman LA. 1994. Oscillating behavior of carbohydrate granule formation and dinitrogen fixation in the cyanobacterium Cyanothece sp. strain ATCC 51142. J. Bacteriol. 176:1586–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wegener KM, et al. 2010. Global proteomics reveal an atypical strategy for carbon/nitrogen assimilation by a cyanobacterium under diverse environmental perturbations. Mol. Cell. Proteomics 9:2678–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ramón-Maiques S, et al. 2010. Substrate binding and catalysis in carbamate kinase ascertained by crystallographic and site-directed mutagenesis studies: movements and significance of a unique globular subdomain of this key enzyme for fermentative ATP production in bacteria. J. Mol. Biol. 397:1261–1275 [DOI] [PubMed] [Google Scholar]
- 33. White AK, Metcalf WW. 2004. Two C–P lyase operons in Pseudomonas stutzeri and their roles in the oxidation of phosphonates, phosphite, and hypophosphite. J. Bacteriol. 186:4730–4739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Robert FO, Pandhal J, Wright PC. 2010. Exploiting cyanobacterial P450 pathways. Curr. Opin. Microbiol. 13:301–306 [DOI] [PubMed] [Google Scholar]
- 35. Schirmer A, Rude MA, Li X, Popova E, del Cardayre SB. 2010. Microbial biosynthesis of alkanes. Science 329:559–562 [DOI] [PubMed] [Google Scholar]
- 36. Toepel J, Welsh E, Summerfield TC, Pakrasi HB, Sherman LA. 2008. Differential transcriptional analysis of the cyanobacterium Cyanothece sp. strain ATCC 51142 during light-dark and continuous-light growth. J. Bacteriol. 190:3904–3913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Elvitigala T, Stöckel J, Ghosh BK, Pakrasi HB. 2009. Effect of continuous light on diurnal rhythms in Cyanothece sp. ATCC 51142. BMC Genomics 10:226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Larsson J, Nylander JA, Bergman B. 2011. Genome fluctuations in cyanobacteria reflect evolutionary, developmental and adaptive traits. BMC Evol. Biol. 11:187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dietz KJ, et al. 2001. Significance of the V-type ATPase for the adaptation to stressful growth conditions and its regulation on the molecular and biochemical level. J. Exp. Bot. 52:1969–1980 [DOI] [PubMed] [Google Scholar]
- 40. Beyenbach KW, Wieczorek H. 2006. The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. J. Exp. Biol. 209:577–589 [DOI] [PubMed] [Google Scholar]
- 41. Eastmond PJ, Graham IA. 2001. Re-examining the role of the glyoxylate cycle in oilseeds. Trends Plant Sci. 6:72–78 [DOI] [PubMed] [Google Scholar]
- 42. Stucken K, et al. 2010. The smallest known genomes of multicellular and toxic cyanobacteria: comparison, minimal gene sets for linked traits and the evolutionary implications. PLoS One 5:e9235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hyatt D, et al. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 11:119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pati A, et al. 2010. GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat. Methods 7:455–457 [DOI] [PubMed] [Google Scholar]
- 45. Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lagesen K, et al. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35:3100–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. 2003. Rfam: an RNA family database. Nucleic Acids Res. 31:439–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 49. Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: signalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 50. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schäffer AA, et al. 2001. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 29:2994–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9:286–298 [DOI] [PubMed] [Google Scholar]
- 53. Han MV, Zmasek CM. 2009. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinform. 10:356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes present in the two largest linear chromosomes of Cyanothece 7822. For many genes in these two chromosomes, no homologous sequences were found in the remaining five Cyanothece strains. Further, many of these genes did not have any paralog sequences in its main chromosome. Some of the functional groups overrepresented in the linear chromosome are highlighted.
Homolog gene groups (1,705) common to six Cyanothece strains. Based on NCBI protein BLAST (See Materials and Methods for more details), 1,705 homologous gene groups shared by six Cyanothece strains (Cyanothece 51142, Cyanothece 7424, Cyanothece 7425, Cyanothece 7822, Cyanothece 8801, and Cyanothece 8802) were identified. Gene identifiers and their annotations corresponding to each homologous group are provided. Representative protein sequences selected from Cyanothece 51142 were BLAST-aligned against all currently sequenced cyanobacterial strains to identify the presence/absence of homologous genes. Genes specific to N2-fixing strains (identified as those genes with homologs in at least 8 unicellular or filamentous N2-fixing strains but at most 6 non-N2-fixing strains) were further classified as those commonly found in both unicellular and filamentous strains (≥8 in unicellular strains and ≥8 in filamentous strains; blue) and those specific to unicellular strains (≥8 in unicellular strains and ≤3 in filamentous strains; green). Remaining genes specific to N2-fixing strains are in red.
Genes specific to N2-fixing organisms, selected using diurnally regulated genes in Cyanothece 51142. Protein sequences of diurnally regulated genes in Cyanothece 51142 (22, 37) were blasted against 61 cyanobacterial genomes to identify the presence of homologous genes. Genes specific to N2-fixing strains were defined as those genes, for which, homologous sequences are present in at least 8 unicellular or filamentous nitrogen-fixing strains and in at most six non-nitrogen-fixing strains. We also identified genes specific to unicellular N2-fixing strains (highlighted in light green) and genes commonly found in both unicellular and filamentous N2-fixing strains (highlighted in light blue).