Abstract
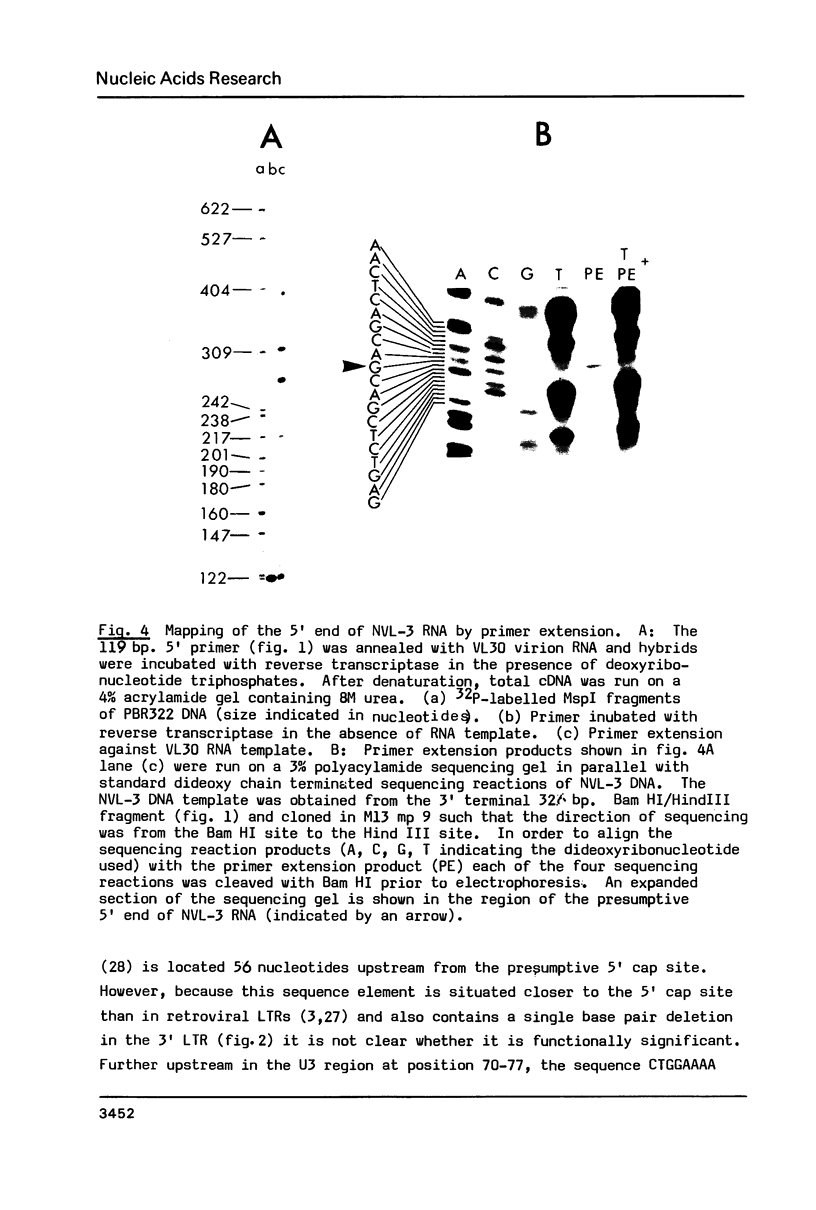
We have determined the nucleotide sequence and mapped the transcriptional boundaries in the long terminal repeats (LTRs) and adjacent regions of a retrovirus transmissible virus-like 30S ( VL30 ) mouse genetic element. The 572 base pair LTRs contain transcriptional regulatory sequences and are bounded by short imperfect repeats, with a minus strand tRNAgly primer binding site and a purine rich plus strand primer site flanking each of their inner boundaries. The 3' end of each LTR consists of an extensive 80 base pair redundancy of tRNA primer site and inverted repeat sequences while 41 and 47 base pair imperfect tandem repeats are present between the 5' capping site and the putative polyadenylation signal. Comparison with other retrovirus-like LTR sequences suggests possible modes of recombination that could occur between VL30 and other genetic elements.
Full text
PDF














Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Besmer P., Olshevsky U., Baltimore D., Dolberg D., Fan H. Virus-like 30S RNA in mouse cells. J Virol. 1979 Mar;29(3):1168–1176. doi: 10.1128/jvi.29.3.1168-1176.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A. T., Norton J. D., Avery R. J. A novel approach to cloning transcriptionally active retrovirus-like genetic elements from mouse cells. Nucleic Acids Res. 1983 Sep 24;11(18):6243–6254. doi: 10.1093/nar/11.18.6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewley J. P., Avery R. J. The virion RNA species of the Kirsten murine sarcoma-leukemia virus complex released from a clonally related series of mouse cells. Arch Virol. 1982;72(1-2):35–46. doi: 10.1007/BF01314448. [DOI] [PubMed] [Google Scholar]
- Cooper G. M., Neiman P. E. Two distinct candidate transforming genes of lymphoid leukosis virus-induced neoplasms. Nature. 1981 Aug 27;292(5826):857–858. doi: 10.1038/292857a0. [DOI] [PubMed] [Google Scholar]
- Corden J., Wasylyk B., Buchwalder A., Sassone-Corsi P., Kedinger C., Chambon P. Promoter sequences of eukaryotic protein-coding genes. Science. 1980 Sep 19;209(4463):1406–1414. doi: 10.1126/science.6251548. [DOI] [PubMed] [Google Scholar]
- Courtney M. G., Elder P. K., Steffen D. L., Getz M. J. Evidence for an early evolutionary origin and locus polymorphism of mouse VL30 DNA sequences. J Virol. 1982 Aug;43(2):511–518. doi: 10.1128/jvi.43.2.511-518.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolberg D., Fan H. Further characterization of virus-like 30S (VL30) RNA of mice: initiation of reverse transcription and intracellular synthesis. J Gen Virol. 1981 Jun;54(Pt 2):281–291. doi: 10.1099/0022-1317-54-2-281. [DOI] [PubMed] [Google Scholar]
- Ellis R. W., Defeo D., Shih T. Y., Gonda M. A., Young H. A., Tsuchida N., Lowy D. R., Scolnick E. M. The p21 src genes of Harvey and Kirsten sarcoma viruses originate from divergent members of a family of normal vertebrate genes. Nature. 1981 Aug 6;292(5823):506–511. doi: 10.1038/292506a0. [DOI] [PubMed] [Google Scholar]
- Fan H., Mueller-Lantzsch N. RNA metabolism of murine leukemia virus. III. Identification and quantitation of endogenous virus-specific mRNA in the uninfected BALB/c cell line JLS-V9. J Virol. 1976 May;18(2):401–410. doi: 10.1128/jvi.18.2.401-410.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan D. J. Retroviruses and transposable elements--which came first? Nature. 1983 Mar 10;302(5904):105–106. doi: 10.1038/302105a0. [DOI] [PubMed] [Google Scholar]
- Foster D. N., Schmidt L. J., Hodgson C. P., Moses H. L., Getz M. J. Polyadenylylated RNA complementary to a mouse retrovirus-like multigene family is rapidly and specifically induced by epidermal growth factor stimulation of quiescent cells. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7317–7321. doi: 10.1073/pnas.79.23.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa E., Mitra S. W., Goff S., Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979 Sep;18(1):93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- Gupta R. C., Roe B. A., Randerath K. The nucleotide sequence of human tRNAGly (anticodon GCC). Nucleic Acids Res. 1979 Oct 25;7(4):959–970. doi: 10.1093/nar/7.4.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howk R. S., Troxler D. H., Lowy D., Duesberg P. H., Scolnick E. M. Identification of a 30S RNA with properties of a defective type C virus in murine cells. J Virol. 1978 Jan;25(1):115–123. doi: 10.1128/jvi.25.1.115-123.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itin A., Keshet E. Nucleotide sequence analysis of the long terminal repeat of murine virus-like DNA (VL30) and its adjacent sequences: resemblance to retrovirus proviruses. J Virol. 1983 Sep;47(3):656–659. doi: 10.1128/jvi.47.3.656-659.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet E., Itin A. Patterns of genomic distribution and sequence heterogeneity of a murine "retrovirus-like" multigene family. J Virol. 1982 Jul;43(1):50–58. doi: 10.1128/jvi.43.1.50-58.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet E., Shaul Y., Kaminchik J., Aviv H. Heterogeneity of "virus-like" genes encoding retrovirus-associated 30S RNA and their organization within the mouse genome. Cell. 1980 Jun;20(2):431–439. doi: 10.1016/0092-8674(80)90629-7. [DOI] [PubMed] [Google Scholar]
- Keshet E., Shaul Y. Terminal direct repeats in a retrovirus-like repeated mouse gene family. Nature. 1981 Jan 1;289(5793):83–85. doi: 10.1038/289083a0. [DOI] [PubMed] [Google Scholar]
- Maisel J., Bender W., Hu S., Duesberg P. H., Davidson N. Structure of 50 to 70S RNA from Moloney sarcoma viruses. J Virol. 1978 Jan;25(1):384–394. doi: 10.1128/jvi.25.1.384-394.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., Berk A. J. Inhibition of RNA cleavage but not polyadenylation by a point mutation in mRNA 3' consensus sequence AAUAAA. Nature. 1983 Oct 13;305(5935):600–605. doi: 10.1038/305600a0. [DOI] [PubMed] [Google Scholar]
- Norton J. D., Carter A. T., Avery R. J. Restriction endonuclease mapping of unintegrated proviral DNA of Kirsten murine sarcoma virus. J Gen Virol. 1982 Jan;58(Pt 1):95–106. doi: 10.1099/0022-1317-58-1-95. [DOI] [PubMed] [Google Scholar]
- Ou C. Y., Boone L. R., Yang W. K. A novel sequence segment and other nucleotide structural features in the long terminal repeat of a BALB/c mouse genomic leukemia virus-related DNA clone. Nucleic Acids Res. 1983 Aug 25;11(16):5603–5620. doi: 10.1093/nar/11.16.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M. Enhancer elements activated by steroid hormones? Nature. 1983 Aug 25;304(5928):687–688. doi: 10.1038/304687a0. [DOI] [PubMed] [Google Scholar]
- Roop D. R., Tsai M. J., O'Malley B. W. Definition of the 5' and 3' ends of transcripts of the ovalbumin gene. Cell. 1980 Jan;19(1):63–68. doi: 10.1016/0092-8674(80)90388-8. [DOI] [PubMed] [Google Scholar]
- Rothenberg E., Donoghue D. J., Baltimore D. Analysis of a 5' leader sequence on murine leukemia virus 21S RNA: heteroduplex mapping with long reverse transcriptase products. Cell. 1978 Mar;13(3):435–451. doi: 10.1016/0092-8674(78)90318-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolnick E. M., Vass W. C., Howk R. S., Duesberg P. H. Defective retrovirus-like 30S RNA species of rat and mouse cells are infectious if packaged by type C helper virus. J Virol. 1979 Mar;29(3):964–972. doi: 10.1128/jvi.29.3.964-972.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwin S. A., Rapp U. R., Benveniste R. E., Sen A., Todaro G. J. Rescue of endogenous 30S retroviral sequences from mouse cells by baboon type C virus. J Virol. 1978 May;26(2):257–264. doi: 10.1128/jvi.26.2.257-264.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotohno K., Mizutani S., Temin H. M. Sequence of retrovirus provirus resembles that of bacterial transposable elements. Nature. 1980 Jun 19;285(5766):550–554. doi: 10.1038/285550a0. [DOI] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Temin H. M. Function of the retrovirus long terminal repeat. Cell. 1982 Jan;28(1):3–5. doi: 10.1016/0092-8674(82)90367-1. [DOI] [PubMed] [Google Scholar]
- Temin H. M. Structure, variation and synthesis of retrovirus long terminal repeat. Cell. 1981 Nov;27(1 Pt 2):1–3. doi: 10.1016/0092-8674(81)90353-6. [DOI] [PubMed] [Google Scholar]
- Tsuda M., Ohshima Y., Suzuki Y. Assumed initiation site of fibroin gene transcription. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4872–4876. doi: 10.1073/pnas.76.10.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beveren C., Rands E., Chattopadhyay S. K., Lowy D. R., Verma I. M. Long terminal repeat of murine retroviral DNAs: sequence analysis, host-proviral junctions, and preintegration site. J Virol. 1982 Feb;41(2):542–556. doi: 10.1128/jvi.41.2.542-556.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. E. Form and function of retroviral proviruses. Science. 1982 May 21;216(4548):812–820. doi: 10.1126/science.6177038. [DOI] [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]
- Wirth T., Glöggler K., Baumruker T., Schmidt M., Horak I. Family of middle repetitive DNA sequences in the mouse genome with structural features of solitary retroviral long terminal repeats. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3327–3330. doi: 10.1073/pnas.80.11.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]